Abstract
Diastrophic dysplasia sulfate transporter (DTDST) is a sulfate/chloride antiporter whose function is impaired in several human chondrodysplasias. We show that DTDST is upregulated by dexamethasone stimulation of HT1080 fibrosarcoma cells and is required for fibronectin (FN) extracellular matrix deposition by these cells. DTDST imports sulfate for the modification of glycosaminoglycans. We find that N-sulfation of these chains is important for FN matrix assembly and that sulfation of cell surface proteoglycans is reduced in the absence of DTDST. Of the candidate HT1080 cell surface proteoglycans, only loss of syndecan-2 compromises FN assembly, as shown by syndecan-2 small interfering RNA knockdown. DTDST is both necessary and sufficient to induce FN matrix assembly in HT1080 cells. Knockdown of DTDST ablates FN matrix, whereas its overexpression increases assembly without dexamethasone stimulation. These results identify a previously unrecognized regulatory pathway for matrix assembly via modulation of a sulfate transporter and proteoglycan sulfation. These data raise the possibility that FN assembly defects contribute to chondrodysplasias.
Introduction
Alterations in the local ECM architecture and composition usually accompany disease progression such as oncogenic transformation. One of the most abundant components of the ECM, the adhesive glycoprotein fibronectin (FN), is lost from the surface of many transformed cells (Hynes, 1990). FN functions predominantly within the context of a fibrillar matrix, and changes in FN fibril assembly occur during tumor progression because of reduced expression and/or insufficient binding to cell surface receptors. Reintroduction of FN into tumor cell cultures increases their adhesivity, restores a morphology characteristic of nontransformed cells (Yamada et al., 1976; Ali et al., 1977; Pasqualini et al., 1996), and reestablishes a fibrillar matrix (Schwarzbauer, 1991). These observations illustrate the detrimental effects of loss of pericellular matrix and underscore the importance of FN expression and deposition into a fibrillar ECM for maintenance of normal tissue organization.
FN matrix assembly is a cell-mediated process, initiated as the integrin receptor α5β1 binds to the RGD sequence and synergy site on FN (Schwarzbauer and Sechler, 1999; Mao and Schwarzbauer, 2005). Clustering of integrin heterodimers brings molecules of FN in close proximity and promotes FN self-association. Inside the cell, adaptor proteins that interact with integrin cytoplasmic domains become juxtaposed and activate their downstream signaling partners, such as FAK and RhoA GTPase (Miranti and Brugge, 2002). Nascent FN fibrils are formed between adjacent cells, and as matrix assembly progresses, these fibrils form a meshwork that is characterized by its insolubility in deoxycholate (DOC) detergent (McKeown-Longo and Mosher, 1983; Chen and Mosher, 1996). Although the major steps in matrix assembly have been well established, little information is available about the gene expression changes that accompany the acquisition of a DOC-insoluble matrix. The human fibrosarcoma cell line HT1080 provides a useful model for FN matrix assembly because these cells do not assemble FN unless stimulated with the transcriptional regulator dexamethasone or the Ras pathway inhibitor PD98059 (McKeown-Longo and Etzler, 1987; Brenner et al., 2000).
We have found that diastrophic dysplasia sulfate transporter (DTDST), a sulfate/choride antiporter (Hastbacka et al., 1994), is up-regulated upon induction of FN matrix assembly in HT1080 cells. Mutations in the human DTDST gene result in a set of autosomal recessive chondrodysplasias that range in severity (Superti-Furga et al., 1996; Haila et al., 2001; Karniski, 2001). Patients show gross skeletal defects, reduced cellular sulfate uptake, and cartilage proteoglycan undersulfation (Rossi et al., 1998). Proteoglycans are characterized by a core protein modified by a heterogeneous number of sulfated glycosaminoglycan (GAG) chains (Esko, 1991). GAG chains are crucial for many proteoglycan functions. For example, CHO cells deficient in xylosyltransferase (Esko et al., 1985), the enzyme responsible for the first step in heparan and chondroitin sulfate GAG chain synthesis, have reduced ability to assemble FN matrix (Chung and Erickson, 1997). Posttranslational addition of sulfate to GAG chains also has a functional role. Elimination of total sulfation using sodium chlorate abolished the coreceptor function of syndecan-4 with basic fibroblast growth factor, thereby preventing mitogenic signaling in Swiss 3T3 cells (Rapraeger et al., 1991). Inhibition of sulfation in C6 glioma cells reduced adhesion to collagen IV, laminin, and FN (Keller et al., 1989; Mendes de Aguiar et al., 2002). Here we show that GAG chain sulfation in HT1080 cells depends on DTDST and, through its role in maintenance of cell surface sulfate levels, DTDST is both necessary and sufficient to stimulate FN matrix assembly.
Results
Up-regulation of DTDST correlates with FN matrix assembly
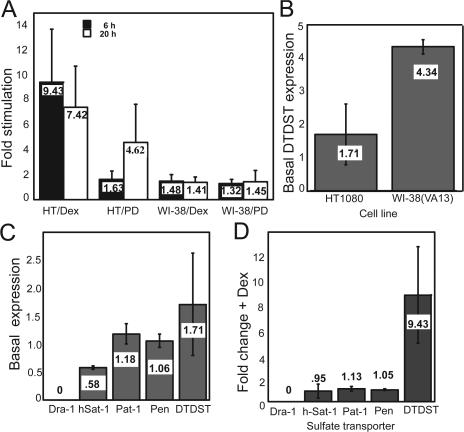
HT1080 human fibrosarcoma cells assemble FN into a matrix only with stimulation by the glucocorticoid dexamethasone (Oliver et al., 1983; McKeown-Longo and Etzler, 1987), activation of cell surface integrins (Brenner et al., 2000), or inhibition of MAP/extracellular signal–related kinase signaling with PD98059 (Brenner et al., 2000). DTDST showed dramatic up-regulation in microarray experiments performed using RNA from HT1080 cells treated with dexamethasone or PD98059. Quantitative RT-PCR analyses confirmed the microarray results and revealed stimulation of DTDST expression in HT1080 cells by dexamethasone at 6 and 20 h and PD98059 at 20 h of treatment (Fig. 1 A). In WI-38(VA13) cells, a transformed lung fibroblast cell line that constitutively assembles an extensive FN matrix, DTDST expression changed less than 1.5-fold with both treatments at both time intervals. Thus, DTDST expression was increased concomitant with HT1080 cells establishing competence to assemble FN into a fibrillar matrix. Basal levels of DTDST expression differ between these two cell lines (Fig. 1 B). Mean relative expression of the transcript is ∼2.5-fold higher in assembly-competent WI-38(VA13) cells, implying that stimulation of DTDST expression in assembly-deficient HT1080 cells may be necessary for sufficient levels of sulfate import.
Figure 1.
