Abstract
Mitotic centromere-associated kinesin (MCAK)/Kif2C is the most potent microtubule (MT)-destabilizing enzyme identified thus far. However, MCAK's function at the centromere has remained mechanistically elusive because of interference from cytoplasmic MCAK's global regulation of MT dynamics. In this study, we present MCAK chimeras and mutants designed to target centromere-associated MCAK for mechanistic analysis. Live imaging reveals that depletion of centromere-associated MCAK considerably decreases the directional coordination between sister kinetochores. Sister centromere directional antagonism results in decreased movement speed and increased tension. Sister centromeres appear unable to detach from kinetochore MTs efficiently in response to directional switching cues during oscillatory movement. These effects are reversed by anchoring ectopic MCAK to the centromere. We propose that MCAK increases the turnover of kinetochore MTs at all centromeres to coordinate directional switching between sister centromeres and facilitate smooth translocation. This may contribute to error correction during chromosome segregation either directly via slow MT turnover or indirectly by mechanical release of MTs during facilitated movement.
Introduction
Live studies on the segregation of chromosomes during mitosis have revealed key principles that describe chromosome behavior in vertebrate cells (Skibbens et al., 1993; Khodjakov and Rieder, 1996). One of these is that chromosomes exhibit directional instability—they oscillate between force-generating poleward translocation and antipoleward movement with rapid switches between persistent movement (Skibbens et al., 1993; Khodjakov and Rieder, 1996). During switching events, kinetochores adjust almost instantaneously from poleward movement, which is synchronized with depolymerizing microtubules (MTs), to antipoleward movement, which is coupled to polymerizing MTs. Furthermore, the linked sister kinetochore responds with precisely the opposite activity within an exceedingly small range of space and time. It is essential that the directional switches be rapid because if the sister kinetochores are not coordinated, the chromosomes will halt, increasing the probability that the flexible vertebrate kinetochore (Dong et al., 2007) will bind inappropriately oriented MTs that could lead to errors in chromosome segregation.
Mitotic centromere-associated kinesin (MCAK) localizes dynamically throughout the inner centromeres, outer kinetochores, at centrosomes, on MT tips, and at the spindle midzone during cell division (Wordeman and Mitchison, 1995; Andrews et al., 2004; Kline-Smith et al., 2004; Moore et al., 2005). MCAK destabilizes MTs from either end (Desai et al., 1999; Hunter et al., 2003), and this activity and localization are under the regulation of mitotic kinases (Andrews et al., 2004; Lan et al., 2004). Because MCAK is localized broadly and dynamically throughout the inner and outer centromere during cell division, we set out to determine precisely what MCAK's MT-destabilizing activity contributes to chromosome segregation. To accomplish this, we engineered a construct that would localize additional ectopic MCAK activity specifically to centromeres by fusing the minimal MT-depolymerizing domain of MCAK to the DNA-binding domain of centromere protein B (CENP-B). The method benefits from the observation that CENP-B depletion has no obvious phenotype (Hudson et al., 1998; Perez-Castro et al., 1998). This clever technique was first used to tether inner CENP irreversibly to the centromere (Eckley et al., 1997). Subsequently, a GFP–CENP-B (DNA-binding domain) chimera was used to study centromere behavior in living cells (Shelby et al., 1996). We combined these techniques to compare the live centromere behavior of MCAK-enriched and -depleted centromeres during mitosis.
Bioriented centromeres depleted of endogenous MCAK exhibited increased tension that was attributable to the lack of coordinated movement between the sister centromeres. In other words, sister centromeres compete with each other for directional dominance. This leads to increases in mean interkinetochore distance while the sisters are both translocating in opposite directions. These effects were reversed by the addition of ectopic MCAK activity to the centromere. Furthermore, we developed a sensitive fluorescent assay based on the accumulation of detyrosinated MTs in the kinetochore fiber (Gundersen and Bulinski, 1986) to establish that turnover of kinetochore fiber MTs was reduced in the absence of MCAK. In contrast, excess MCAK added to the centromere simultaneously suppressed MT flux while subtly enhancing MT turnover by a nonflux-related mechanism. Thus, MCAK may not specifically target aberrant MTs for detachment but instead facilitates generalized detachment and turnover of kinetochore MTs from all centromeres during chromosome movement. This activity promotes directional synchrony between translocating sister chromosomes and assists in the preservation of genetic fidelity.
Results
Constructs used to modify centromeric MCAK levels and track centromere behavior
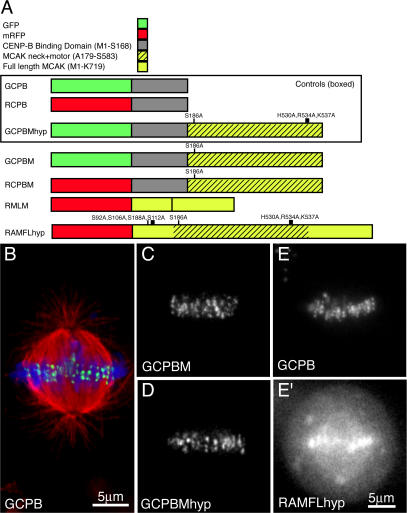
Table I and Fig. 1 A diagram and describe, respectively, the chimeric constructs used in this study to enrich or deplete MCAK on the centromere and to assay centromere behavior. Table I can be used for quick reference, whereas the constructs are described in more detail below. Sister centromeres were tracked in living cells via a construct consisting of EGFP fused to the centromere-binding domain of CENP-B (Pluta et al., 1992). This construct is referred to as GCPB (GFP–CENP-B–binding domain). The fusion protein expressed by this construct localizes specifically to centromeres (Fig. 1 B). HeLa cells were preferentially chosen for this study because the constructs rarely, if ever, overexpressed to the point that fluorescent protein appeared in the cytoplasm, providing us greater numbers of cells for live imaging. However, all of our constructs produce the same effects in CHO cells as they do in HeLa cells, although fewer CHO cells are available for live analysis. A monomeric red (monomeric RFP [mRFP] 1.0l; Campbell et al., 2002) version of this construct (RCPB [RFP–CENP-B–binding domain]) was used in conjunction with photoactivatable GFP-tubulin and green fluorescent counterstains in fixed cells.
Table I.
