Abstract
The mitochondrial outer membrane contains two preprotein translocases: the general translocase of outer membrane (TOM) and the β-barrel–specific sorting and assembly machinery (SAM). TOM functions as the central entry gate for nuclear-encoded proteins. The channel-forming Tom40 is a β-barrel protein, whereas all Tom receptors and small Tom proteins are membrane anchored by a transmembrane α-helical segment in their N- or C-terminal portion. Synthesis of Tom precursors takes place in the cytosol, and their import occurs via preexisting TOM complexes. The precursor of Tom40 is then transferred to SAM for membrane insertion and assembly. Unexpectedly, we find that the biogenesis of α-helical Tom proteins with a membrane anchor in the C-terminal portion is SAM dependent. Each SAM protein is necessary for efficient membrane integration of the receptor Tom22, whereas assembly of the small Tom proteins depends on Sam37. Thus, the substrate specificity of SAM is not restricted to β-barrel proteins but also includes the majority of α-helical Tom proteins.
Introduction
Mitochondria from the baker's yeast Saccharomyces cerevisiae are residence to ∼1,000 different proteins (Sickmann et al., 2003; Prokisch et al., 2004; Reinders et al., 2006). The vast majority of mitochondrial proteins is nuclear encoded, synthesized as precursors on cytosolic ribosomes, and must subsequently be transported into the organelle. This challenging task of precursor delivery and integration into one of the four mitochondrial subcompartments (mitochondrial outer membrane, intermembrane space, inner membrane, and matrix) is essential for organelle biogenesis and ultimately for eukaryotic cell viability (Dolezal et al., 2006; Neupert and Hermann, 2007). Accordingly, the presence of dynamic translocation and assembly machineries within all four mitochondrial subcompartments permits the effective recognition, translocation, and sorting of mitochondrial precursors (Jensen and Johnson, 2001; Hoogenraad et al., 2002; Endo et al., 2003; Koehler, 2004; Rehling et al., 2004; Oka and Mihara, 2005).
A crossroads in the import of all nuclear-encoded precursors takes place at the level of the outer membrane, where they encounter the translocase of outer membrane (TOM) complex (Gabriel et al., 2003; Pfanner et al., 2004; Rapaport, 2005; for review see Ryan, 2004). This multisubunit machine effectively translocates precursors across the outer membrane, upon which there is a specific segregation of import pathways induced by sorting elements within precursors. In yeast, the 450-kD TOM complex consists of the general protein import channel, which is formed by the β-barrel protein Tom40, the import receptors Tom20 and Tom22, as well as three small Tom proteins (Tom5, Tom6, and Tom7), which are involved in assembly and stability of the TOM complex (Hill et al., 1998; Meisinger et al., 2001, 2004; Model et al., 2001, 2002). In addition, the receptor Tom70 forms a homodimer that transiently interacts with the TOM complex and facilitates the transfer of hydrophobic precursor proteins from cytosolic chaperones to the TOM complex (Wiedemann et al., 2001; Young et al., 2003). All Tom precursors are encoded by the nuclear genome and must also be imported into mitochondria and assembled into the TOM complex, a process for which they require the presence of preexisting TOM machinery (Keil and Pfanner, 1993; Dembowski et al., 2001; Model et al., 2001; Rapaport et al., 2001; Wiedemann et al., 2003; Nakamura et al., 2004).
Although the TOM complex is involved in the import of several hundred different mitochondrial precursor proteins, it is not capable of integrating β-barrel precursors into the mitochondrial outer membrane, such as Tom40 and the most abundant outer membrane protein, porin. Membrane insertion of β-barrel proteins requires the sorting and assembly machinery (SAM) complex of the outer membrane. The SAM complex is composed of three core constituents: Sam37 (Mas37), which was the first identified component (Wiedemann et al., 2003), and the two essential proteins Sam50 (Tob55/Omp85; Kozjak et al., 2003; Paschen et al., 2003; Gentle et al., 2004) and Sam35 (Tob38/Tom38; Ishikawa et al., 2004; Milenkovic et al., 2004; Waizenegger et al., 2004). Sam50 is an integral membrane protein conserved from bacteria to humans. Its bacterial counterpart, Omp85/YaeT, is required for the insertion of β-barrel proteins into the outer membrane of Gram-negative bacteria (Voulhoux et al., 2003; Bos and Tommassen, 2004; Humphries et al., 2005; Schleiff and Soll, 2005; Dolezal et al., 2006; Ruiz et al., 2006; Kozjak-Pavlovic et al., 2007; for review see Ryan, 2004). Sam35 and Sam37 behave as peripheral membrane proteins that are anchored to the outer membrane by their tight association with Sam50.
Tom40 is the only β-barrel Tom protein, whereas the other Tom subunits are anchored in the outer membrane by a transmembrane α helix. Tom20 and Tom70 contain a membrane anchor at the N terminus, whereas Tom22 and the small Tom proteins are anchored in the outer membrane by a membrane anchor that is located at the C terminus (Tom5, Tom6, and Tom7) or in the C-terminal half of the protein (Tom22). The targeting signals of the α-helical Tom proteins are typically contained in the transmembrane segment and hydrophilic flanking regions (Cao and Douglas, 1995; Egan et al., 1999; Kanaji et al., 2000; Dembowski et al., 2001; Allen et al., 2002; Habib et al., 2003; Horie et al., 2003; Waizenegger et al., 2003). Import of the precursors of α-helical Tom proteins into mitochondria was shown to require various components of preexisting TOM complexes (Schneider et al., 1991; Keil and Pfanner, 1993; Dembowski et al., 2001; Nakamura et al., 2004; Ahting et al., 2005; Rapaport, 2005). Late steps of the assembly of Tom40 as well as of α-helical Tom proteins are promoted by mitochondrial distribution and morphology (Mdm) proteins (Meisinger et al., 2004, 2006, 2007). Based on the similarity to the bacterial Omp85 machinery, it was concluded that the SAM complex played a selective role in the biogenesis of β-barrel proteins (Pfanner et al., 2004; Paschen et al., 2005; Dolezal et al., 2006; for review see Ryan, 2004). This conclusion was underscored by the finding that the import of Tom20 was not impaired by inactivation of SAM components (Paschen et al., 2003; Milenkovic et al., 2004; Waizenegger et al., 2004). The current view thus includes that α-helical Tom proteins are imported by preexisting TOM complexes and assembled with the help of Mdm proteins, whereas the SAM complex is dedicated to the biogenesis of β-barrel proteins and is not relevant for α-helical proteins.
In this study, we report the surprising observation that the SAM complex is involved in the biogenesis of several α-helical subunits of the TOM complex. All three SAM subunits are required for the efficient membrane integration of Tom22, whereas the small Tom members display a requirement on Sam37 for their assembly into the TOM complex. These results point to a novel function for the SAM complex in the biogenesis of alternative mitochondrial outer membrane precursors and expand the substrate specificity of this machinery beyond that of β-barrel precursors.
Results
The membrane-integral SAM subunit Sam50 is involved in the biogenesis of Tom22
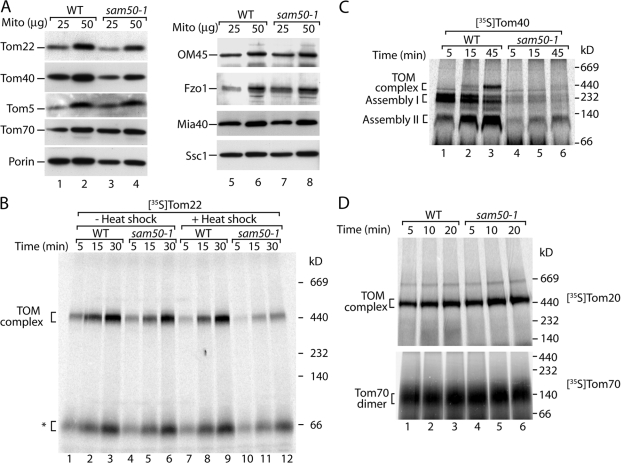
We used a temperature-sensitive yeast mutant of SAM50 (sam50-1) that has been used in defining the role of the essential protein Sam50 in the biogenesis of β-barrel precursors (Kozjak et al., 2003). To probe for the spectrum of proteins affected by a defect of Sam50 in vivo, we shifted the cells to the nonpermissive temperature of 37°C for 10 h before isolation of mitochondria. Analysis of the steady-state protein levels of sam50-1 mitochondria by immunodecoration revealed a surprising reduction in the levels of Tom22 in comparison with wild-type mitochondria (Fig. 1 A). The reduction occurred to a comparable level as that of the β-barrel proteins Tom40 and porin, whereas Tom5, Tom70, OM45, and Fzo1 of the outer membrane, the intermembrane space resident Mia40, and the matrix-located Ssc1 remained unaffected (Fig. 1 A).