Up-regulation of DTDST correlates with induction of FN matrix assembly. (A) RNA was isolated from HT1080 (HT) or WI-38(VA13) cells (WI-38), treated with 0.1 μM dexamethasone (Dex) or 2.5 μM PD98059 (PD) for 6 or 20 h. cDNA was prepared using random hexamers and DTDST levels were determined by real-time RT-PCR. Each sample was normalized to a housekeeping gene, ubiquitin C, and fold stimulation relative to unstimulated cells was determined. (B) RNA from untreated HT1080 or WI-38(VA13) cells was used for quantitative RT-PCR to assess the relative DTDST expression level in each cell line. (C) Quantitative RT-PCR on RNA from untreated HT1080 cells was performed using primers for the five human sulfate/anion transporters (down-regulated in adenoma [Dra-1], h-Sat, Pat-1, Pendrin, and DTDST). (D) RNA from HT1080 cells treated or not with dexamethasone for 6 h was used for real time RT-PCR using primer pairs for each of the sulfate transporters. Values were normalized and fold stimulation was determined as in A. In all cases, the mean of three independent trials was calculated and the value is listed on each bar. Error bars represent standard deviation.
DTDST is one of the sulfate transporters that shuttles extracellular sulfate into the cell in exchange for chloride. Five Na+-independent sulfate/anion transporters have been identified in the human genome (Markovich, 2001). Four of the five sulfate transporters (h-Sat-1, Pat-1, DTDST, and Pendrin) are expressed to varying degrees by HT1080 cells (Fig. 1 C), but only DTDST was up-regulated by dexamethasone stimulation (Fig. 1 D).
Inhibition of sulfation ablates FN matrix assembly
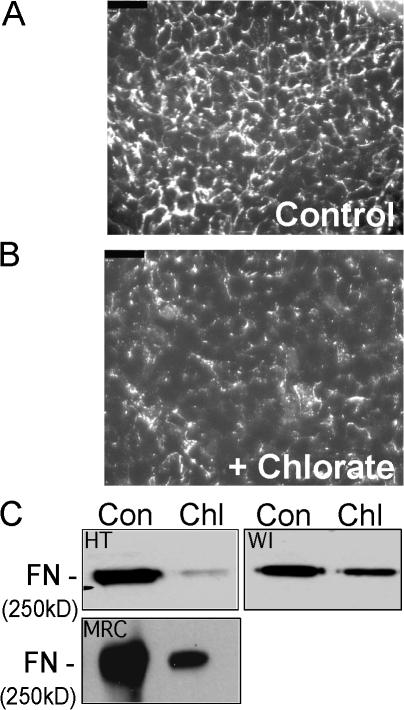
Cells isolated from diastrophic dysplasia patients show reduced sulfate uptake and defective sulfation of GAG chains (Rossi et al., 1996; Superti-Furga et al., 1996). To determine whether sulfation is required for FN matrix assembly in HT1080 cells, we treated cells with sodium chlorate to block sulfate addition. Chlorate is a competitive inhibitor of ATP sulfurylase, one of the enzymes involved in generation of adenosine 3′-phosphate 5′-sulfatophosphate, the universal sulfate donor to intracellular proteins and carbohydrate moieties (Klaassen and Boles, 1997). Moreover, chlorate treatment diminishes heparan sulfate sulfation by 92%, and these unsulfated GAG chains have a dramatically reduced affinity for FN (Keller et al., 1989). HT1080 cells were treated for 48 h with sodium chlorate and matrix assembly was then stimulated with dexamethasone for 20 h. Chlorate treatment did not affect cell viability and did not alter cell morphology or spreading in response to FN. Immunofluorescence imaging of matrix on dexamethasone-stimulated chlorate-treated cells revealed a decrease in FN fibrils compared with buffer-treated cells (Fig. 2, A and B). Consistent with this, immunoblot analysis of DOC lysates showed a dramatic decrease in DOC-insoluble FN upon chlorate treatment (Fig. 2 C). Isolation of DOC-insoluble material from dexamethasone-stimulated and chlorate-treated WI-38(VA13) cells revealed a similar, although less dramatic, inhibition of matrix formation (Fig. 2 C). Proteins, including FN, can be sulfated on tyrosine residues through the action of tyrosylprotein sulfotransferase (Lee and Huttner, 1983). Inhibition of tyrosylprotein sulfotransferase with 2-chloroadenosine, however, had no effect on FN matrix formation (unpublished data), indicating that the essential sulfation for matrix assembly is on moieties other than tyrosine.
Figure 2.
Inhibition of sulfation ablates FN matrix assembly. HT1080 cells plated on coverslips were control treated (A) or treated for 48 h with 50 mM sodium chlorate (B), and 0.1 μM dexamethasone was added for the last 20 h. Cells were fixed and stained with anti-FN monoclonal antibody HFN7.1. Bars, 50 μm. (C) DOC lysates were prepared from HT1080, WI-38(VA13) cells, or MRC-5 human embryonic fibroblasts (MRC), either untreated (Con) or treated with sodium chlorate (Chl) as in B. DOC-insoluble material proportional to either 10 μg (HT and WI) or 0.67 μg (MRC) of total protein was electrophoresed on a 5% polyacrylamide-SDS gel. FN was detected with HFN7.1 monoclonal antibody.
FN has been implicated in the early stages of bone and cartilage formation during mesenchymal cell condensation and differentiation (Dessau et al., 1980; Mackie et al., 1987; Tavella et al., 1997). MRC-5 primary human fibroblasts differentiate into osteoblastic cells when treated with dexamethasone or BMP-2 (Almeida et al., 2001). To determine whether chlorate inhibition affected FN matrix assembly by cells that have the potential to become osteoblasts, DOC-insoluble material from chlorate-treated MRC-5 cells was analyzed and showed dramatic reduction compared with that in untreated cells (Fig. 2 C). These data show that sulfation plays an important role in FN matrix formation in assembly-competent WI-38(VA13) cells, in assembly-deficient HT1080 fibrosarcoma cells, and in nononcogenic cells capable of bone differentiation.