Constructs used to modify MCAK activity at live centromeres
| Name | Description | Purpose |
|---|---|---|
| GCPB | The centromere-binding domain of CENP-B fused to the C terminus of GFP |
A control construct to mark centromeres for live imaging and for use with mRFP-fused constructs |
| RCPB | The centromere-binding domain of CENP-B fused to mRFP |
A control construct to mark centromeres in conjunction with GFP constructs or photoactivatable GFP |
| GCPBM | GCPB fused to the N terminus of the neck + motor minimal functional MT-depolymerizing domain of MCAK |
Designed to anchor ectopic MT-depolymerizing activity to the centromere |
| RCPBM | RCPB fused to the neck + motor of MCAK | A red version of GCPBM for use with photoactivatable GFP |
| GCPBMhyp | GCPBM with three point mutations in the motor domain that eliminate the MT-depolymerizing activity of MCAK |
Controls for any effects from anchoring GCPBM at the centromere. We found this construct to be indistinguishable in appearance and behavior from GCPB |
| RMLM | mRFP fused to the N terminus of motorless MCAK | A dominant-negative construct designed to deplete endogenous MCAK on centromeres |
| RAMFLhyp | mRFP fused to full-length MCAK inactivated with three point mutations in the motor domain and five aurora B phosphorylation sites mutated to alanine |
A more potent dominant-negative construct designed to deplete endogenous active MCAK on centromeres |
Figure 1.
Diagram of constructs and localization in mitotic cells. (A) EGFP (green) was fused to the N terminus of the minimal centromere-binding domain of CENP-B (gray) to mark centromeres (GCPB). This construct was fused to the minimal neck plus motor domain of MCAK (yellow/black crosshatch), which possessed an S186A mutation preventing phosphorylation by aurora B kinase (GCPBM). The control for this construct possesses three point mutations inactivating the motor domain (GCPBMhyp). Finally, full-length MCAK (yellow) with all aurora B phosphorylation sites mutated to alanine and the motor domain inactivated was linked to mRFP for use as a dominant-negative construct for endogenous active MCAK (RAMFLhyp). (B) GCPB localizes specifically to centromeres in HeLa cells colabeled with antitubulin (red) and Hoechst (blue). (C–E) Z projections of live metaphase HeLa cells expressing the constructs GCPBM (C) and GCPBMhyp (D), which localize specifically to centromeres in live HeLa cells. (E and E′) A live HeLa cell cotransfected with GCPB (E) and RAMFLhyp (E′). Centromere behavior is scored by observing the GCPB channel in dual-transfected cells.
Ectopic MCAK activity was added to the centromere using a construct consisting of the centromere-binding domain of EGFP–CENP-B (GCPB) fused to the neck plus motor domain (Fig. 1 A, yellow and black stripes) of MCAK (GCPBM [GCPB-MCAK neck + motor]). This construct localized to precisely the same domain as GCPB because all of the centromere-targeting domains of MCAK have been removed (Fig. 1 C). In HeLa cells, the construct was rarely, if ever, observed to overexpress to the point at which the centromeres were saturated and excess protein appeared in the cytoplasm. In contrast, in select CHO cells, the expression levels were high enough that we were able to confirm and quantify that the GCPBM fusion protein exhibited robust MT-destabilizing activity when overexpressed to levels that overflow into the cytoplasm of CHO cells (unpublished data). MCAK neck + motor has been previously shown to correspond to the minimal domain capable of full-strength MT-depolymerizing activity under the physiological conditions found in cell cytoplasm (Maney et al., 2001; Ovechkina et al., 2002). The potential negative regulation of depolymerizing activity caused by phosphorylation by aurora B kinase is obviated by mutating site S186 to alanine (Andrews et al., 2004). MCAK neck + motor fused to GFP–CENP-B (GCPBM) localized specifically to centromeres when expressed at levels used in this study (Fig. 1 C) and was immune to the regulatory effects of aurora B kinase both with respect to activity and localization. Disruption of aurora B kinase activity with either dominant-negative mutants or the aurora B–specific inhibitor ZM447439 does not displace CENP-B or any of our CENP-B chimeric proteins from centromeres (unpublished data).
A monomeric red version of this construct was used with photoactivatable GFP-tubulin (RCPBM [RCPB-MCAK neck + motor]). GCPBM was controlled for any effects on centromere structure using an identical construct that was triply mutated within the core motor domain to eliminate MT-depolymerizing activity (H530A, R534A, and K537A). This construct is referred to as GCPBMhyp because the mutation corresponds to our previously described hypir mutation (Moore et al., 2005), which was originally characterized in conventional kinesin (Woehlke et al., 1997). Identical to GCPBM, the protein coded by this construct localizes specifically to centromeres (Fig. 1 D). A key advantage of the CENP-B–binding domain fusion is that it isolates the ectopic activity of MCAK specifically to centromeres so that its effect on centromere behavior can be analyzed in isolation. Endogenous MCAK can be found dynamically located to the inner centromere, the outer face of the centromere, and any location in between (Maney et al., 1998; Andrews et al., 2004). Ectopic active MCAK anchored to the CENP-B–binding location is within the spatial confines of MCAK's distribution, so the effect is to enrich the centromere with additional active MCAK beyond the more tightly regulated endogenous complement.
Because MCAK is dynamically regulated during mitosis (Andrews et al., 2004; Lan et al., 2004), we engineered a construct designed to specifically interfere with activated MCAK. We prepared an mRFP version of full-length MCAK in which the motor domain is inactivated with the hypir triple mutation and all five of the aurora B phosphorylation sites are mutated to alanine. Because this construct is full length with respect to MCAK, it localizes to all of the same regions as endogenous MCAK. Unfortunately, we have no markers to differentiate between RAMFLhyp and endogenous MCAK, so we cannot quantify the extent of displacement of endogenous protein by the mutant. However, the mutant robustly localizes to all regions where we normally find endogenous MCAK. As expected, it localizes strongly to centromeres and less avidly to centrosomes (Fig. 1 E′). MCAK can be found at any time either at the inner centromere, outer centromere, or both during chromosome movement (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200707120/DC1; Andrews et al., 2004). For this reason, we coexpress GCPB with RAMFLhyp and image only the green channel to track centromeres over time (Fig. 1 E) after scoring the level of expression and localization of RAMFLhyp with a single initial exposure (Fig. 1 E′). To confirm the effects of dominant-negative RAMFLhyp expression, we also performed global MCAK depletion using siRNA and overexpression of a different, previously described motorless MCAK construct (Maney et al., 1998) fused to mRFP. We determined that these constructs did not affect the amount or distribution of other CENPs such as CENP-E (not depicted) or Hec1 (Fig. S1). Our polyclonal antiserum against endogenous MCAK does not cross react with MCAK neck + motor, allowing us to assay the level of endogenous MCAK on centromeres in the presence of GCPBM and GCPBMhyp. The CENP-B chimeric protein did not affect the localization of endogenous MCAK (Fig. S1).