Figure 1.
Sam50 is required for the biogenesis of Tom22. (A) Mitochondria were isolated from wild-type and sam50-1 yeast cells after a 10-h in vivo heat shock at 37°C. Mitochondrial proteins were subjected to SDS-PAGE and immunodecoration. (B) Wild-type (WT) and sam50-1 cells were grown at 23°C. Mitochondria were isolated and preincubated at 25°C for 2 min (lanes 1–6) or for 37°C for 15 min (lanes 7–12). Import of the 35S-labeled precursor of Tom22 was performed at 25°C for the indicated times. Mitochondria were isolated, lysed in digitonin-containing buffer, and subjected to blue native electrophoresis and digital autoradiography. The asterisk indicates the low molecular weight form of the Tom22 assembly pathway. (C) The experiment was performed as described for B except that the 35S-labeled Tom40 precursor was used. (D) The 35S-labeled Tom20 (top) and Tom70 (bottom) precursors were imported in mitochondria from wild-type and sam50-1 as described for B.
To minimize indirect pleiotropic effects, we grew the yeast cells at the permissive temperature of 23°C, isolated mitochondria, and induced the mutant phenotype in vitro by incubation of the mitochondria for 15 min at 37°C. Under these conditions, the levels of preexisting TOM complex as well as of individual Tom subunits were comparable between sam50-1 and wild-type mitochondria, and the mutant mitochondria were fully competent in transporting precursor proteins through TOM to internal mitochondrial compartments (Kozjak et al., 2003). Tom22 was synthesized as radiolabeled precursor in rabbit reticulocyte lysate and imported into the isolated mitochondria. Assembly into the TOM complex was directly monitored by blue native electrophoresis upon lysis of the mitochondria with digitonin (Model et al., 2001; Meisinger et al., 2004). Assembly of Tom22 was inhibited in mitochondria from sam50-1 cells in comparison with that of the corresponding wild-type mitochondria (Fig. 1 B, lanes 7–12), indeed indicating an involvement of Sam50 in the biogenesis of Tom22. The defect in Tom22 assembly depended on induction of the mutant phenotype, as in the absence of an in vitro heat shock, import and assembly of Tom22 in sam50-1 mitochondria was indistinguishable from that of wild-type mitochondria (Fig. 1 B, lanes 1–6). As controls, we imported the radiolabeled precursors of Tom40, Tom20, and Tom70 into heat-treated mitochondria. In wild-type mitochondria, Tom40 assembly occurred via intermediates of 250 kD (assembly I, corresponding to the SAM complex) and 100 kD (assembly II, reflecting the association of Tom5 with Tom40) before maturation into the TOM complex (Wiedemann et al., 2003; Meisinger et al., 2004), whereas in sam50-1 mitochondria, the assembly was strongly inhibited (Fig. 1 C; Kozjak et al., 2003). In contrast, Tom20 assembly into the TOM complex and Tom70 assembly to the mature homodimer occurred with similar efficiency in wild-type and sam50-1 mitochondria (Fig. 1 D). We conclude that mitochondria with a defective Sam50 are not only impaired in the assembly pathway of the β-barrel protein Tom40 but also the α-helical protein Tom22.
Both peripheral SAM proteins Sam35 and Sam37 affect the assembly pathway of Tom22
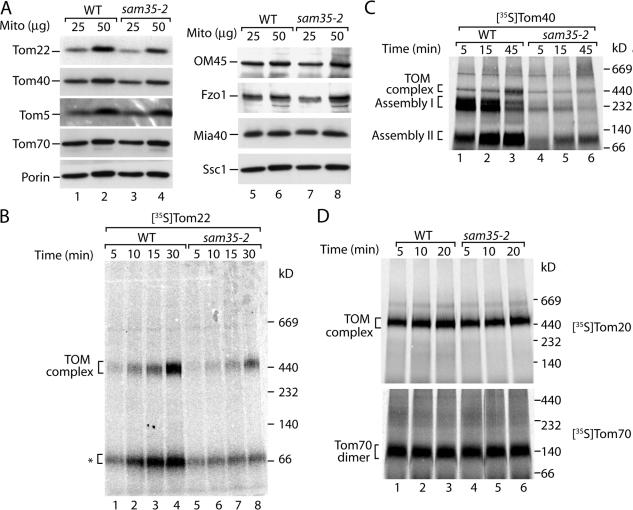
We asked whether the two peripheral SAM proteins Sam35 and Sam37, which expose domains at the cytosolic side, also affect the biogenesis pathway of Tom22. The essential protein Sam35 was addressed through use of the temperature-sensitive yeast mutant sam35-2 (Milenkovic et al., 2004). The cells were exposed to an in vivo heat shock at 37°C for 10 h, and mitochondria were isolated and analyzed for steady-state protein levels. Remarkably, the level of Tom22 in the sam35-2 mutant mitochondria was reduced to a greater extent than that of Tom40, whereas further proteins analyzed were present in similar amounts as in wild-type mitochondria (Fig. 2 A). For protein import experiments, we used sam35-2 mitochondria that were isolated from cells grown at permissive conditions. Before incubation with radiolabeled precursor proteins, the isolated mitochondria were subjected to a short-term shift to nonpermissive conditions (37°C). Similar to the situation with sam50-1 mitochondria, the sam35-2 mitochondria retain wild-type levels of the TOM complex in addition to all of the mitochondrial marker proteins analyzed and efficiently import precursor proteins to internal mitochondrial compartments (Milenkovic et al., 2004). However, the assembly of Tom22 was strongly inhibited in sam35-2 mitochondria (Fig. 2 B). As controls, the import of Tom40 displayed the classic defect in β-barrel protein assembly, whereas the import of Tom20 and Tom70 remained unaffected (Fig. 2, C and D; Milenkovic et al., 2004). Thus, the second essential SAM protein, Sam35, also affects the assembly pathway of the precursor of Tom22.
Figure 2.
Sam35 is required for the biogenesis of Tom22. (A) Mitochondria were isolated from wild-type and sam35-2 yeast cells after a 10-h in vivo heat shock at 37°C. Mitochondrial proteins were subjected to SDS-PAGE and immunodecoration. (B) Wild-type (WT) and sam35-2 cells were grown at 23°C. Mitochondria were isolated and preincubated at 37°C for 15 min, and import of 35S-labeled Tom22 was performed at 25°C for the indicated times. Mitochondria were isolated, lysed in digitonin-containing buffer, and subjected to blue native electrophoresis and digital autoradiography. The asterisk indicates the low molecular weight form of the Tom22 assembly pathway. (C) The experiment was performed as outlined for B, but 35S-labeled Tom40 was used. (D) Import of 35S-labeled Tom20 (top) and Tom70 (bottom) precursors in mitochondria from wild-type and sam35-2 was performed as described for B.