FN assembly requires sulfated GAG chains
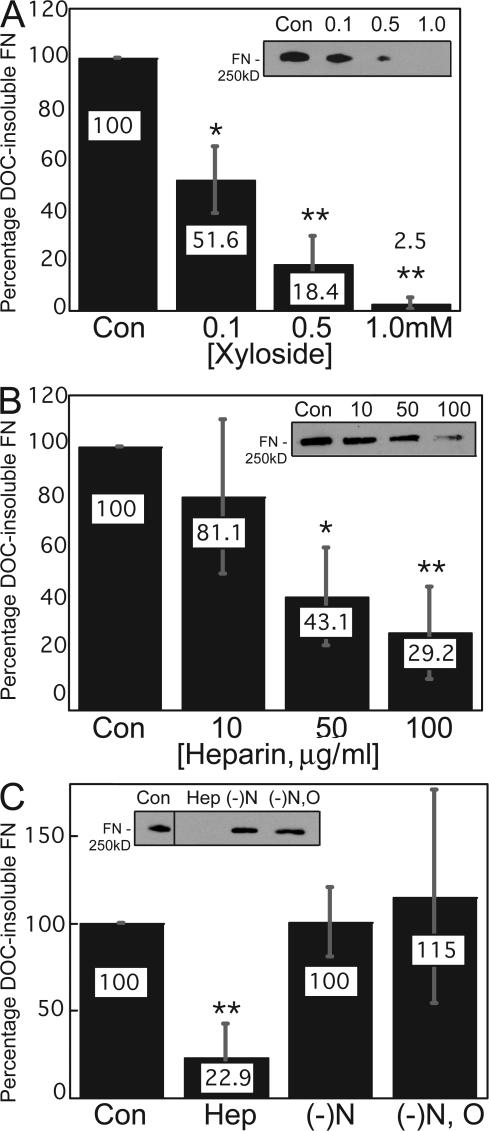
Cells isolated from DTD patients (Superti-Furga et al., 1996; Rossi et al., 1998) or from a DTDST mutant mouse model (Forlino et al., 2005) show reduced sulfate uptake, and the primary biochemical defect is undersulfation of proteoglycans (Rossi et al., 1998). However, characterization of proteoglycans from DTD patient fibroblasts has revealed normal synthesis of core proteins and normal initiation and elongation of GAG chains (Rossi et al., 1996). To determine whether GAG chains or their sulfation play a role in FN matrix assembly by HT1080 cells, we used 4-methylumbelliferyl-β-D-xyloside, an inhibitor of GAG chain initiation (Fritz and Esko, 2001). Increasing concentrations of xylosides caused a dosage-dependent and statistically significant decrease in dexamethasone-induced DOC- insoluble FN matrix (Fig. 3 A). Treatment of BHK cells with 200 μM xylosides did not reduce matrix assembly (Chung and Erickson, 1997). This concentration of xylosides is on the low end of our dosage response, so it is possible that treatment of these cells with higher concentrations may have an inhibitory effect. BHK cell matrix assembly was also less sensitive to inhibition by soluble heparin and required treatment with GAG chains purified from BHK cells themselves, supporting the idea that HT1080 and BHK cells have different sensitivities to inhibitors of matrix assembly.
Figure 3.
Sulfated GAG chains contribute to FN matrix assembly. DOC-insoluble material was isolated from HT1080 cells grown in medium containing 0.1 μM dexamethasone plus the indicated treatments for 20 h. DOC-insoluble material proportional to 10 μg of total protein was resolved and FN was detected in immunoblots using HFN7.1 antibody (A–C, insets). Treatments included increasing amounts of 4-methyl-umbelliferyl-β-D- xyloside (A), increasing concentrations of soluble heparin (B), or 250 μg/ml fully sulfated heparin (Hep), heparin that was N-desulfated ((-)N), or heparin that was entirely devoid of sulfate groups ((-)N, O). Con, cells treated with vehicle only. In all cases, values are percentages as compared with the respective controls and are normalized to GAPDH expression. *, P < 0.04; **, P < 0.02. All error bars represent standard deviations based on the mean of three independent trials.
HT1080 FN matrix was also reduced by the addition of soluble heparin to the culture medium (Fig. 3 B). GAG chains are multiply sulfated by N and O linkages (Esko, 1991). Unlike soluble sulfated heparin, addition of soluble N-desulfated or fully desulfated heparin did not block FN matrix assembly (Fig. 3 C). The inability of soluble desulfated heparin to significantly inhibit assembly indicates that sulfation is a critical modification for the function of GAG chains in this process.
Sulfation of cell surface molecules depends on DTDST
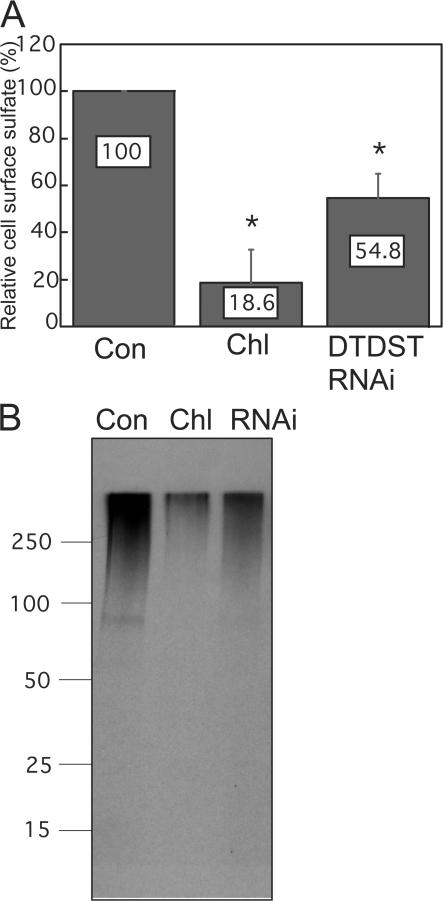
Metabolic labeling and cell fractionation experiments have shown that a significant proportion of total sulfated macromolecules is localized to the membrane fraction (Sjoberg and Malmstrom, 1982; Wang et al., 1999; Shworak, 2001). We found that the matrix assembly defect of chlorate-treated cells was not restored by HT1080 cell conditioned medium (unpublished data), indicating that cell-associated, rather than secreted, sulfated molecules play a role in FN matrix assembly. To test the requirement for DTDST, its expression was knocked down by treatment with a DTDST siRNA SMARTpool or with individual oligonucleotides from the pool. A significant reduction in transcript levels, nearly 13-fold, was detected by quantitative RT-PCR, whereas the expression of FN and of the other sulfate/anion transporters showed only slight variations with DTDST siRNA treatment (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200707150/DC1). To determine the effect of DTDST knockdown on sulfation of cell-associated molecules, biotinylated cell surface proteins from [35S]sulfate-labeled cells were isolated and analyzed. Compared with control membranes, the amount of sulfated molecules from chlorate-treated cells was reduced a dramatic fivefold and from DTDST siRNA-treated cells, it was twofold lower (Fig. 4 A). SDS-PAGE of these proteins confirmed that the label was in a high molecular mass smear, indicative of GAG-bearing proteoglycans (Fig. 4 B). These results suggest an important role for this transporter in controlling levels of sulfated cell surface GAGs on HT1080 cells.
Figure 4.