Centromeres enriched with MCAK exhibit decreased sister centromere separation
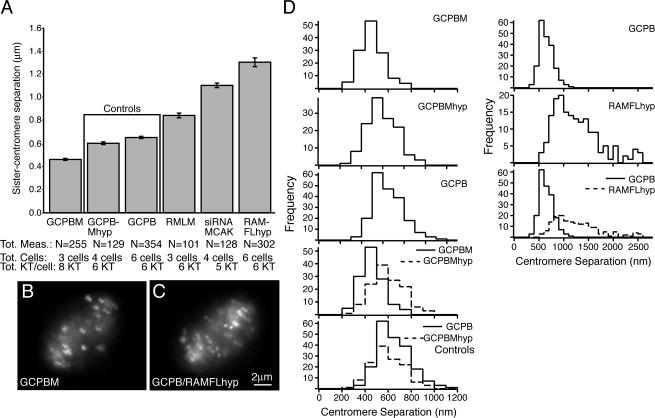
The intercentromere distance was measured over time in live, bioriented centromeres oscillating in the vicinity of the metaphase plate. Quantification of the effect of manipulations of MCAK activity on sister centromere separation is shown in Fig. 2 A. Live sister centromeres were tracked frame by frame, and the centroid to centroid distance between the sisters was measured. Only bioriented metaphase centromeres were scored. Ectopic localization of excess MCAK to centromeres decreased the mean sister centromere separation from ∼0.6 to 0.45 μm. In contrast, displacement of endogenous MCAK from centromeres either with motorless MCAK (Maney et al., 1998), RAMFLhyp, or siRNA significantly (P < 0.0001 for both treatments) increased sister centromere separation (Fig. 2 A). In the case of RAMFLhyp, the separation was almost twofold greater (1.2 μm) than control centromeres. The number of cells from which these live measurements (between 150 and 200 per treatment) were recorded is indicated in Fig. 2 A. For each cell, between three and six kinetochores were tracked for between 5 and 15 20-s frames. The differences between GCPBM and GCPBMhyp and GCPB and RAMFLhyp were significantly different whether the data were pooled per frame (P = 0.02 and P < 0.001), per kinetochore (P = 0.02 and P < 0.001, respectively), or per cell (P = 0.10 and P = 0.10). Histograms of these compiled measurements are shown in Fig. 2 D.
Figure 2.
MCAK suppresses tension in a dose-dependent manner. (A) Addition of exogenous MCAK (GCPBM) to centromeres decreases centromere separation significantly from controls (GCPB and GCPBMhyp) in living cells (P < 0.0001). Expression of mRFP motorless MCAK (RMLM), RAMFLhyp, or depletion of MCAK with siRNA significantly increases sister centromere separation (as visualized by GCPB; P < 0.0001). Between five and eight live sister centromeres per cell were measured for 5–15 successive frames (20 s per frame). Each bar (SEM [error bars] is shown) represents three to six different cells from at least two different experiments on different days. (B) Sister centromeres in metaphase cells expressing GCPBM appear quite close together. See Video 1 (available at http://www.jcb.org/cgi/content/full/jcb.200707120/DC1). (C) Sister centromeres in metaphase cells coexpressing GCPB and RAMFLhyp appear to be under significantly more tension (P < 0.0001). See Video 2. (D) Histograms of sister centromere separation for GCPB, GCPBM, GCPBMhyp, and RAMFLhyp. Overlaid comparisons are shown between GCPBM and GCPBMhyp, the two controls (which are not significantly different; P = 0.40), and GCPB- and GCPB/RAMFLhyp-coexpressing cells.
The tension effects are obvious by visual inspection of still micrographs taken from live videos of oscillating centromeres bound either to GCPBM (Fig. 2 B) or transfected with RAMFLhyp and visualized with GFPB (Fig. 2 C). The difference in the means between the GCPBM and either GCPBMhyp or GCPB (the controls) is significant (P < 0.0001). The same is true for RFLMhyp and the controls (P < 0.0001). However, the controls were not significantly different (P = 0.40). In fact, through the course of this study, we observed no substantive differences between GCPB and GCPBMhyp. Therefore, in some experiments, only one control is shown for simplicity. In contrast to motorless MCAK, which localizes preferentially to the inner centromere (Maney et al., 1998), RAMFLhyp localizes similarly to wild-type MCAK with a slight preference for the outer centromere face (Andrews et al., 2004). It is also more potent than siRNA depletion of MCAK, for which it is difficult to achieve >80% reduction in protein levels by Western blotting (unpublished data). Nevertheless, each of these complementary but technically distinct methods of depleting centromere-associated MCAK resulted in an increase in sister centromere separation. Furthermore, there was a slight but measureable increase in BubR1 label on MCAK-enriched kinetochores exhibiting decreased tension (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200707120/DC1).
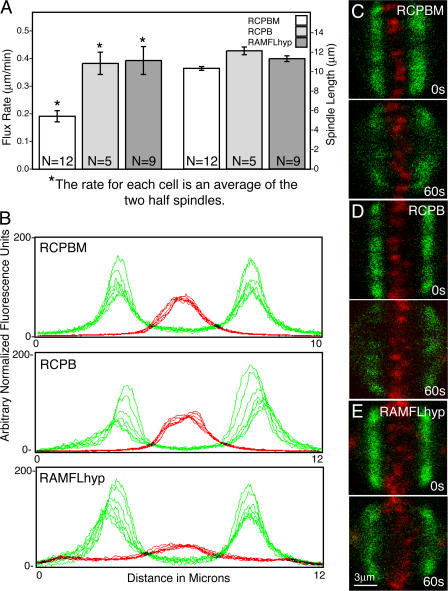
Centromeric MCAK suppresses kinetochore fiber flux
HeLa cells simultaneously expressing photoactivatable GFP-tubulin and either RCPB, RCPBM, or RAMFLhyp were photoactivated and imaged live at 1-s intervals to measure the rate at which tubulin translocates from the distal face of the centromere to the poles. The rate of flux was measured as the distance from the center of the oscillating centromeres to the midpoint of the front of the fluorescent tubulin. In cells expressing ectopic centromeric MCAK activity, the mean rate of flux was decreased by half (Fig. 3 A, left axis) and, in some cells, was halted altogether (Fig. 3 B, RCPBM). A slight reduction in overall spindle length was also evident in cells expressing RCPBM (Fig. 3 A, right axis). Still micrographs at the time of photoactivation and 60 s later of control cells expressing mRCPBM (Fig. 3 C), RCBP (Fig. 3 D), and RAMFLhyp (Fig. 3 E) are shown in conjunction with photoactivatable tubulin (Fig. 3 E, green). In contrast, FRAP showed no difference in the half-time of nonkinetochore fiber recovery between the treatments (Fig. S2 B).