Yeast cells lacking Sam37 are viable but are impaired in growth at elevated temperature (Gratzer et al., 1995; Wiedemann et al., 2003). sam37Δ cells grown at 30°C not only showed reduced levels of Tom40 as reported previously (Wiedemann et al., 2003) but the levels of Tom22 were also strongly reduced (Fig. 3 A). For analysis of protein import, we grew the cells at 23°C, at which the endogenous levels of the TOM complex remained unaffected (Wiedemann et al., 2003). Import of radiolabeled Tom22 into isolated sam37Δ mitochondria revealed a similar assembly defect (Fig. 3 B) as observed for sam50-1 and sam35-2 mitochondria. Assembly of Tom40 displayed an arrest at a smaller form of assembly intermediate I that reflected the crippled SAM complex lacking Sam37 (Fig. 3 C; Wiedemann et al., 2003), whereas assembly of Tom20 and Tom70 proceeded as efficiently as in wild-type mitochondria (Fig. 3 D).
Figure 3.
Sam37 is required for the biogenesis of Tom22. (A) Mitochondria were isolated from wild-type and sam37Δ yeast cells grown at 30°C and analyzed by SDS-PAGE and immunodecoration. (B) Wild-type (WT) and sam37Δ cells were grown at 23°C, and mitochondria were isolated. 35S-labeled Tom22 was imported at 25° for the indicated times and analyzed by blue native electrophoresis and digital autoradiography. The asterisk indicates the low molecular weight form of the Tom22 assembly pathway. (C) Import of the 35S-labeled Tom40 precursor was performed as described for B. (D) The experiment was performed as outlined for B, but the 35S-labeled precursors of Tom20 (top) and Tom70 (bottom) were imported into isolated mitochondria.
Thus, mutants of each of the three SAM proteins display a similar defect in the biogenesis of Tom22 in vivo and in vitro, whereas the assembly of Tom20 and Tom70 is not affected. Therefore, these α-helical Tom proteins use different assembly pathways. The pathway of Tom22 possesses some parallels to that of β-barrel precursors, displaying a requirement on all three components of the SAM complex for import and assembly into the outer membrane. β-barrel precursors are transferred from the TOM complex to the SAM complex with the help of TIM (translocase of inner membrane) chaperone complexes of the intermembrane space (Hoppins and Nargang, 2004; Wiedemann et al., 2004). Thus, we asked whether Tom22 displayed a similar dependence on intermembrane space chaperones during its biogenesis. Isolated mitochondria were subjected to swelling to generate mitoplasts, permitting the release of soluble intermembrane space components, including the TIM chaperone complexes (Wiedemann et al., 2004). The assembly pathway of Tom22 was not inhibited by the swelling of mitochondria (Fig. 4 A), whereas the biogenesis pathway of Tom40 was blocked, and assembly intermediate I (SAM intermediate) was not formed as has been reported previously (Fig. 4 B; Wiedemann et al., 2004). We conclude that the biogenesis pathway of Tom22 resembles that of β-barrel precursors with regard to the dependence on SAM components; however, its transfer to the SAM complex does not require the soluble chaperone complexes of the intermembrane space.
Figure 4.
Swelling of mitochondria blocks the assembly pathway of Tom40 but not of Tom22. Mitochondria isolated from wild-type yeast cells were preincubated in isotonic buffer or hypotonic (swelling) buffer for 30 min on ice. The mitochondria/mitoplasts were isolated and incubated with the 35S-labeled precursors of Tom22 (A) and Tom40 (B) at 25°C for the indicated times. Mitochondria were reisolated, lysed in digitonin-containing buffer, and subjected to blue native electrophoresis and digital autoradiography. The asterisk indicates the low molecular weight form of the Tom22 assembly pathway.
Integration of Tom22 into the outer membrane is impaired in mutants of Sam50, Sam37, and Sam35
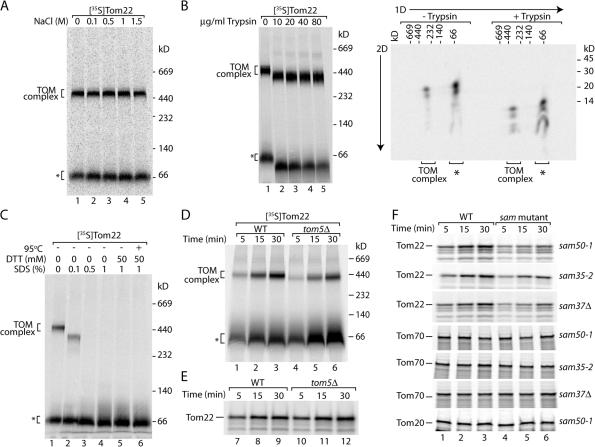
We noticed that the blue native assays for assembly of Tom22 into the TOM complex also revealed a low molecular weight form that was reduced in mitochondria from all three sam mutants (Figs. 1 B, 2 B, and 3 B; asterisks). It was possible that this low molecular weight species represented an intermediate stage in the biogenesis of Tom22, and, therefore, we characterized it further. Like the mature TOM complex (Meisinger et al., 2001), the low molecular weight species was resistant to treatment of mitochondria with high concentrations of salt (Fig. 5 A), suggesting that it was stably associated with the mitochondrial membranes. Upon treatment of mitochondria with trypsin, the mature TOM complex and the low molecular weight form of Tom22 migrated faster on the blue native gels (Fig. 5 B, left). To determine whether the low molecular weight form of Tom22 was correctly oriented within the lipid bilayer, we performed a 2D analysis of trypsin-treated mitochondria (i.e., blue native separation followed by denaturing SDS-PAGE). An identical profile consisting of three characteristic fragments was observed for Tom22 from both the mature TOM complex and the low molecular weight form (Fig. 5 B, right), whereas Tom22 was completely degraded when the membranes were lysed with detergent before the protease treatment (Keil and Pfanner, 1993). Thus, the low molecular weight species of Tom22 was inserted into the outer membrane in the same orientation as Tom22 in the mature TOM complex. We used antibodies directed against various mitochondrial outer membrane and intermembrane space proteins, but none of them recognized this species of Tom22 (unpublished data). Indeed, the mobility of this species on blue native gels was not altered by harsh treatments, including heating in the presence of SDS and reductant (Fig. 5 C) that leads to the dissociation of all known mitochondrial translocase complexes, indicating that it represented monomeric Tom22 (the mobility of membrane proteins in the low molecular weight range of blue native gels is often slower than that of soluble marker proteins; Wiedemann et al., 2001, 2003). These results suggest that the monomeric form of Tom22 was correctly integrated into the outer membrane of wild-type mitochondria.
Figure 5.
The SAM complex is involved in the integration of Tom22 into the outer membrane. (A) 35S-labeled Tom22 precursor was imported into wild-type mitochondria. Mitochondria were isolated and resuspended in SEM buffer containing the indicated NaCl concentration and incubated on ice for 10 min. Mitochondria were reisolated, washed in SEM buffer, lysed in digitonin-containing buffer, and subjected to blue native electrophoresis and digital autoradiography. (B) 35S-labeled Tom22 precursor was imported into isolated wild-type mitochondria and, after import, was incubated with the indicated concentration of trypsin for 10 min on ice. Trypsin was inhibited by the addition of 30-fold excess soybean trypsin inhibitor and incubation on ice for a further 10 min. Mitochondria were isolated, washed in SEM buffer, and treated for blue native electrophoresis (left). After electrophoresis, samples treated with 0 and 50 μg/ml trypsin were analyzed by SDS-PAGE in the second dimension (right). (C) 35S-labeled Tom22 precursor was imported into wild-type mitochondria. After import, mitochondria were isolated and lysed in digitonin-containing buffer with the indicated amounts of SDS and DTT. Samples were incubated for 15 min on ice (lanes 1–5) or at 95°C for 5 min (lane 6). (D) 35S-labeled Tom22 precursor was imported into isolated mitochondria from wild-type (WT) and tom5Δ cells (grown at 23°C) for the indicated times. The mitochondria were reisolated, lysed in digitonin-containing buffer, and subjected to blue native electrophoresis and digital autoradiography. (A–D) Asterisks indicate the low molecular weight forms of the Tom22 assembly pathway. (E) Import of Tom22 was performed as described for D. The reisolated mitochondria were treated with 0.1 M Na2CO3, pH 11.5, for 30 min on ice. Membrane sheets were isolated by ultracentrifugation, solubilized in laemmli buffer, and analyzed by Tris-tricine gel electrophoresis and digital autoradiography. (F) 35S-labeled precursors of Tom22, Tom70, or Tom20 were imported into isolated mitochondria from wild-type, sam50-1, sam35-2, and sam37Δ yeast for the indicated times. The reisolated mitochondria were treated with Na2CO3 and analyzed as described for E.