Cell surface–sulfated molecules are decreased in the absence of DTDST. (A) Cultures were stimulated with dexamethasone for 20 h and labeled for the final 12 h with 50 μCi/ml [35S]sulfate, cell surface proteins were biotinylated, and biotin-labeled molecules were isolated from whole cell lysates with streptavidin-agarose beads. Equal amounts of protein were counted, the means were calculated, and data are presented as percentages relative to control cells. Error bars are standard deviation based on the mean of three trials. *, P < 0.02. (B) Material eluted from streptavidin beads was separated on a 4–20% gradient SDS gel and the dried gel was exposed to film for 35 d. Molecular mass standards in kilodaltons are indicated on the left. Con, control; Chl, chlorate-treated; RNAi, DTDST siRNA-treated.
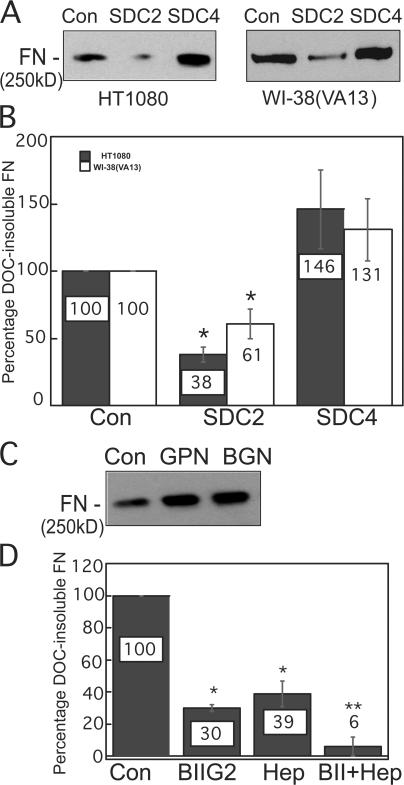
Knockdown of syndecan-2, but not of other proteoglycans, blocks FN matrix assembly
HT1080 cells express the cell surface proteoglycans syndecan-1, -2, and -4, glypican-1, and betaglycan as determined by quantitative RT-PCR. Knockdown of each transmembrane proteoglycan using specific siRNAs was effective at reducing both mRNA and protein levels (Fig. S2, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200707150/DC1). Knockdown of syndecan-2 had a significant impact on matrix formation in both HT1080 and WI-38(VA13) cells (Fig. 5, A and B), but syndecan-2 RNAi did not alter transcript levels of the other transmembrane proteoglycans (Fig. S2 C). Knockdown of syndecan-4, betaglycan, or glypican-1 consistently showed a modest increase in DOC-insoluble FN (Fig. 5, A and C), but this difference was not statistically significant (Fig. 5 B). Down-regulation of syndecan-1 induces a fibroblastic phenotype (Kato et al., 1995), which fits with the slight increase in FN matrix we observed with syndecan-1 siRNA treatment (unpublished data). Syndecan-2 has been previously suggested to be an FN-binding proteoglycan (Itano et al., 1993, 1996), primarily via its three heparan sulfate GAG chains, and has been implicated in matrix assembly (Klass et al., 2000). Our results single out syndecan-2 as an important component of the FN assembly machinery in HT1080 cells.
Figure 5.
Syndecan-2 knockdown diminishes FN matrix assembly. (A) HT1080 or WI-38(VA13) cells transfected with control, syndecan-2 (SDC2), or syndecan-4 (SDC4) siRNAs were dexamethasone stimulated, and FN levels in DOC-insoluble lysates proportional to 5 μg of total protein were analyzed with HFN7.1 antibody. (B) Immunoblots developed with ECL Plus reagent were used to quantify FN levels in samples that were normalized to GAPDH. Control transfected samples were set at 100% and percentages of DOC-insoluble FN with syndecan-2 or -4 siRNA treatments are expressed relative to the control. Error bars represent standard deviation from three independent experiments. *, P < 0.03. (C) DOC-insoluble lysates from HT1080 cells transfected with control, glypican-1 (GPN), or betaglycan (BGN) siRNAs were immunoblotted with HFN7.1 antibody. (D) Cells were pretreated with an integrin α5 function-blocking antibody, BIIG2, at 1:250 dilution, 100 μg/ml of soluble heparin, or both. FN levels in DOC-insoluble fractions from three independent trials were analyzed and quantified as in B. *, P < 0.007 compared with control; **, P < 0.009 compared with single treatments.
Integrin α5β1 is the primary receptor for FN matrix assembly by fibroblasts (Wu et al., 1993). Activation of α5β1 is sufficient to stimulate matrix assembly by HT1080 cells (Brenner et al., 2000). We assessed the contributions of proteoglycans and integrins in this process by combining an anti-α5β1 function- blocking antibody, BIIG2, with soluble fully sulfated heparin. DOC-insoluble matrix was reduced with either BIIG2 or soluble heparin alone, but simultaneous inhibition of both integrin and GAG binding eliminated dexamethasone-induced matrix formation (Fig. 5 D). These results show that integrins work in cooperation with sulfated proteoglycans at the cell surface to mediate FN matrix assembly.
DTDST function is necessary and sufficient to stimulate FN matrix assembly
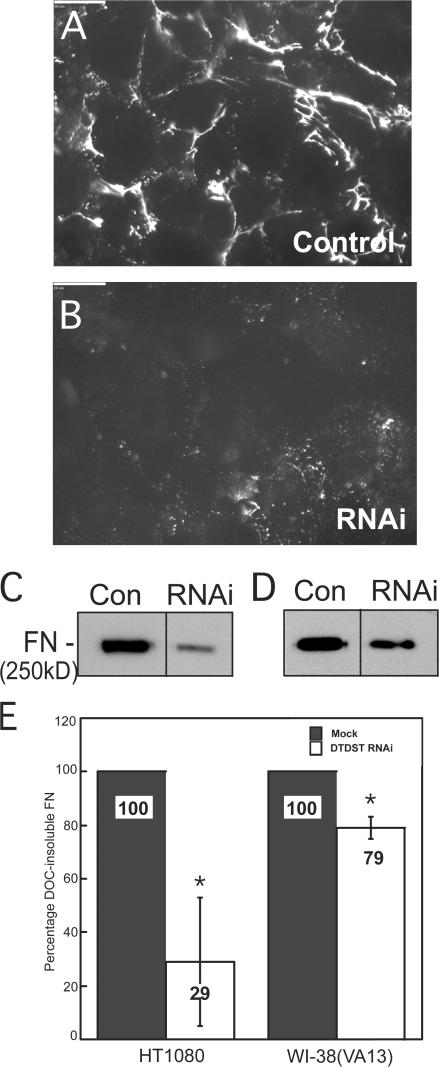
The requirement for DTDST in FN assembly was demonstrated using siRNA knockdown of DTDST in dexamethasone-treated HT1080 cells. Reduced DTDST levels led to an obvious reduction in mature FN fibrils by immunofluorescence (Fig. 6, A and B) and a quantitative decrease in the amount of DOC-insoluble FN in both HT1080 and WI-38(VA13) cells (Fig. 6, C–E). Therefore, DTDST expression is necessary to enhance FN fibril assembly by HT1080 cells and also contributes to matrix formation by assembly-competent WI-38(VA13) cells.