Figure 3.
MCAK enrichment on centromeres antagonizes MT flux. (A) Addition of MCAK to centromeres significantly decreases the flux rate of kinetochore fibers (left axis; P = 0.0003). Depletion of centromeric MCAK does not affect the rate of flux (P = 0.85). Overall spindle length is also slightly decreased in MCAK-enriched centromeres (right axis). MCAK depletion did not significantly alter spindle length (P = 0.18). Error bars represent SEM. (B) Successive 10-s line scans of photoactivated tubulin (green) in control (RCPB) and MCAK-enriched (RCPBM) cells. Flux is inhibited in cells possessing MCAK-enriched centromeres. (C) A representative RCPBM cell at 0 s (right after photoactivation) and 60 s later. (D) A representative RCPB cell at t = 0 and t = 60 s. (E) A representative RAMFLhyp cell at t = 0 and t = 60 s.
MCAK increases sister kinetochore coordination
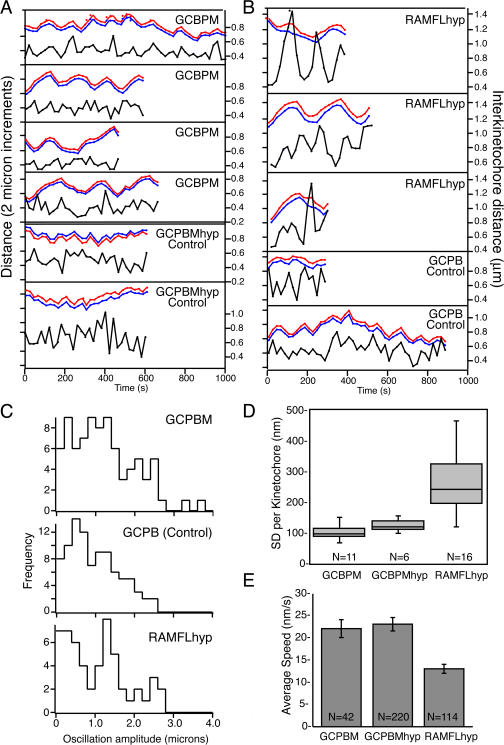
Fig. 4 (A and B) shows traces of individual sister centromere pairs (red and blue) plotted over time. The right axes in Fig. 4 (A and B) show the sister centromere separation (black line) over time for the tracked sisters. A general trend is that centromeres bound to GCPBM exhibit more persistent movement and longer oscillations relative to controls (GCPB or GCPBMhyp). The pooled oscillation amplitudes defined as the distance in micrometers between turnarounds are plotted in the histograms in Fig. 4 C. Centromeres depleted of MCAK were under generally higher tension with large peaks in separation that correlated to a lack of coordination between the sister centromeres (Fig. 4 B). This trend is obvious when the SD per kinetochore for each construct is graphed in a box plot (Fig. 4 D). The box plots consist of the SD of the sister centromere separation over successive live images (20 s) for between 6 and 15 individual kinetochores from between three and six cells per treatment. The global increase in tension exhibited by cells lacking active centromere-associated MCAK appears to be partly caused by increased time spent in a state in which both sisters of a single sister centromere pair are simultaneously engaged with MTs and attempting to move poleward at the same time. In contrast, both control (GCPB) cells and cells expressing ectopic MCAK (GCPBM) are well coordinated in directionality. Furthermore, centromeres with GCPBM exhibit more persistent directionality of movement that manifests in a greater mean oscillation amplitude (distance between turnarounds) than is typical for control centromeres. For consistency, only bioriented metaphase centromeres were evaluated. Histograms of pooled oscillation data suggest that the oscillation amplitude of MCAK-depleted (RAMFLhyp) centromeres tends toward a bimodal distribution consisting of both very short excursions punctuated by frequent reversals and also rather long excursions (Fig. 4 C). Live imaging suggests that this is caused by the tendency of battling sister centromeres to suddenly reverse direction as they lose the competition for poleward engagement.
Figure 4.
Tracks of sister centromere oscillations over time. (A) Representative traces of a pair of sister centromeres (red and blue) tracked over time. Each tick (left axis) corresponds to a distance of 2 μm. The right axis shows sister centromere separation plotted below (black line). GCPBM centromeres exhibit more directional persistence and greater oscillation amplitude than control (GCBMhyp) centromeres. (B) Similar traces of sister centromeres (red and blue) tracked from cells expressing RAMFLhyp versus GCPB controls. MCAK-depleted centromeres (RAMFLhyp) are not as well coordinated in directionality. (C) Histograms of the oscillation amplitudes of pooled data from three to six centromeres each in three to six different cells filmed and transfected on different days. The oscillation amplitude of GCPBM is significantly greater than GCPB (control) at the 95% confidence interval (P = 0.033). The mean oscillation amplitude of RAMFLMhyp is not substantially different from controls because the distribution is bimodal. (D) Box plots of the SD of the sister centromere separation of individual sister centromere pairs over 8–15 frames. N, number of kinetochores. (E) Mean distance over time of oscillating metaphase centromeres in control (GCPBMhyp), MCAK-enriched (GCPBM), or MCAK-depleted (RAMFLhyp) cells. Error bars represent SEM.
The utility of sister centromere coordination is shown in Fig. 4 E. The mean distance traveled per 20 s of MCAK-depleted centromere pairs is half that of control or MCAK-enriched centromeres. The maximum velocity that the centromeres are capable of obtaining is similar for all of the treatments. The decrease in distance traveled per 20-s time interval for RAMFLhyp cells in Fig. 4 E reflects the tendency for centromere pairs to remain paused while battling for directional dominance.