Thus, we asked why the levels of the low molecular weight form of Tom22 were reduced in all three sam mutants and wondered whether this indicated an early preassembly function of the SAM complex in the biogenesis pathway of Tom22. We searched for an outer membrane mutant that differentially affected assembly of Tom22 into the mature TOM complex and formation of the low molecular weight form. We found that mitochondria lacking Tom5 displayed a differential effect. Tom5 has been shown to associate with Tom40 at a post-SAM stage (assembly II; Wiedemann et al., 2003; Meisinger et al., 2004). In tom5Δ mitochondria, assembly of Tom22 into the TOM complex was reduced, whereas the low molecular weight form accumulated in increased amounts (Fig. 5 D). To probe for the membrane integration of Tom22, we treated the mitochondria at alkaline pH, conditions that lead to the extraction of soluble and peripheral membrane proteins, whereas integral membrane proteins remain membrane inserted (Fujiki et al., 1982; Burri et al., 2006; Stojanovski et al., 2007). The precursor of Tom22 was imported into isolated mitochondria that were subsequently treated with sodium carbonate, pH 11.5. Membrane-integrated species were isolated through ultracentrifugation and separated by SDS-PAGE. Fig. 5 E shows that the membrane integration of Tom22 occurred with similar efficiency in wild-type and tom5Δ mitochondria, indicating that Tom22, which was accumulated in the low molecular weight form in tom5Δ mitochondria, was already integrated into the outer membrane but not yet assembled into the TOM complex.
We asked whether the SAM complex participated in the biogenesis of Tom22 in a pre- or postmembrane integration manner. 35S-labeled Tom22 was imported into sam50-1, sam35-2, and sam37Δ mitochondria followed by treatment at alkaline pH. We observed a decrease in the efficiency of Tom22 membrane integration in all three mutants; Tom22 insertion was not blocked completely but proceeded at a reduced level (Fig. 5 F). For comparison, the membrane integration of Tom20 and Tom70 was not affected by the sam mutant mitochondria (Fig. 5 F), excluding the possibility that the sam mutants indirectly altered the extractability of Tom proteins from mitochondria at alkaline pH. Thus, the three SAM proteins are required for the biogenesis of Tom22 at a different stage than Tom5. Although Tom5 is not required for membrane integration of Tom22 but only for the subsequent assembly reaction, the subunits of the SAM complex increase the efficiency of the membrane insertion of Tom22.
We asked whether the SAM complex may also be involved in the biogenesis of further α-helical outer membrane proteins and monitored the import of various proteins into sam50-1, sam35-2, and sam37Δ mitochondria by extraction with alkaline pH. Additionally, blue native electrophoresis was used for proteins that migrated as complexes on native gels. We tested the following proteins: C tail–anchored Fis1 (Mozdy et al., 2000; Tieu and Nunnari, 2000) and Gem1 (Fig. 6 A; Frederick et al., 2004), N-terminally anchored OM45 (Fig. 6 B; Yaffe et al., 1989; Waizenegger et al., 2004), and proteins that have more than one transmembrane domain, Fzo1 (Hermann et al., 1998; Rapaport et al., 1998) and Ugo1 (Fig. 6, C and D; Sesaki and Jensen, 2001; Coonrod et al., 2007). None of these proteins showed a dependence on a functional SAM complex for integration into the outer membrane. Thus, the results obtained so far indicated that the biogenesis of Tom22 but not of other α-helical outer membrane proteins required the SAM complex.
Figure 6.
Import and assembly of non-Tom outer membrane precursors occurs independently of the SAM complex. (A) 35S-labeled precursors of Fis1 (top) and Gem1 (bottom) were imported into mitochondria from wild-type (WT), sam50-1, sam35-2, and sam37Δ yeast cells for the indicated times. After, import-isolated mitochondria were treated with 0.1 M Na2CO3, pH 11.5, for 30 min on ice. Membranes were isolated by ultracentrifugation, solubilized in laemmli buffer, and analyzed by Tris-tricine gel electrophoresis and digital autoradiography. (B) 35S-labeled OM45 precursor was imported as described in A. Mitochondria were solubilized in digitonin-containing buffer for blue native electrophoresis (top) or treated with 0.1 M Na2CO3, pH 11.5, for SDS-PAGE analysis (bottom). (C) 35S-labeled Fzo1 precursor was imported, and samples were treated as described for A. (D) The 35S-labeled precursor of Ugo1 was imported as described for A. Mitochondria were solubilized in digitonin-containing buffer for blue native electrophoresis (top) or treated with 0.1 M Na2CO3, pH 11.5, for SDS-PAGE analysis (bottom).
A role for Sam37 in the assembly of small Tom proteins
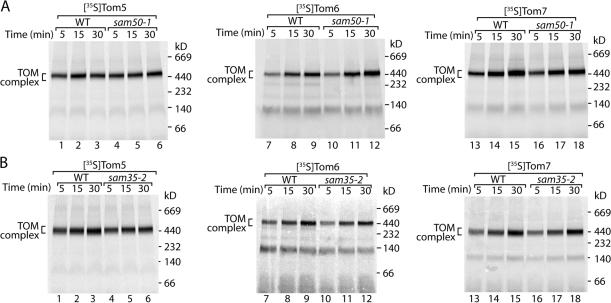
Finally, we asked whether the assembly pathway of the remaining three α-helical subunits of the TOM complex (the small proteins Tom5, Tom6, and Tom7) depended on SAM. The 35S-labeled precursors were imported into isolated mitochondria from wild-type and sam50-1 yeast cells, and assembly was monitored by blue native electrophoresis. Contrary to that of Tom22, the small Tom members displayed no major assembly defect in sam50-1 mitochondria (Fig. 7 A). In a similar manner, import of the small Tom proteins into mitochondria isolated from sam35-2 yeast cells also revealed no substantial assembly defect (Fig. 7 B). Thus, mutants of the two essential SAM proteins did not inhibit the biogenesis of small Tom proteins.
Figure 7.
Assembly of Tom5, Tom6, and Tom7 is independent of Sam50 and Sam35. (A) 35S-labeled Tom5 (lanes 1–6), Tom6 (lanes 7–12), and Tom7 (lanes 13–18) precursors were imported into isolated mitochondria from wild-type (WT) and sam50-1 yeast cells for the indicated times. Mitochondria were isolated, lysed in digitonin-containing buffer, and subjected to blue native electrophoresis and digital autoradiography. (B) 35S-labeled Tom5 (lanes 1–6), Tom6 (lanes 7–12), and Tom7 (lanes 13–18) precursor proteins were imported into isolated mitochondria from wild-type and sam35-2 yeast for the indicated times. Mitochondria were isolated, lysed in digitonin-containing buffer, and subjected to blue native electrophoresis.