Figure 6.
DTDST expression is necessary to stimulate FN matrix assembly. HT1080 cells were transfected with control siRNA oligonucleotides (A) or DTDST siRNAs (RNAi; B) and stimulated with 0.1 μM dexamethasone. After 48 h, cells were fixed and stained with anti-FN HFN7.1 monoclonal antibody. Scale bars represent 20 μm. DOC-insoluble material from HT1080 (C) or WI-38(VA13) (D) cells transfected with siCONTROL or DTDST siRNA was immunoblotted with HFN7.1 antibody. (E) DOC-insoluble material from three independent trials was immunoblotted and developed with ECL Plus. Values represent percent of FN signal relative to the control condition for each cell line. Error bars represent standard deviation. *, P < 0.04.
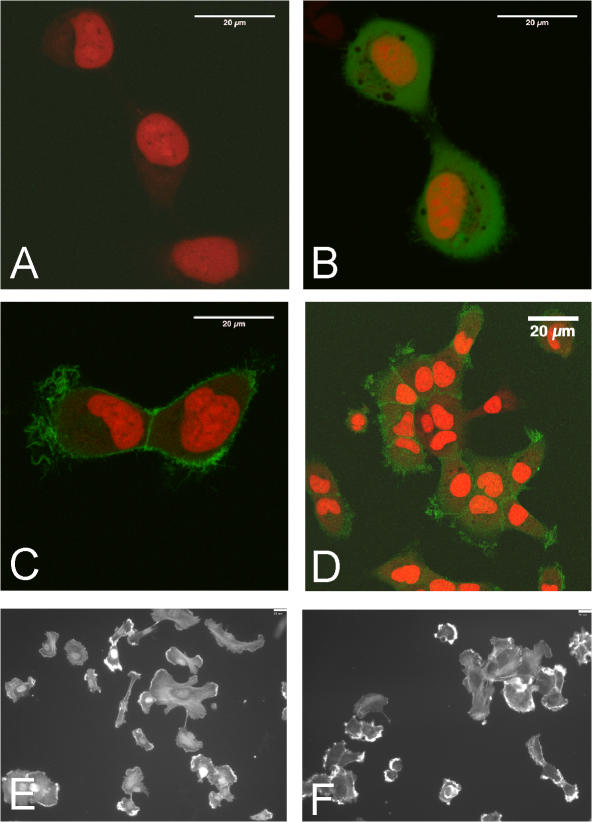
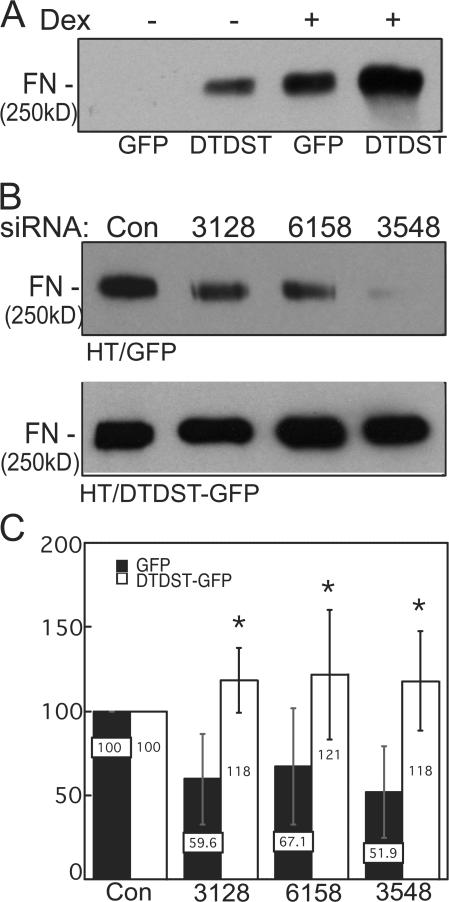
Dexamethasone-induced DTDST expression in HT1080 cells increases concomitantly with competence for FN matrix assembly. To determine whether DTDST overexpression was sufficient to induce assembly in the absence of dexamethasone treatment, we generated HT1080 cells expressing a translational fusion of DTDST cDNA with GFP under control of the cytomegalovirus promoter (HT/DTDST-GFP) or expressing the GFP vector alone (HT/GFP). DTDST-GFP localized to membrane ruffles (Fig. 7, C and D). This distribution differed from that of GFP alone (Fig. 7 B) and was significantly above background HT1080 autofluorescence (Fig. 7 A). No differences in cell morphology (Fig. 7, E and F) or endogenous FN secretion (not depicted) were observed between HT/GFP and HT/DTDST-GFP cells. Overexpression of DTDST-GFP increased accumulation of DOC-insoluble matrix in the absence of dexamethasone treatment (Fig. 8 A). In three independent experiments, we observed at least 15-fold more DOC-insoluble FN with DTDST overexpression than in control HT/GFP cells. These results demonstrate that up-regulation of this sulfate transporter is sufficient to induce FN assembly by HT1080 cells. Dexamethasone stimulation increased the amount of FN matrix in all cells but the effect was much more pronounced in HT/DTDST-GFP cells compared with HT/GFP cells (Fig. 8 A). Furthermore, sulfated cell surface proteins were increased with DTDST-GFP overexpression (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200707150/DC1) but up- or down-regulation of DTDST did not affect the level of syndecan-2 protein (Fig. S3 B). Similar results were obtained with two different HT/DTDST-GFP cell clones.
Figure 7.
A DTDST-GFP fusion protein localizes to the plasma membrane. HT1080 cells were mock transfected (A) or stably transfected with either pEGFP-N1 vector (B) or pEGFP-N1/DTDST (C and D). Live cells were stained with DRAQ5 and plated onto glass-bottom culture dishes for confocal imaging. Images in A–C were captured with a zoom factor of three. (D) Image of live HT/DTDST-GFP cells (as in C) was collected with a zoom factor of 1. Bars, 20 μm. HT/GFP (E) and HT/DTDST-GFP (F) cells on FN-coated coverslips were fixed, permeabilized, and stained with rhodamine-phalloidin and DAPI to visualize cell shapes.
Figure 8.
Overexpression of DTDST is sufficient to stimulate FN matrix assembly in the absence of dexamethasone. (A) HT/DTDST-GFP (DTDST) or HT-GFP (GFP) cells were stimulated, or not, with 0.1 μM dexamethasone as indicated. FN was detected in DOC-insoluble fractions using HFN7.1 antibody. Blot is representative of three independent experiments on a single transfectant and similar results were obtained with two other cell clones. (B) HT/GFP (top) or HT/DTDST-GFP (bottom) cells stimulated with dexamethasone were treated with siRNAs directed against three remote locations within the 3′ UTR of endogenous DTDST (3128, 6158, and 3548). DOC-insoluble material was analyzed and the immunoblots are representative of three independent trials. (C) DOC-insoluble FN amounts were normalized to siCONTROL-transfected amounts (set at 100) from either HT/GFP or HT/DTDST-GFP cells. Error bars represent standard deviation. *, P < 0.02 for comparison of two cell lines treated with the same oligonucleotide.