MCAK partially rescues the effects of aurora B kinase inhibition
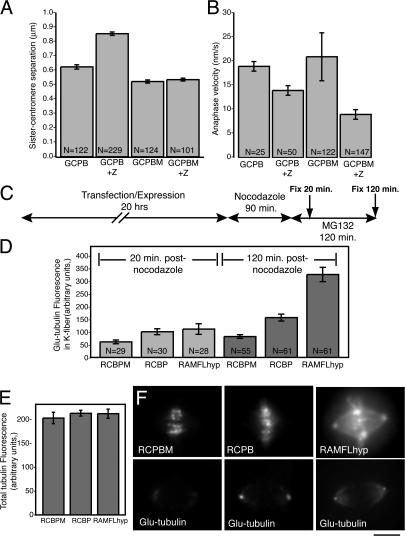
To determine whether the increased tension could be attributable to increased numbers of aberrant (merotelic) attachments, we inhibited aurora B kinase with a pulse of 5 μM ZM447439 to decrease the turnover of kinetochore MTs and also to promote the accumulation of merotelic connections. We did not administer ZM447439 to specifically influence MCAK but to lock kinetochores more tightly onto kinetochore fibers (possibly via Ndc80 dephosphorylation), as has been shown previously (Cimini et al., 2006; DeLuca et al., 2006). We found that ZM447439 increased sister centromere tension in live, bioriented centromeres (Fig. 5 A). This is consistent with an induction of stable attachments at the centromere (Cimini et al., 2003; Pinsky et al., 2006). ZM447439 also decreased the mean rate of anaphase chromosome movement. We hypothesize that this is the result of an increase in merotelic connections that results in global lagging of anaphase chromosomes (Cimini et al., 2006). Interestingly, MCAK enrichment at the centromere rescued the increase in tension triggered by ZM447439 (Fig. 5 A), whereas the addition of GCPBM to the centromere did not rescue the rate of anaphase velocity (Fig. 5 B). Neither MCAK enrichment nor MCAK depletion (unpublished data) had a substantial effect on mean anaphase segregation velocity, which is consistent with a previous study (Ganem et al., 2005).
Figure 5.
MCAK opposes attachment at the kinetochore. (A) Additional MCAK on the centromere rescues the increase in tension of bioriented kinetochores caused by transient aurora B kinase inactivation. N, number of individual live measurements resulting from six bioriented kinetochores in each of three metaphase cells. (B) Additional MCAK on the centromere does not rescue the decrease in anaphase chromosome segregation velocity caused by aurora B kinase inhibition. This suggests that the increase in tension is not solely caused by increased numbers of lateral merotelic MT–kinetochore interactions. N, number of individual live measurements resulting from five kinetochores in each of three to five cells. (C) Method for retyrosinating CHO tubulin to baseline before spindle assembly. Release of cells from nocodazole permits MT assembly and detyrosination (increase in glu-tubulin). (D) Cells exhibit significantly more kinetochore fiber detyrosinated tubulin (glu-tubulin) when depleted of MCAK (P < 0.0001). N, number of cells. (E) Total tubulin fluorescence measured from the same region that the glu-tubulin fluorescence was measured (D, dark bars). (F) Glu-tubulin labeling of representative cells transfected with RCBPM, RCPB, or RAMFLhyp after 120-min nocodazole reversal. Error bars represent SEM. Bar, 0.5 μm.
MCAK increases kinetochore MT turnover
The effects of changing MCAK levels on kinetochore attachment were too subtle to register using standard techniques of calcium or cold stability to assess attachment. We used a more sensitive technique to monitor global attachment and turnover. We used the appearance of detyronsinated tubulin (glu-tubulin) in kinetochore fibers as a read-out for kinetochore fiber stability (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200707120/DC1). The assay allows different experimental treatments of the centromere to register a relative change in kinetochore MT stability but does not enable us to directly measure the half-life of MT turnover. Because HeLa cells have little glu-tubulin in their kinetochore fibers (Gundersen and Bulinski, 1986), we used CHO cells for this experiment. The experiment is diagrammed in Fig. 5 C. Cells expressed our experimental chimeric constructs for 20 h. They were then incubated in 2.5 μM nocodazole for 90 min to disassemble all of the MTs and permit all glu-tubulin dimers to be retyrosinated (Webster et al., 1987). The cells were then washed free of nocodazole to allow the reformation of MTs and were incubated in MG132 for either 20 or 120 min to arrest the cells in metaphase. After 20 min, glu-tubulin could be detected in kinetochore fibers of metaphase cells, but there were no obvious differences between control cells and MCAK-depleted cells (Fig. 5 D, left). By 120 min, there was significantly more glu-tubulin in MCAK-depleted cells relative to controls (P < 0.0001; Fig. 5 D, right). Furthermore, there was significantly less glu-tubulin in MCAK-enriched cells despite the suppression of kinetochore fiber flux in these cells (P < 0.005). This suggests that MCAK promotes the nonflux-dependent turnover of MTs. Representative images are shown in Fig. 5 E.
Discussion
Mitotic sister chromosomes individually attach to and harness the power of dynamic MT ends while maintaining an intercentromere elastic linkage that enables them to translocate jointly on the mitotic spindle and orient facing opposite spindle poles (Shelby et al., 1996). Robust MT attachments and proper orientation facing the opposite spindle poles maximizes the elastic pull on the sister centromeres and signals the cell that anaphase may safely commence. We have found that ectopically increasing the level of MCAK activity on centromeres decreases sister centromere tension, although not to the point that the spindle checkpoint is triggered. Conversely, decreased levels of MCAK on centromeres substantially increased tension across sister centromeres. Our data contradicts two other studies that suggest that the depletion of MCAK has no effect on tension (Ganem et al., 2005) or decreases tension (Kline-Smith et al., 2004). We believe that this discrepancy may be caused by the inclusion of nonbioriented centromeres in the data pool. We specifically only measure bioriented metaphase centromeres because that is the status of most, if not all, of the centromeres in our cells and also because for this study, the relationship between tension and attachment of syntelic and monotelic centromeres is not subject to mechanistic interpretation.