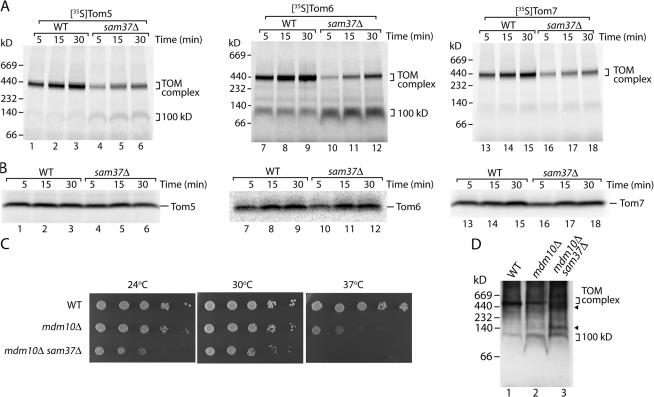
Surprisingly, the corresponding analysis in mitochondria isolated from sam37Δ yeast cells disclosed a noticeable assembly defect for all three small Tom proteins relative to that of mitochondria isolated from wild-type cells (Fig. 8 A). The small Tom proteins were previously shown to assemble via an intermediate stage of ∼100 kD, likely reflecting the assembly intermediate II of Tom40; defects in late steps of TOM assembly in mitochondria lacking Mdm10 or individual small Tom proteins led to an accumulation of the 100-kD intermediate (Model et al., 2001; Meisinger et al., 2004). For Tom6 and, to a smaller extent, also for Tom5, we indeed observed increased amounts of the precursors in the 100-kD form in sam37Δ mitochondria compared with wild-type mitochondria (Fig. 8 A), raising the possibility that Sam37 was required at a late stage in the assembly of small Tom proteins. This view was supported by the unaltered levels of Tom5 in mitochondria isolated from sam50-1 (Fig. 1 A), sam35-2 (Fig. 2 A), and sam37Δ yeast (Fig. 3 A) after in vivo growth at elevated temperature.
Figure 8.
Assembly of Tom5, Tom6, and Tom7 is dependent on Sam37. (A) 35S-labeled Tom5 (lanes 1–6), Tom6 (lanes 7–12), and Tom7 (lanes 13–18) precursors were imported into isolated mitochondria from wild-type (WT) and sam37Δ yeast cells for the indicated times. Reisolated mitochondria were lysed in digitonin-containing buffer and separated by blue native electrophoresis, and radiolabeled proteins were detected by digital autoradiography. (B) 35S-labeled Tom5 (lanes 1–6), Tom6 (lanes 7–12), and Tom7 (lanes 13–18) precursors were imported into isolated mitochondria from wild-type and sam37Δ yeast cells for the indicated times. The reisolated mitochondria were resuspended in 0.1 M Na2CO3, pH 11.5, and incubated on ice for 30 min. Membrane sheets were isolated by ultracentrifugation, solubilized in laemmli buffer, and separated by Tris-tricine gel electrophoresis. (C) Growth of wild-type yeast, mdm10Δ, and mdm10Δ sam37Δ deletion strains on YPD at 24, 30, and 37°C. (D) 50 μg mitochondria from wild-type, mdm10Δ, and mdm10Δ sam37Δ yeast cells were solubilized in digitonin-containing buffer, separated by blue native electrophoresis, and subsequently analyzed by immunoblotting with antibodies directed against Tom5. Arrowheads indicate intermediate complexes.
Thus, we addressed the role of Sam37 in the integration of small Tom proteins into the membrane by sodium carbonate treatment. The 35S-labeled small Tom precursors were imported into mitochondria from the sam37Δ strain, and then the mitochondria were treated at pH 11.5 for the separation of integral and peripheral membrane proteins. The integration of Tom5, Tom6, and Tom7 took place to a comparable extent in mitochondria from wild-type and sam37Δ cells (Fig. 8 B), indicating that Sam37 was required for the assembly of small Tom proteins in a postmembrane integration manner.
The lack of Sam37 has been shown to impair the association of Mdm10 with the SAM complex (Meisinger et al., 2006). Given that mitochondria lacking Mdm10 display a similar assembly defect for the small Tom proteins (Meisinger et al., 2004), we wondered whether the assembly defect observed for the small Tom proteins in sam37Δ mitochondria could be solely attributed to the loss of Mdm10 association with SAM. To address this issue, we generated a strain that was lacking both Sam37 and Mdm10. At all temperatures tested, growth of the double deletion strain was more compromised than that of mdm10Δ, with a complete inability for growth at 37°C (Fig. 8 C), indicating that the lack of both proteins leads to a stronger phenotype than the loss of Mdm10 alone. Mitochondria were isolated from both mdm10Δ and mdm10Δ sam37Δ yeast cells that were grown at low temperature to minimize indirect effects. Blue native electrophoresis and subsequent immunoblotting monitored assembly of the small Tom protein Tom5. In mdm10Δ mitochondria, the amount of Tom5 in the mature 450-kD TOM complex was reduced compared with wild-type mitochondria, and the 100-kD form could be observed (Fig. 8 D, lane 2). The pattern observed in mitochondria from the mdm10Δ sam37Δ double deletion differed considerably from that of mdm10Δ mitochondria, revealing strongly reduced amounts of the mature TOM complex and the presence of additional intermediate complexes (Fig. 8 D, lane 3). We conclude that both Sam37 and Mdm10 are involved in the late assembly steps of small Tom proteins.
Discussion
We report a new function of the SAM complex. To date, this mitochondrial SAM was considered to be a β-barrel–specific machinery, which is also reflected in the alias name TOB (topogenesis of outer membrane β-barrel proteins). We show that the majority of α-helical subunits of the TOM complex depend on functional SAM for proper assembly.
The previous view that SAM was exclusively dedicated to β-barrel proteins was based on several observations. First, all β-barrel precursors analyzed depended on SAM (Tom40, porin, Sam50, and Mdm10). Second, Tom20, which carries an N-terminal α-helical transmembrane segment, was efficiently imported and assembled in mutants of SAM proteins (Paschen et al., 2003; Milenkovic et al., 2004; Waizenegger et al., 2004; this study). Third, the similarity to the bacterial Omp85/YaeT system of protein assembly suggested that the mitochondrial Sam50 machinery was also dedicated to the assembly of β-barrel proteins only. However, in the bacterial outer membrane, β-barrel proteins are the predominant membrane-integral proteins, and, thus, α-helical proteins have not been studied (Schulz, 2002; Ruiz et al., 2006). In contrast, the mitochondrial outer membrane contains more α-helical proteins than β-barrel proteins (Rapaport, 2003; Burri et al., 2006; Schmitt et al., 2006; Zahedi et al., 2006).
The first indication that SAM may be involved in the biogenesis of α-helical proteins was obtained in vivo by determining the steady-state levels of Tom22 upon the growth of yeast mutants of Sam50, Sam35, and Sam37 at elevated temperature. In each case, a decrease in the level of Tom22 was observed comparable with the decrease of Tom40. In agreement with these observations, Hoppins et al. (2007) disclosed a reduction in Tom22 levels in a SAM50 sheltered knockout strain in Neurospora crassa, although the effect was attributed as secondary because of reduced Tom40 levels. However, the use of a temperature-conditional SAM50 mutant in our analysis clearly dismisses Tom40 levels as the causative factor for the reduction in Tom22 levels. A direct analysis of Tom22 assembly in isolated mitochondria by blue native electrophoresis demonstrated a requirement on each of the three SAM proteins for incorporation of the receptor into the TOM complex. We dissected the biogenesis pathway of Tom22 into two consecutive steps: (1) membrane insertion observed as a low molecular weight form on blue native electrophoresis and (2) assembly into the 450-kD TOM complex. The small Tom protein Tom5 permitted separation of both steps, as mutant mitochondria lacking Tom5 were only impaired in the second (assembly) stage and, thus, accumulated the membrane-inserted low molecular weight form. However, mutants of each of the SAM proteins not only inhibited the assembly of Tom22 into the 450-kD complex but also reduced the efficiency of the membrane insertion of Tom22. Thus, the import pathway of the precursor of Tom22 involves initial targeting via the TOM complex (Keil and Pfanner, 1993) and, thus, a loose association with the mitochondrial surface followed by SAM-stimulated insertion into the outer membrane.