To rescue the effects of DTDST siRNA, we exploited the fact that the DTDST-GFP construct lacks DTDST 3′ untranslated region (UTR) sequences and designed siRNA oligonucleotides that target this region of the transcript. Treatment with these oligonucleotides should knock down endogenous DTDST expression, whereas the DTDST-GFP transcript would be resistant to this inhibition. Three 3′ UTR siRNA oligonucleotides were transfected individually into HT/GFP and HT/DTDST-GFP cell lines and DOC-insoluble lysates were analyzed. All 3′ UTR oligonucleotides reduced FN matrix levels in HT/GFP cells to varying degrees (Fig. 8, B and C). In contrast, the HT/DTDST-GFP cell line assembled FN matrix even in the presence of these oligonucleotides (Fig. 8, B and C). Therefore, expression of DTDST-GFP compensates for the RNAi-mediated knockdown of endogenous DTDST transcripts and rescues the defect in FN matrix assembly. Collectively, our results demonstrate that DTDST expression is both necessary and sufficient to enhance FN matrix assembly through a process that depends on production of sulfated HT1080 cell surface proteins.
Discussion
We have identified an important regulatory role for the sulfate/chloride antiporter DTDST in FN matrix assembly. DTDST gene expression increases significantly under conditions that promote matrix formation. RNAi knockdown and enforced expression of DTDST had opposite effects on FN matrix levels, indicating that this transporter is both necessary and sufficient to enhance HT1080 matrix assembly. GAG chains are known to participate in cell–FN interactions, but our results show that GAG chain sulfation also affects these interactions. Furthermore, knockdown of DTDST reduced cell surface sulfate levels, reinforcing the notion that DTDST functions to provide sufficient intracellular sulfate to maintain sulfated GAGs at the cell surface. Of the known membrane-associated proteoglycans on HT1080 cells, syndecan-2 is the only one involved in the stimulation of FN assembly. Therefore, regulation of DTDST expression and sulfate transport provides a novel mechanism to modulate levels of matrix assembly through sulfation of cell surface molecules.
Although there are several ways to obtain intracellular sulfate, cells rely primarily on active transport mechanisms for intracellular sulfate homeostasis (Markovich, 2001). Of the five known sulfate/anion transporters in humans, DTDST is the most ubiquitous and was the only one up-regulated in conditions that induced FN matrix assembly in HT1080 cells. DTDST is integral for the maintenance of physiological sulfate levels. Cartilage from human DTD patients or a mouse knockin mutant model has low staining with alcian blue and toluidine blue (Rossi et al., 1996, 1998; Forlino et al., 2005), which are both cationic dyes with an affinity for negatively charged sulfate groups (Karlsson and Bjornsson, 2001). Undersulfation was confirmed by analyses of isolated cartilage proteoglycans. The extent of this undersulfation varied with the clinical severity of the disorder (Karniski, 2001), indicating a direct connection between DTDST function and sulfation of proteoglycans.
DTDST is expressed in a wide range of tissues including colon, various glands, pancreas, and kidney, among others (Haila et al., 2001). The most notable defects in DTD patients, however, are in cartilage architecture because of the high sulfate demand of this tissue. Fibroblasts isolated from these patients also show reduced activity in sulfate uptake assays and a high resistance to chromate, which is lethal to cells only when it is imported via functional sulfate transporters (Rossi et al., 1996). Although there are several different mutations so far identified in the human DTDST gene (Superti-Furga et al., 1996; Karniski, 2001), changes in activity of this transporter have not previously been linked to altered behaviors of tumor cells. There is precedent for down-regulation of other sulfate transporters in cancer. Down-regulated in adenoma, a mammalian sulfate/chloride antiporter expressed exclusively in the colon, was isolated by subtractive hybridization comparing normal colon with colon carcinoma cells (Schweinfest et al., 1993), and reduced levels of pendrin have been reported in thyroid carcinoma (Bidart et al., 2000). Furthermore, reduced amounts and sulfation of heparan sulfate proteoglycans have been observed in SV40-transformed cells (Winterbourne and Mora, 1981), HT1080 fibrosarcoma cells (Timar and Paterson, 1990), E1A immortalized rat intestinal cells (Levy et al., 1990), and transformed rat kidney cells (Romaris et al., 1994). Therefore, sulfate import may be necessary for maintenance of a normal, nontransformed phenotype, possibly through regulation of cell surface proteoglycans and FN matrix.
In chondrodysplasia patients as well as in the mouse model of DTD, proteoglycan sulfation was significantly reduced by mutations in DTDST. In general, of the total intracellular sulfate content, more than two thirds is consumed by the sulfation of GAG chains (Shworak, 2001). These molecules would therefore be most dramatically affected by a deficiency in sulfate import. It has been shown that some cells localize 70% of total 35SO4-labeled heparan sulfate chains to the plasma membrane (Wang et al., 1999). Deficiencies in sulfate import would therefore primarily affect sulfated GAGs at the cell surface and this, in fact, fits with what we observed with chlorate or DTDST RNAi treatment. An important role for DTDST in GAG sulfation is further supported by the enhanced cell surface sulfate content in overexpressing cells.
Our finding that syndecan-2 is important for matrix assembly by HT1080 cells supports previous work that showed that a truncated form of this proteoglycan had a dominant- negative effect on deposition of FN and laminin during ECM assembly by CHO cell transfectants (Klass et al., 2000). Syndecans bind to the HepII domain of FN via electrostatic interactions mediated by heparan sulfate chains (Itano et al., 1993; Busby et al., 1995). Syndecan-4 has been shown to work with integrins for full cell spreading and focal adhesion formation (Woods and Couchman, 1994; Saoncella et al., 1999; Midwood et al., 2004). Thus, it was somewhat surprising that syndecan-4 RNAi did not inhibit matrix assembly and, in fact, seemed to promote a modest increase in matrix levels. One possible explanation for this difference is that syndecan-2 and -4 cytoplasmic tails interact with different effectors that potentially have distinct roles in cell adhesion and FN assembly. In that case, knockdown of expression of syndecan-4 would reduce competition between these proteoglycans for binding to the HepII site, allowing increased syndecan-2 engagement by FN and enhancing its associated intracellular signaling and matrix assembly. This explanation would also suggest that the relative proportions of proteoglycans, rather than their absolute levels, affect cell phenotypes. Glypican-1 reportedly also interacts with the HepII domain on FN (Tumova et al., 2000). Therefore, it is conceivable that increased FN matrix in the absence of glypican-1, and possibly also betaglycan, could result from a similar loss of a competitive inhibitor that can bind to a common site on FN.