Landmark studies have previously shown that lagging chromosomes during anaphase can be attributed to the erroneous establishment of MT connections from one kinetochore to both spindle poles (merotelic connections) before anaphase (Cimini et al., 2001). MCAK has been widely implicated in the correction of merotelic attachments during cell division (Kline-Smith et al., 2004; Knowlton et al., 2006). However, the large number of merotelic attachments that appear in cells during the course of cell division requiring correction (Cimini et al., 2003) suggests that MCAK cannot be the major mechanism for error correction in mammalian cells. Numerous studies support this conclusion (Lampson et al., 2004; Ganem et al., 2005), including those that showcase the mild effects of MCAK depletion (Maney et al., 1998; Cassimeris and Morabito, 2004; Ganem and Compton, 2004; Stout et al., 2006). Here, we demonstrate that MCAK contributes to error correction (as measured by lagging chromosomes) in a modest but dose-dependent manner and provide a mechanistic explanation for this activity.
Rather than being specialized for the specific detachment of merotelic connections, which occur abundantly but stochastically, we propose a model in which MCAK modestly enhances both the motility and the kinetochore fiber turnover of all kinetochores simultaneously. Modestly increased, kinetochore fiber turnover would lead to the eventual, albeit nonspecific, loss of errant connections, whereas the facilitation of movement might result in the mechanical detachment or pruning of errant connections. This model resembles that proposed for the Drosophila melanogaster orthologue of MCAK, KLP59C (Rogers et al., 2004), with the exception that we believe MCAK facilitates movement in vertebrate cells not via pacman-like activity but by loosening the kinetochores slightly from the MTs of the kinetochore fiber. We propose that MCAK accomplishes this by modestly increasing the proportion of MTs detaching and reattaching within the kinetochore fiber. It may be that the observation in Drosophila that the velocity of kinetochore movement decreases to the velocity of flux upon KLP59C inhibition (Rogers et al., 2004) is compatible with this hypothesis as well because inhibiting KLP59C might increase the extent of kinetochore MT attachment, which would simultaneously impair chromosome movement that is coordinated with MT end dynamics but would facilitate minus end flux-based chromosome translocation.
Cimini et al. (2003) have shown in cells experiencing delayed metaphase in MG132 that there is a time-dependent error correction process going on before anaphase that reduces the percentage of lagging chromosomes in anaphase many fold while only reducing the percentage of merotelic connections by about one third. Presently, it is not known what is taking place during this metaphase delay to eliminate and ameliorate these merotelic connections so that they do not result in lagging chromosomes. They originally hypothesized that MCAK may play a role in this process, as its MT-destabilizing activity would come in handy for pruning errant MT connections. Our data presented here support this hypothesis, as both MCAK-dependent random kinetochore MT turnover and oscillatory mechanical detachment would require time to manifest their potential for error correction.
Previous studies have established a relationship between kinetochore MT attachment and tension across bioriented kinetochores (for review see Pinsky and Biggins, 2005). Kinetochore attachment is not an all or none phenomenon, and many experimental treatments can compromise attachment without eliminating it. Conversely, interfering with aurora B regulation of the NDC80 complex, an essential complex that contributes the majority of the kinetochore-MT attachment activity, locks kinetochores onto MTs. This increase in attachment results in a striking increase in tension across bioriented centromeres, which is facilitated by a putatively flux-based shortening of kinetochore fibers at the poles (DeLuca et al., 2006).
This positive relationship between attachment and tension suggests that MCAK may use its MT-destabilizing activity to detach a small number of kinetochore fiber MTs and, by extension, transiently decrease the number of binding sites between the kinetochore and the kinetochore fiber (Fig. 6). By promoting dynamic instability without severely compromising kinetochore attachment to MTs, MCAK in effect loosens the kinetochores so that bioriented sisters are able to translocate more rapidly and easily. Our observations have two implications for kinetochore function: one is that dynamic instability within the kinetochore fiber may be a more prevalent mechanism of MT turnover than previously appreciated in some cells, and, second, kinetochores must achieve a balance between stable attachment and movement to translocate coordinately. It is formally possible that MCAK's effect on error correction is caused by its ability to promote coordinated movement, kinetochore fiber turnover, or both.
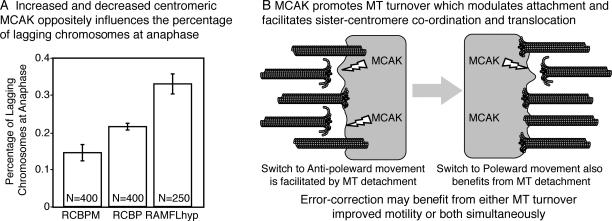
Figure 6.
MCAK facilitates error correction. (A) Addition of RCPBM to the centromere significantly reduced the percentage of lagging chromosomes in anaphase CHO cells, whereas expression of dominant-negative RFLMhyp significantly increased the percentage of lagging chromosomes (P = 0.02). Error bars represent SEM. (B) Model for the role of MCAK in regulating MT attachment in translocating kinetochores. MT-dependent chromosome movement must, by necessity, balance the breakage of binding sites and the reformation of new sites to maintain attachment. MCAK may use its depolymerizing activity to release a small proportion of MTs from the kinetochore. This facilitates and synchronizes movement by modulating the number of attachment sites between the kinetochore and the MTs. We believe that MCAK provides this activity to both the leading and trailing kinetochore. Improved centromere synchronization, increased MT turnover, or both activities together may facilitate error correction.
What is MCAK's true function at the centromere? It definitely but subtly contributes to error correction, MT detachment, and MT turnover. However, although MCAK can easily reduce the tension on aurora B–inhibited centromeres, it cannot correct anaphase segregation errors in the face of the loss of this kinase (Fig. 5, A and B). Thus, MCAK appears not to be the primary error correcting machinery regulated by aurora B. This confirms a previous study by Lampson et al. (2004). However, our study does demonstrate that sister centromere tension and coordination are substantially and globally affected across all kinetochores by modulating centromeric MCAK levels. Live imaging of sister centromere pairs that have been depleted of MCAK protein reveals that the increased tension results primarily from an increase in the time that sister centromeres spend simultaneously translocating poleward against each other. Classic experiments of live newt lung cells during cell division (Skibbens et al., 1993) demonstrated that sister centromeres oscillate in a surprisingly regular saw-tooth pattern and that reversals in direction by one centromere resulted in a rapid reversal by the attached sister centromere. The identity of the signal to reverse direction is still not known. However, we propose that MCAK is an important component of the machinery for assuring that the sister centromeres translocate in lockstep.