A different picture was observed for the three small Tom proteins. Import and assembly of Tom5, Tom6, and Tom7 were not inhibited in mutants of the two essential SAM proteins Sam50 and Sam35. Surprisingly, mitochondria lacking Sam37 displayed defects in assembly of the small Tom proteins into the 450-kD TOM complex, whereas the preceding step of membrane insertion was not impaired in the mutant mitochondria. This late function of Sam37 in TOM assembly resembles that of Mdm10. Mitochondria lacking Mdm10 are impaired in the final steps of assembly of Tom40, Tom22, and small Tom proteins (Meisinger et al., 2004). Cells lacking Mdm10 are viable at low temperature but are impaired in growth at elevated temperature, like cells lacking Sam37 (Gratzer et al., 1995; Wiedemann et al., 2003; Meisinger et al., 2004). A fraction of Mdm10 has been shown to interact with the SAM complex in a Sam37-dependent manner to promote the final maturation steps of the TOM complex (Meisinger et al., 2004, 2006, 2007). However, the role of Sam37 cannot solely be attributed to a recruitment of Mdm10 to the SAM complex, as a double mutant lacking both Sam37 and Mdm10 shows a stronger defect both in cell growth and in small Tom assembly. For the assembly pathway of Tom22, the function of Sam37 is required earlier than that of Mdm10; Sam37 together with Sam50 and Sam35 promote membrane insertion of monomeric Tom22, whereas mitochondria lacking Mdm10 can insert Tom22 into the outer membrane and are impaired in the subsequent assembly steps (Meisinger et al., 2004). Collectively, we conclude that both Sam37 and Mdm10 function in TOM assembly, affecting the β-barrel protein Tom40 and the α-helical proteins Tom22, Tom5, Tom6, and Tom7. However, this function of Sam37 and Mdm10 does not include all α-helical Tom proteins because Tom20 and Tom70 with N-terminal membrane anchors are independent of the SAM/Mdm10 machinery. Moreover, several more α-helical outer membrane proteins analyzed did not depend on an active SAM complex for import into the outer membrane. Only the subunits of the TOM complex with a C-terminal membrane anchor require the help of Sam37 and Mdm10.
Why does the early step of the membrane insertion of Tom22 depend on the SAM complex, whereas the simple tail-anchored proteins Tom5, Tom6, and Tom7 can be membrane inserted in the absence of a functional SAM complex? Rodriguez-Cousiño et al. (1998) and Nakamura et al. (2004) showed that Tom22 is not simply inserted into the outer membrane by a C-terminal membrane anchor but contains several distinct import elements, which are separated at the level of its primary structure: a segment in the cytosolic domain, the transmembrane α helix, and a segment in the intermembrane space domain. The cytosolic segment interacted with the other import elements, suggesting that Tom22 is not simply inserted as a linear polypeptide chain but is targeted in a hairpin structure. The involvement of more than one segment of a polypeptide chain in membrane insertion is a hallmark of β-barrel proteins, and the SAM complex is thought to provide a scaffold for this insertion process at the protein–lipid interphase (Gentle et al., 2004; Pfanner et al., 2004; Gentle et al., 2005; Habib et al., 2005; Paschen et al., 2005; for review see Ryan, 2004). The SAM mutants used in this study did not completely block but reduced the efficiency of the membrane insertion of Tom22, which is in agreement with the view that SAM may function as a scaffold that facilitates the insertion process of partially folded precursor proteins. As β-barrel precursors apparently contain a considerable amount of partially folded elements (Kleinschmidt and Tamm, 1996; Eppens et al., 1997; Rapaport and Neupert, 1999), it is conceivable that their membrane insertion shows a strict dependence on a functional SAM complex.
Irrespective of these speculations, the findings presented here clearly extend the substrate spectrum of the mitochondrial SAM complex to several α-helical proteins. We conclude that the SAM complex is essential for the biogenesis of β-barrel proteins but, in addition, facilitates the biogenesis of α-helical TOM proteins that contain a membrane anchor in their C-terminal portion.
Materials and methods
Isolation of mitochondria and in vitro protein import
S. cerevisiae were grown on YPG medium (1% [wt/vol] yeast extract, 2% [wt/vol] bactopeptone, and 3% [wt/vol] glycerol). The strains used were described previously (Dietmeier et al., 1997; Model et al., 2001; Kozjak et al., 2003; Wiedemann et al., 2003; Milenkovic et al., 2004). The full open reading of MDM10 was disrupted in YPH499 and sam37Δ yeast cells with a kanamycin (kanMX4) cassette. Mitochondria were isolated by differential centrifugation and adjusted to a protein concentration of 10 mg/ml in SEM buffer (250 mM sucrose, 1 mM EDTA, and 10 mM MOPS-KOH, pH 7.2) and stored in aliquots at −80°C. Radiolabeled precursor proteins were generated by in vitro transcription/translation in the presence of [35S]methionine using rabbit reticulocyte lysate (GE Healthcare; Stojanovski et al., 2007). Mitochondria from temperature-sensitive mutants and the corresponding wild-type mitochondria in import buffer (3% [wt/vol] fatty acid–free BSA, 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 5 mM methionine, 2 mM KH2PO4, and 10 mM MOPS-KOH, pH 7.2) were preincubated at 37°C for 15 min. The samples were transferred to 25°C for 2 min, and 4 mM ATP, 2 mM NADH, 100 μg/ml creatine kinase, and 5 mM creatine phosphate were added. Mitochondria not requiring an initial heat shock were added to import buffer supplemented with an energy-regenerating system (4 mM ATP, 2 mM NADH, 100 μg/ml creatine kinase, and 5 mM creatine phosphate) and were preincubated at 25°C for 2 min. The import was initiated by the addition of reticulocyte lysate (5–10% [vol/vol] of import reaction). After the indicated times, mitochondria were isolated by centrifugation and washed in SEM buffer. Samples to be treated for alkaline extraction were resuspended in freshly prepared 0.1 M Na2CO3 and were incubated on ice for 30 min. Membranes were isolated by centrifugation at 100,000 g for 1 h at 4°C and subsequently solubilized in laemmli buffer and separated by Tris-tricine PAGE.
Blue native electrophoresis
Mitochondrial pellets (50 μg of protein) were resuspended in 45 μl of ice-cold digitonin-containing buffer (0.5–1% digitonin, 20 mM Tris-Cl, pH 7.4, 0.1 mM EDTA, 50 mM NaCl, and 10% [wt/vol] glycerol) and were incubated on ice for 10–15 min (Stojanovski et al., 2007). Samples were clarified by centrifugation at 12,000 g for 15 min at 4°C, and 5 μl of sample buffer (5% [wt/vol] Coomassie brilliant blue G-250, 100 mM Bis-Tris, pH 7.0, and 500 mM ɛ-amino-n-caproic acid) was added to the clarified supernatant (Meisinger et al., 2001). Samples were separated on a 4–16% polyacrylamide gradient gel at 4°C. The mobility of molecular weight markers was determined on parallel lanes/gels run under identical conditions. The radiolabeled proteins were detected by digital autoradiography.
Miscellaneous
Western transfers were performed on polyvinylidene difluoride membranes, and immunodecoration was performed according to standard techniques. Enhanced chemiluminescence was used for detection (GE Healthcare).
Acknowledgments
We thank Drs. N. Wiedemann, T. Becker, and A. Chacinska for discussion.
This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 388 and 746, Gottfried Wilhelm Leibniz Program, Max Planck Research Award, Bundesministerium für Bildung und Forschung, and the Fonds der Chemischen Industrie. D. Stojanovski and V. Kozjak-Pavlovic are recipients of Alexander von Humboldt research fellowships.
V. Kozjak-Pavlovic's present address is Max-Planck-Institut für Infektionsbiologie, D-10117 Berlin, Germany.
Abbreviations used in this paper: Mdm, mitochondrial distribution and morphology; SAM, sorting and assembly machinery; TOM, translocase of outer membrane.