Because FN matrix assembly is induced by up-regulation of DTDST and the downstream effects on proteoglycan sulfation and function, is it possible that perturbation of this process is an underlying cause of the skeletal defects in patients with DTD-related chondrodysplasias? FN is a major component of the chondrocyte microenvironment during cartilage development (Hedman et al., 1982; Poole et al., 1992) and, in adult tissue, colocalizes with some forms of collagen (Chang et al., 1997). Pericellular FN likely promotes chondrocyte adhesion and interacts with cell surface sulfated proteoglycans, which may be essential for initial deposition of ECM. Collagen fibril formation is dependent on the presence of FN matrix in some cell types (Velling et al., 2002), and FN is replaced by a more collagen-rich matrix during cartilage differentiation (Dessau et al., 1978; Dessau et al., 1980, 1981), raising the possibility that FN matrix may present a framework for collagen deposition during mesenchyme condensation and cartilage development. β1 integrin receptors for collagen and FN are expressed in chondrocytes, and β1-null chondrocytes are defective in skeletal development with reduced adhesion on collagen II and FN and development of chondrodysplasias (Aszodi et al., 2003). Clearly, there is interdependence between assembly of FN and collagen matrices and integrin–ECM interactions in cartilage formation. FN has also been detected in the ossification centers and growth plates of developing and mature bones (Nordahl et al., 1995; unpublished data) and has been implicated in early events in osteogenesis, where it is thought to promote osteoblast adhesion (Mackie et al., 1987; Moursi et al., 1996). Evidence supporting a role for FN matrix in differentiation of cartilage and bone, along with our data on the reduction of FN matrix in the absence of DTDST expression, provides an intriguing explanation for the skeletal defects observed in the diastrophic dysplasias.
Materials and methods
Cell culture
HT1080, WI-38(VA13), and MRC-5 cells (from T. Shenk, Princeton University, Princeton, NJ) were grown in DME with 10% FBS (Thermo Fisher Scientific) and antibiotic–antimycotic cocktail (Invitrogen). For experiments in low sulfate conditions, cells were grown in Ham's F12 (Invitrogen) with 10% PBS-dialyzed FBS and no sulfate salt–containing antibiotics.
Construction of a DTDST-overexpressing cell line (HT/DTDST-GFP)
PCR was performed to generate XhoI and XmaI sites flanking DTDST cDNA from the pGEMHE-DTD plasmid (from L. Karniski, University of Iowa, Iowa City, IA) using the primers 5′-ATTCCTCTCGAGCTGTCGGCGCCGCG-3′ and 5′-CCAGTTCCCGGGTTCC-GCTTCCGCTTCCATCACTACTAAGACTCAGACCATTGGC-3′ and Elongase DNA polymerase (Invitrogen) under the following conditions for 25 cycles: 94°C for 1.5 min, 52°C for 1 min, and 68°C for 3 min. pEGFP-N1 (BD Biosciences; Clontech Laboratories, Inc.) was digested and ligated to the XhoI–XmaI PCR product to generate pEGFPN1/DTDST with GFP cDNA fused at the 3′ end of DTDST cDNA. 4 μg pEGFPN1/DTDST DNA or the corresponding empty vector was transfected into HT1080 cells using Lipofectamine 2000 (Invitrogen). Individual clones were selected with 1 mg/ml geneticin sulfate (Invitrogen) and serially expanded until stable cell lines expressing the construct or carrying the empty vector were established. Five clones were expanded with DTDST expression levels that ranged between 4.8- and 16-fold above HT1080 cells carrying the empty vector (HT/GFP), as determined by quantitative RT-PCR. The three highest expressors with levels at 9.8-, 10.5-, and 16-fold above control clones were analyzed for matrix assembly.
Real time RT-PCR
RNA from subconfluent cells was isolated using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. cDNA was made from the isolated RNA template using Superscript II Reverse transcriptase, 2.5 mM of random hexamer mix, and 1 μg of total RNA. The reverse transcription reaction was performed with the following conditions: 37°C for 60 min, 99°C for 5 min, and 5°C for 5 min. The SYBR Green PCR Master Mix (Applied Biosystems) was used with 800 nM of each primer. The real-time reaction was done in triplicate in an ABI Prism 7900 HT detection system (Applied Biosystems) using the following reaction profile: 2 min at 50°C and 10 min at 95°C for one cycle, followed by 15 s at 95°C and 1 min at 60°C for 40 cycles. SDS 2.1 software (Applied Biosystems) was used to plot the standard curve and to determine the relative expression levels of each mRNA. Values for each experimental condition were normalized to the level of ubiquitin C in a parallel RNA sample.
Metabolic labeling and cell surface biotinylation
Cells were lifted in versene (Invitrogen) and 6 × 105 cells per well were plated in 6-well plates. After attachment, the medium was replaced with Ham's F12 low sulfate medium plus any treatments as indicated. Na2-[35S]-SO4 (MP Biomedicals) was reconstituted in water. 50 μCi/ml was added to each well and cells were labeled for 12 h.
Cultures were washed thoroughly with cold PBS and incubated for 30 min at 4°C with 1mg/ml Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific). Biotinylation reactions were quenched with five washes of 50 mM NH4Cl in PBS. Cells were lysed using radioimmunoprecipitation assay buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM EGTA, 100 U/ml aprotinin, 10 μg/ml leupeptin, and 1 mM PMSF). Streptavidin immobilized on 6% agarose beads was washed and resuspended in binding buffer (0.1% SDS in PBS). 100 μl of the 50% slurry was added to 200 μl of whole cell lysate. Samples were incubated for 2 h with rotation at 4°C. Streptavidin beads were washed five times with binding buffer and boiled for 5 min in SDS sample buffer (2% SDS, 62.5 mM Tris-HCl, pH 6.8, 10% glycerol, and 2.5% 2-mercaptoethanol). Aliquots were removed before the addition of 2-mercaptoethanol to determine total protein concentration by bicinchoninic acid assay (Thermo Fisher Scientific). Equivalent amounts of total protein were counted in a liquid scintillation counter (TriCarb 2800TR; PerkinElmer), and mean counts per minute were obtained over 20-min periods. Material eluted from streptavidin beads was separated on a 4–20% gradient SDS gel. The dried gel was exposed to film for 35 d.