Materials and methods
Construction of DNA plasmids
GCPB (pGFPCPB1) was constructed by PCR amplification of codons 1–167 of the C. griseus CENP-B gene (a gift of M. Valdivia, Universidad de Cádiz, Cádiz, Spain) and subcloning into the EcoRI and Xba sites of pEGFP-C1 (Clontech Laboratories, Inc.). To prepare RCPB (pMX234), the EGFP gene of pEGFP-N1 (Clontech Laboratories, Inc.) was replaced by PCR-amplified mRFP1.0 (Campbell et al., 2002) and codons 1–167 of CENP-B to generate an EcoRI–NotI fragment bearing the mRFP–CENP-B fusion. To prepare GCPBM (pMX240), the aforementioned GCPB construct was fused to residues 186–583 of MCAK incorporating the AAAAA mutations (Andrews et al., 2004). GCPBMhyp (pMX241) incorporates the additional mutations H530A/R534A/K537A inactivating the motor domain. These were added to the aforementioned GCPBM construct. To prepare RAMFLhyp (pMX215), the EGFP gene of pEGFP-N1 was replaced by PCR-amplified mRFP1.0 (Campbell et al., 2002) and AAAAA-MCAK (Andrews et al., 2004) in combination with the additional mutations H530A/R534A/K537A. To construct photoactivatable GFP-tubulin, Homo sapiens α-tubulin was amplified by PCR from EGFP-tubulin (Clontech Laboratories, Inc.) and inserted in pPAGFP-C1 (Patterson and Lippincott-Schwartz, 2002).
Cell culture and transfections
HeLa or CHO cells were transfected using either Lipofectamine or Nucleofector II (Amaxa). For live imaging, HeLa cells were cultured for 36–48 h in MEMalpha (Invitrogen) medium with 10% FBS (Hyclone) and 5% CO2 on 35-mm2 dishes coated with poly-l-lysine (MatTek Corp.). Before filming, the cells were switched to 37°C CO2-independent media (Invitrogen).
Immunofluorescence analysis
HeLa or CHO cells were transfected and fixed as previously described (Maney et al., 1998) after 48 h or 24 h, respectively. Cells were labeled with anti-Hec1 (Abcam, Inc.) or affinity-purified rabbit anti–glu-tubulin (Chemicon) and appropriately conjugated secondary antibodies (Jackson Immunochemicals). For glu-tubulin quantification, cells expressed chimeric proteins for 20 h and were incubated in 2.5 μM nocodazole (Sigma-Aldrich) for 90 min, a time which has been demonstrated to be sufficient for the retyrosination of tubulin in CHO cells (Webster et al., 1987). The cells were reversed into nocodazole-free culture media containing 5 μM MG132 (Calbiochem) for either 20 or 120 min. The cells were fixed, and the mean fluorescence intensity of glu-tubulin staining was measured adjacent to the pole using ImageJ (National Institutes of Health). There was no significant difference between controls and MCAK-depleted cells at 20 min, although the MCAK-enriched cells exhibited a significant suppression in the appearance of glu-tubulin (P < 0.005). By 120 min, mitotic cells with MCAK-depleted centromeres had a significant increase in glu-tubulin (P < 0.0001) relative to controls. MCAK-enriched centromeres were significantly slowed in the accumulation of glu-tubulin (P < 0.005).
Live imaging
Transfected cells were imaged using an inverted microscope (Diaphot 200; Nikon) with a 60× 1.4 NA lens (Nikon) and either a cooled CCD camera (Micromax CCD; Photometrics) or a spinning disk unit (CARV; Kinetic Imaging) and camera (ORCA ER; Hamamatsu). Images were acquired using MetaMorph (MDS Analytical Technologies). Temperature was maintained at 36°C using a thermoelectric stage. Some cells were imaged on a deconvolution microscope system (DeltaVision-RT; Applied Precision) equipped with a 60× 1.4 NA lens (Olympus) and a 37°C environmental chamber (Applied Precision). Photoactivation and FRAP studies were performed on a confocal microscope (LSM 510; Carl Zeiss, Inc.). For the flux measurements, images were collected every 10 s. For the aurora B inhibition, ZM447439 (AstraZeneca) was added to 5 mM, and cells were imaged 1 h later.
Analysis
Individual kinetochores were tracked manually on spindles rotated so the longitudinal spindle axis was along the x axis. Tracking was accomplished using ImageJ, and y axis movement was subtracted out to simplify scoring oscillatory movement. None of the treatments appear to increase y axis (lateral) movement above control cells. Speed corresponded to distance along the x axis per time interval. Sister centromere separation was calculated by measuring the centroid to centroid distance of GCPB-, GCPBM-, or GCPBMhyp-labeled centromeres in ImageJ for as many frames as both remained in focus. Data consisted of successive frames (usually 30) for between five and eight centromere pairs per cell for at least three different cells. The measurements were averaged, and means were reported. There was no difference between averaging all of the measurements and pooling the measurements per centromere. Histograms and tracking data were plotted in IGOR Pro (WaveMetrics). For the flux measurements, distance-calibrated fluorescence profiles measured in ImageJ were overlaid, and the distance between the center of the oscillating metaphase centromeres to the moving front of the bulk photoactivated bar was measured over time. In effect, this measured the maximum rate of flux but did not distinguish between differences in heterogeneous behavior within the kinetochore fiber.
Online supplemental material
Fig. S1 shows that Hec1 label is normal in cells expressing chimeric and mutant MCAK constructs. Fig. S2 shows that BubR1 levels are slightly elevated on centromeres with increased MCAK and decreased tension (A) and that recovery of nonkinetochore MTs is not significantly different between control cells and MCAK-enriched or MCAK-depleted mitotic cells (B). Fig. S3 shows a diagram of the tubulin tyrosination and detyrosination cycle (A) and that detyrosinated tubulin (glu-tubulin) is increased in stable subsets of MTs in interphase and mitotic cells (B). Video 1 shows a metaphase HeLa cell expressing GCPBM. Video 2 shows a metaphase HeLa cell coexpressing RAMFLhyp and GCPB. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200707120/DC1.
Acknowledgments
Greg Martin (Keck Center for Neural Imaging, San Fransisco, CA) and Paulette Brunner (Center for Cell Dynamics, Friday Harbor Laboratories, Friday Harbor, WA) provided invaluable assistance with live imaging. We thank Jason Stumpff for many helpful discussions. We are indebted to Chip Asbury and Andrew Franck for the IGOR tracking macro.