References
- Ahting, U., T. Waizenegger, W. Neupert, and D. Rapaport. 2005. Signal-anchored proteins follow a unique insertion pathway into the outer membrane of mitochondria. J. Biol. Chem. 280:48–53. [DOI] [PubMed] [Google Scholar]
- Allen, R., B. Egan, K. Gabriel, T. Beilharz, and T. Lithgow. 2002. A conserved proline residue is present in the transmembrane-spanning domain of Tom7 and other tail-anchored protein subunits of the TOM translocase. FEBS Lett. 514:347–350. [DOI] [PubMed] [Google Scholar]
- Bos, M.P., and J. Tommassen. 2004. Biogenesis of the Gram-negative bacterial outer membrane. Curr. Opin. Microbiol. 7:610–616. [DOI] [PubMed] [Google Scholar]
- Burri, L., K. Vascotto, I.E. Gentle, N.C. Chan, T. Beilharz, D.I. Stapleton, L. Ramage, and T. Lithgow. 2006. Integral membrane proteins in the mitochondrial outer membrane of Saccharomyces cerevisiae. FEBS J. 273:1507–1515. [DOI] [PubMed] [Google Scholar]
- Cao, W., and M.G. Douglas. 1995. Biogenesis of ISP6, a small carboxy-terminal anchored protein of the receptor complex of the mitochondrial outer membrane. J. Biol. Chem. 270:5674–5679. [DOI] [PubMed] [Google Scholar]
- Coonrod, E.M., M.A. Karren, and J.M. Shaw. 2007. Ugo1p is a multipass transmembrane protein with a single carrier domain required for mitochondrial fusion. Traffic. 8:500–511. [DOI] [PubMed] [Google Scholar]
- Dembowski, M., K.P. Künkele, F.E. Nargang, W. Neupert, and D. Rapaport. 2001. Assembly of Tom6 and Tom7 into the TOM core complex of Neurospora crassa. J. Biol. Chem. 276:17679–17685. [DOI] [PubMed] [Google Scholar]
- Dietmeier, K., A. Hönlinger, U. Bömer, P.J.T. Dekker, C. Eckerskorn, F. Lottspeich, M. Kübrich, and N. Pfanner. 1997. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 388:195–200. [DOI] [PubMed] [Google Scholar]
- Dolezal, P., V. Likic, J. Tachezy, and T. Lithgow. 2006. Evolution of the molecular machines for protein import into mitochondria. Science. 313:314–318. [DOI] [PubMed] [Google Scholar]
- Egan, B., T. Beilharz, R. George, S. Isenmann, S. Gratzer, B. Wattenberg, and T. Lithgow. 1999. Targeting of tail-anchored proteins to yeast mitochondria in vivo. FEBS Lett. 451:243–248. [DOI] [PubMed] [Google Scholar]
- Endo, T., H. Yamamoto, and M. Esaki. 2003. Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J. Cell Sci. 116:3259–3267. [DOI] [PubMed] [Google Scholar]
- Eppens, E.F., N. Nouwen, and J. Tommassen. 1997. Folding of a bacterial outer membrane protein during passage through the periplasm. EMBO J. 16:4295–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick, R.L., J.M. McCaffery, K.W. Cunningham, K. Okamoto, and J.M. Shaw. 2004. Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J. Cell Biol. 167:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki, Y., A.L. Hubbard, S. Fowler, and P.B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, K., B. Egan, and T. Lithgow. 2003. Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. EMBO J. 22:2380–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle, I., K. Gabriel, P. Beech, R. Waller, and T. Lithgow. 2004. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle, I.E., L. Burri, and T. Lithgow. 2005. Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 58:1216–1225. [DOI] [PubMed] [Google Scholar]
- Gratzer, S., T. Lithgow, R.E. Bauer, E. Lamping, F. Paltauf, S.D. Kohlwein, V. Haucke, T. Junne, G. Schatz, and M. Horst. 1995. Mas37p, a novel receptor subunit for protein import into mitochondria. J. Cell Biol. 129:25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib, S.J., A. Vasiljev, W. Neupert, and D. Rapaport. 2003. Multiple functions of tail-anchor domains of mitochondrial outer membrane proteins. FEBS Lett. 555:511–515. [DOI] [PubMed] [Google Scholar]
- Habib, S.J., T. Waizenegger, M. Lech, W. Neupert, and D. Rapaport. 2005. Assembly of the TOB complex of mitochondria. J. Biol. Chem. 280:6434–6440. [DOI] [PubMed] [Google Scholar]
- Hermann, G.J., J.W. Thatcher, J.P. Mills, K.G. Hales, M.T. Fuller, J. Nunnari, and J.M. Shaw. 1998. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143:359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, K., K. Model, M.T. Ryan, K. Dietmeier, F. Martin, R. Wagner, and N. Pfanner. 1998. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature. 395:516–521. [DOI] [PubMed] [Google Scholar]
- Hoogenraad, N.J., L.A. Ward, and M.T. Ryan. 2002. Import and assembly of proteins into mitochondria of mammalian cells. Biochim. Biophys. Acta. 1592:97–105. [DOI] [PubMed] [Google Scholar]
- Hoppins, S.C., and F.E. Nargang. 2004. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 279:12396–12405. [DOI] [PubMed] [Google Scholar]
- Hoppins, S.C., N.E. Go, A. Klein, S. Schmitt, W. Neupert, D. Rapaport, and F.E. Nargang. 2007. Alternative splicing gives rise to different isoforms of the Neurospora crassa Tob55 protein that vary in their ability to insert β-barrel proteins into the outer mitochondrial membrane. Genetics. 177:137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie, C., H. Suzuki, M. Sakaguchi, and K. Mihara. 2003. Targeting and assembly of mitochondrial tail-anchored protein Tom5 to the TOM complex depend on a signal distinct from that of tail anchored proteins dispersed in the membrane. J. Biol. Chem. 278:41462–41471. [DOI] [PubMed] [Google Scholar]
- Humphries, A.D., I.C. Streimann, D. Stojanovski, A.J. Johnston, M. Yano, N.J. Hoogenraad, and M.T. Ryan. 2005. Dissection of the mitochondrial import and assembly pathway for human Tom40. J. Biol. Chem. 280:11535–11543. [DOI] [PubMed] [Google Scholar]
- Ishikawa, D., H. Yamamoto, Y. Tamura, K. Moritoh, and T. Endo. 2004. Two novel proteins in the mitochondrial outer membrane mediate β-barrel protein assembly. J. Cell Biol. 166:621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, R.E., and A.E. Johnson. 2001. Opening the door to mitochondrial protein import. Nat. Struct. Biol. 8:1008–1010. [DOI] [PubMed] [Google Scholar]
- Kanaji, S., J. Iwahashi, Y. Kida, M. Sakaguchi, and K. Mihara. 2000. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151:277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil, P., and N. Pfanner. 1993. Insertion of MOM22 into the mitochondrial outer membrane strictly depends on surface receptors. FEBS Lett. 321:197–200. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt, J.H., and L.K. Tamm. 1996. Folding intermediates of a β-barrel membrane protein: kinetic evidence for a multi-step membrane insertion mechanism. Biochemistry. 35:12993–13000. [DOI] [PubMed] [Google Scholar]
- Koehler, C.M. 2004. New developments in mitochondrial assembly. Annu. Rev. Cell Dev. Biol. 20:309–335. [DOI] [PubMed] [Google Scholar]
- Kozjak, V., N. Wiedemann, D. Milenkovic, C. Lohaus, H.E. Meyer, B. Guiard, C. Meisinger, and N. Pfanner. 2003. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 278:48520–48523. [DOI] [PubMed] [Google Scholar]
- Kozjak-Pavlovic, V., K. Ross, N. Benlasfer, S. Kimming, A. Karlas, and T. Rudel. 2007. Conserved roles of Sam50 and metaxins in VDAC biogenesis. EMBO Rep. 8:576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger, C., M.T. Ryan, K. Hill, K. Model, J.H. Lim, A. Sickmann, H. Müller, H.E. Meyer, R. Wagner, and N. Pfanner. 2001. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small Tom proteins, and import receptors. Mol. Cell. Biol. 21:2337–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger, C., M. Rissler, A. Chacinska, D. Milenkovic, V. Kozjak, B. Schönfisch, C. Lohaus, H.E. Meyer, M.P. Yaffe, B. Guiard, et al. 2004. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell. 7:61–71. [DOI] [PubMed] [Google Scholar]