This work was supported by the National Institutes of Health (grant GM69429 to L. Wordeman). The Center for Cell Dynamics is a National Institutes of Health Center for Excellence (grant GM066050).
Abbreviations used in this paper: CENP, centromere protein; MCAK, mitotic centromere-associated kinesin; mRFP, monomeric RFP; MT, microtubule.
References
- Andrews, P.D., Y. Ovechkina, N. Morrice, M. Wagenbach, K. Duncan, L. Wordeman, and J.R. Swedlow. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., O. Tour, A.E. Palmer, P.A. Steinbach, G.S. Baird, D.A. Zacharias, and R.Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris, L., and J. Morabito. 2004. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell. 15:1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini, D., B. Howell, P. Maddox, A. Khodjakov, F. Degrassi, and E.D. Salmon. 2001. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 153:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini, D., B. Moree, J.C. Canman, and E.D. Salmon. 2003. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 116:4213–4225. [DOI] [PubMed] [Google Scholar]
- Cimini, D., X. Wan, C.B. Hirel, and E.D. Salmon. 2006. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 16:1711–1718. [DOI] [PubMed] [Google Scholar]
- DeLuca, J.G., W.E. Gall, C. Ciferri, D. Cimini, A. Musacchio, and E.D. Salmon. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982. [DOI] [PubMed] [Google Scholar]
- Desai, A., S. Verma, T.J. Mitchison, and C.E. Walczak. 1999. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 96:69–78. [DOI] [PubMed] [Google Scholar]
- Dong, Y., K.J. Vanden Beldt, X. Meng, A. Khodjakov, and B.F. McEwen. 2007. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9:516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley, D.M., A.M. Ainsztein, A.M. Mackay, I.G. Goldberg, and W.C. Earnshaw. 1997. Chromosomal proteins and cytokinesis: patterns of cleavage furrow formation and inner centromere protein positioning in mitotic heterokaryons and mid-anaphase cells. J. Cell Biol. 136:1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, N.J., and D.A. Compton. 2004. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 166:473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, N.J., K. Upton, and D.A. Compton. 2005. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 15:1827–1832. [DOI] [PubMed] [Google Scholar]
- Gundersen, G.G., and J.C. Bulinski. 1986. Distribution of tyrosinated and nontyrosinated α-tubulin during mitosis. J. Cell Biol. 102:1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, D.F., K.J. Fowler, E. Earle, R. Saffery, P. Kalitsis, H. Trowell, J. Hill, N.G. Wreford, D.M. de Kretser, M.R. Cancilla, et al. 1998. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 141:309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, A.W., M. Caplow, D.L. Coy, W.O. Hancock, S. Diez, L. Wordeman, and J. Howard. 2003. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 11:445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and C.L. Rieder. 1996. Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J. Cell Biol. 135:315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith, S.L., A. Khodjakov, P. Hergert, and C.E. Walczak. 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 15:1146–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton, A.L., W. Lan, and P.T. Stukenberg. 2006. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 16:1705–1710. [DOI] [PubMed] [Google Scholar]
- Lampson, M.A., K. Renduchitala, A. Khodjakov, and T.M. Kapoor. 2004. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6:232–237. [DOI] [PubMed] [Google Scholar]
- Lan, W., X. Zhang, S.L. Kline-Smith, S.E. Rosasco, G.A. Barrett-Wilt, J. Shabanowitz, D.F. Hunt, C.E. Walczak, and P.T. Stukenberg. 2004. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 14:273–286. [DOI] [PubMed] [Google Scholar]
- Maney, T., A.W. Hunter, M. Wagenbach, and L. Wordeman. 1998. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142:787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney, T., M. Wagenbach, and L. Wordeman. 2001. Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J. Biol. Chem. 276:34753–34758. [DOI] [PubMed] [Google Scholar]
- Moore, A.T., K.E. Rankin, G. von Dassow, L. Peris, M. Wagenbach, Y. Ovechkina, A. Andrieux, D. Job, and L. Wordeman. 2005. MCAK associates with the tips of polymerizing microtubules. J. Cell Biol. 169:391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovechkina, Y., M. Wagenbach, and L. Wordeman. 2002. K-loop insertion restores microtubule depolymerizing activity of a “neckless” MCAK mutant. J. Cell Biol. 159:557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, G.H., and J. Lippincott-Schwartz. 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 297:1873–1877. [DOI] [PubMed] [Google Scholar]
- Perez-Castro, A.V., F.L. Shamanski, J.J. Meneses, T.L. Lovato, K.G. Vogel, R.K. Moyzis, and R. Pedersen. 1998. Centromeric protein B null mice are viable with no apparent abnormalities. Dev. Biol. 201:135–143. [DOI] [PubMed] [Google Scholar]
- Pinsky, B.A., and S. Biggins. 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15:486–493. [DOI] [PubMed] [Google Scholar]
- Pinsky, B.A., C. Kung, K.M. Shokat, and S. Biggins. 2006. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 8:78–83. [DOI] [PubMed] [Google Scholar]
- Pluta, A.F., N. Saitoh, I. Goldberg, and W.C. Earnshaw. 1992. Identification of a subdomain of CENP-B that is necessary and sufficient for localization to the human centromere. J. Cell Biol. 116:1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, G.C., S.L. Rogers, T.A. Schwimmer, S.C. Ems-McClung, C.E. Walczak, R.D. Vale, J.M. Scholey, and D.J. Sharp. 2004. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 427:364–370. [DOI] [PubMed] [Google Scholar]
- Shelby, R.D., K.M. Hahn, and K.F. Sullivan. 1996. Dynamic elastic behavior of α-satellite DNA domains visualized in situ in living human cells. J. Cell Biol. 135:545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens, R.V., V.P. Skeen, and E.D. Salmon. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122:859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout, J.R., R.S. Rizk, S.L. Kline, and C.E. Walczak. 2006. Deciphering protein function during mitosis in PtK cells using RNAi. BMC Cell Biol. 7:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, D.R., G.G. Gundersen, J.C. Bulinski, and G.G. Borisy. 1987. Assembly and turnover of detyrosinated tubulin in vivo. J. Cell Biol. 105:265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlke, G., A.K. Ruby, C.L. Hart, B. Ly, N. Hom-Booher, and R.D. Vale. 1997. Microtubule interaction site of the kinesin motor. Cell. 90:207–216. [DOI] [PubMed] [Google Scholar]
- Wordeman, L., and T.J. Mitchison. 1995. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 128:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]