- Meisinger, C., N. Wiedemann, M. Rissler, A. Strub, D. Milenkovic, B. Schönfisch, H. Müller, V. Kozjak, and N. Pfanner. 2006. Mitochondrial protein sorting: differentiation of β-barrel assembly by Tom7-mediated segregation of Mdm10. J. Biol. Chem. 281:22819–22826. [DOI] [PubMed] [Google Scholar]
- Meisinger, C., S. Pfannschmidt, M. Rissler, D. Milenkovic, T. Becker, D. Stojanovski, M.J. Young, R.E. Jensen, A. Chacinska, B. Guiard, et al. 2007. The morphology proteins Mdm12/Mmm1 function in the major β-barrel assembly pathway of mitochonria. EMBO J. 26:2229–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic, D., V. Kozjak, N. Wiedemann, C. Lohaus, H.E. Meyer, B. Guiard, N. Pfanner, and C. Meisinger. 2004. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J. Biol. Chem. 279:22781–22785. [DOI] [PubMed] [Google Scholar]
- Model, K., C. Meisinger, T. Prinz, N. Wiedemann, K.N. Truscott, N. Pfanner, and M.T. Ryan. 2001. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol. 8:361–370. [DOI] [PubMed] [Google Scholar]
- Model, K., T. Prinz, T. Ruiz, M. Radermacher, T. Krimmer, W. Kühlbrandt, N. Pfanner, and C. Meisinger. 2002. Protein translocase of the outer mitochondrial membrane: role of import receptors in the structural organization of the TOM complex. J. Mol. Biol. 316:657–666. [DOI] [PubMed] [Google Scholar]
- Mozdy, A.D., J.M. McCaffery, and J.M. Shaw. 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane protein Fis1p. J. Cell Biol. 151:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y., H. Suzuki, M. Sakaguchi, and K. Mihara. 2004. Targeting and assembly of rat mitochondrial translocase of outer membrane 22 (TOM22) into the TOM complex. J. Biol. Chem. 279:21223–21232. [DOI] [PubMed] [Google Scholar]
- Neupert, W., and J.M. Hermann. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723–749. [DOI] [PubMed] [Google Scholar]
- Oka, T., and K. Mihara. 2005. A railroad switch in mitochondrial protein import. Mol. Cell. 18:145–146. [DOI] [PubMed] [Google Scholar]
- Paschen, S.A., T. Waizenegger, T. Stan, M. Preuss, M. Cyrklaff, K. Hell, D. Rapaport, and W. Neupert. 2003. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature. 426:862–866. [DOI] [PubMed] [Google Scholar]
- Paschen, S.A., W. Neupert, and D. Rapaport. 2005. Biogenesis of β-barrel membrane proteins of mitochondria. Trends Biochem. Sci. 30:575–582. [DOI] [PubMed] [Google Scholar]
- Pfanner, N., N. Wiedemann, C. Meisinger, and T. Lithgow. 2004. Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 11:1044–1048. [DOI] [PubMed] [Google Scholar]
- Prokisch, H., C. Scharfe, D.G. Camp, W. Xiao, L. David, C. Andreoli, M.E. Monroe, R.J. Moore, M.A. Gritsenko, C. Kozany, et al. 2004. Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol. 2:e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, D. 2003. Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep. 4:948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, D. 2005. How does the TOM complex mediate insertion of precursor proteins into the mitochondrial outer membrane? J. Cell Biol. 171:419–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, D., and W. Neupert. 1999. Biogenesis of Tom40, core component of the TOM core complex of mitochondria. J. Cell Biol. 146:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport, D., M. Brunner, W. Neupert, and B. Westermann. 1998. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273:20150–20155. [DOI] [PubMed] [Google Scholar]
- Rapaport, D., R.D. Taylor, M. Käser, T. Langer, W. Neupert, and F.E. Nargang. 2001. Structural requirements of Tom40 for assembly into preexisting complexes of mitochondria. Mol. Biol. Cell. 12:1189–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehling, P., K. Bradner, and N. Pfanner. 2004. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 5:519–530. [DOI] [PubMed] [Google Scholar]
- Reinders, J., R.P. Zahedi, N. Pfanner, C. Meisinger, and A. Sickmann. 2006. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 5:1543–1554. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Cousiño, N., F.E. Nargang, R. Baardman, W. Neupert, R. Lill, and D.A. Court. 1998. An import signal in the cytosolic domain of the Neurospora mitochondrial outer membrane protein TOM22. J. Biol. Chem. 273:11527–11532. [DOI] [PubMed] [Google Scholar]
- Ruiz, N., D. Kahne, and T.J. Silhavy. 2006. Advances in understanding outer-membrane biogenesis. Nat. Rev. Microbiol. 4:57–66. [DOI] [PubMed] [Google Scholar]
- Ryan, M.T. 2004. Chaperones: inserting β-barrels into membranes. Curr. Biol. 14:R207–R209. [DOI] [PubMed] [Google Scholar]
- Schleiff, E., and J. Soll. 2005. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 6:1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, S., H. Prokisch, T. Schlunck, D.G. Camp, U. Ahting, T. Waizenegger, C. Scharfe, T. Meitinger, A. Imhof, W. Neupert, et al. 2006. Proteome analysis of mitochondrial outer membrane from Neurospora crassa. Proteomics. 6:72–80. [DOI] [PubMed] [Google Scholar]
- Schneider, H., T. Söllner, K. Dietmeier, C. Eckerskorn, F. Lottspeich, B. Trülzsch, W. Neupert, and N. Pfanner. 1991. Targeting the master receptor MOM19 to mitochondria. Science. 254:1659–1662. [DOI] [PubMed] [Google Scholar]
- Schulz, G.E. 2002. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta. 1565:308–317. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., and R.E. Jensen. 2001. UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 152:1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann, A., J. Reinders, Y. Wagner, C. Joppich, R. Zahedi, H.E. Meyer, B. Schönfisch, I. Perschil, A. Chacinska, B. Guiard, et al. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA. 100:13207–13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski, D., N. Pfanner, and N. Wiedemann. 2007. Import of proteins into mitochondria. Methods Cell Biol. 80:783–806. [DOI] [PubMed] [Google Scholar]
- Tieu, Q., and J. Nunnari. 2000. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151:353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux, R., M.P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 299:262–265. [DOI] [PubMed] [Google Scholar]
- Waizenegger, T., T. Stan, W. Neupert, and D. Rapaport. 2003. Signal-anchor domains of proteins of the outer membrane of mitochondria. J. Biol. Chem. 278:42064–42071. [DOI] [PubMed] [Google Scholar]
- Waizenegger, T., S.J. Habib, M. Lech, D. Mokranjac, S.A. Paschen, K. Hell, W. Neupert, and D. Rapaport. 2004. Tob38, a novel essential component in the biogenesis of β-barrel proteins of mitochondria. EMBO Rep. 5:704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann, N., N. Pfanner, and M.T. Ryan. 2001. The three modules of the ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 20:951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann, N., V. Kozjak, A. Chacinska, B. Schönfisch, S. Rospert, M.T. Ryan, N. Pfanner, and C. Meisinger. 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 424:565–571. [DOI] [PubMed] [Google Scholar]
- Wiedemann, N., K.N. Truscott, S. Pfannschmidt, B. Guiard, C. Meisinger, and N. Pfanner. 2004. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 279:18188–18194. [DOI] [PubMed] [Google Scholar]
- Yaffe, M.P., R.E. Jensen, and E.C. Guido. 1989. The major 45-kDa protein of the yeast mitochondrial outer membrane is not essential for cell growth or mitochondrial function. J. Biol. Chem. 264:21091–21096. [PubMed] [Google Scholar]
- Young, J.C., N.J. Hoogenraad, and F.U. Hartl. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 112:41–50. [DOI] [PubMed] [Google Scholar]
- Zahedi, R.P., A. Sickmann, A.M. Boehm, C. Winkler, N. Zufall, B. Schönfisch, B. Guiard, N. Pfanner, and C. Meisinger. 2006. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell. 17:1436–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]