Pretreatments
Cells were pretreated with various agents in normal growth media and 25 mM Hepes, pH 7.2, for 30 min with tumbling at 37°C. Treatments included 50 mM sodium chlorate (Sigma-Aldrich), 4-methylumbelliferyl-β-D-xyloside (Sigma-Aldrich), and soluble heparin or chondroitin sulfate-A (Sigma-Aldrich). Concentrations are indicated in the figure legends. For sulfation pattern experiments, cells were incubated with 250 μg/ml N-acetyl-de-O-sulfated heparin or de-N-sulfated heparin (Sigma-Aldrich). For α5β1 integrin blocking experiments, cells were allowed to adhere to tissue culture plates for 2 h. The medium was then replaced with anti-α5 integrin monoclonal antibody BIIG2 at a 1:250 dilution (obtained from C.H. Damsky, University of California, San Francisco, San Francisco, CA). After the 30-min preincubation, cells were stimulated with 0.1 μM dexamethasone and plated onto tissue culture plates. In all cases, after 20 h of dexamethasone stimulation, DOC lysates were harvested.
Immunofluorescence
For matrix assembly experiments, 6 × 105 cells per well were seeded onto 6-well tissue culture plates and allowed to attach and spread on glass coverslips with dexamethasone for 20 h. Cells were fixed in 3.7% formaldehyde in PBS plus 0.5 mM MgCl2 for 15 min. Samples were incubated with anti–human FN monoclonal antibody HFN7.1 (Developmental Studies Hybridoma Bank) at a 1:100 dilution in 2% ovalbumin (Sigma-Aldrich) in PBS for 30 min at 37°C, followed by a 1:600 dilution of fluorescein-conjugated goat anti–mouse secondary antibody (Invitrogen). Images were acquired on a microscope (Eclipse TE2000-U; Nikon) with 2,000-ms exposure times. Brightness was adjusted to +8 and contrast to +48 in Photoshop (Adobe). To analyze cell shapes, cells were allowed to attach and spread on FN-coated coverslips for 20 h, and then were fixed, permeabilized with 0.5% NP-40 (EMD), and stained with DAPI (Invitrogen) at a 1:1,000 dilution to visualize nuclei and a 1:1,000 dilution of rhodamine-phalloidin (Invitrogen) for actin filaments. Images were acquired with a 20×/0.45 objective. Image brightness/contrast was adjusted in Photoshop to +2/+54 and +31/+24 for HT/GFP and HT/DTDST-GFP, respectively. For live confocal image acquisition, cells were stained with DRAQ5 (Qbiogene) at a final concentration of 5 μM for 30 min at 37°C, and then plated onto glass-bottom culture dishes (MatTek) for confocal analysis. All confocal images were captured on a confocal system (LSM 510; Carl Zeiss, Inc.) using a water immersion objective (C-Apochromat 40× 1.2 NA; Nikon) and LSM 510 software (version 3.2). Brightness and contrast for images collected at zoom factor 3 were identically modified in Photoshop to +22. Images collected at zoom factor 1 were adjusted in Photoshop to +22 brightness and +18 contrast.
DOC insolubility assay and immunoblotting
Cells were plated onto 6-well dishes, pretreated as indicated, and stimulated with dexamethasone for 20 h. After this period, DOC-soluble and -insoluble fractions were prepared as described previously (Sechler et al., 1996). Volumes were scaled up accordingly based on the surface area of the dish. Protein concentrations of DOC-soluble fractions were determined using the bicinchoninic acid assay, and amounts of DOC-insoluble material proportional to 5 or 10 μg of DOC-soluble material were run on a 5% polyacrylamide-SDS gel, transferred to a nitrocellulose membrane (GE Healthcare), and blocked in 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Tween-20. Human FN was detected using HFN7.1 antibody at 1:300 dilution in the blocking buffer, and anti-GAPDH (ab9484; Abcam) was used at 1:10,000 dilution. Goat anti–mouse conjugated to HRP (Thermo Fisher Scientific) was used as a secondary antibody at 1:10,000. Blots from three independent trials were developed using the ECL reagent (Thermo Fisher Scientific). When quantification was necessary, blots were developed using ECL Plus Western Detection System GE Healthcare), a scanner (STORM 860; GE Healthcare), and ImageQuant TL v2005.04 software (GE Healthcare). Values were normalized to GAPDH levels.
Normalization and statistics
Quantitative RT-PCR was performed in triplicate for each RNA preparation. Raw values were averaged and normalized to the mean value for ubiquitin C. Experimental values were then divided by control values to give a fold change. To express values as a percent, the control value was set at 100%. All error bars represent standard deviations based on the mean of three independent trials. Probability of a significant difference between two values was determined by a paired, two-tailed Student's t test. Values were considered to be statistically significant when P < 0.05.
siRNA treatment
Cells were trypsinized and 1.8 × 105 cells were plated in 6-well tissue culture dishes in DME and 10% FBS without antibiotics. After 24 h, 100 nM SMARTPOOL siRNAs (Dharmacon, Inc.) for DTDST or individual proteoglycans were transfected as described in the Lipofectamine RNAiMAX protocol (Invitrogen). This procedure was also used with each individual siRNA oligonucleotide at 100 nM (coding region siRNAs: 800, 1263, 1824, 2124; 3′ UTR siRNAs: 3128, 6158, 3548 [all are named for the first base position in the DTDST cDNA]). The siCONTROL nontargeting siRNA pool composed of four oligonucleotides experimentally shown not to target mammalian RNAs was transfected in parallel. 24 h after transfection, cells were trypsinized and replated at 6 × 105 cells with 0.1 μM dexamethasone.
Detection of proteoglycans by immunoblotting
A procedure provided by A. Rapraeger (University of Wisconsin Medical School, Madison, WI) was used. Whole cell lysates were prepared using radioimmunoprecipitation assay buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and a protease inhibitor cocktail). Lysates were methanol precipitated overnight at −20°C. Samples were spun at 14,000 rpm for 5 min at 4°C and 1 vol of 100% chilled acetone was added. Lyophilized GAGase enzymes were reconstituted in PBS containing 10−5 M CaCl2 and MgCl2, 1 g/liter glucose, and 1 mg/ml BSA. For GAGase treatment, pellets were resuspended in heparitinase I buffer (50 mM Hepes, pH 6.5, 50 mM sodium acetate, 150 mM NaCl, and 5 mM CaCl2). Heparitinase I (Seikagaku Inc.) at a final concentration of 2.4 × 10−3 IU/ml and chondroitinase ABC (Sigma-Aldrich) at a final concentration of 0.1 U/ml were added to each sample. Samples were incubated at 37°C for 2 h, and then 1 μl of fresh enzyme was added and the 2-h incubation was repeated. Reactions were stopped with the addition of electrophoresis sample buffer. Proteins were transferred to a methanol prewetted Immobilon-P polyvinylidene fluoride membrane (Millipore). Blots were fixed in 0.25% glutaraldehyde for 30 min, rinsed several times in distilled H2O, rinsed in TBS (50 mM Tris-HCl, pH 7.5, and 200 mM NaCl), and blocked in TBS with 3% BSA. Blots were hybridized with 3G10 anti–heparan sulfate delta antibody (Seikagaku Inc.) at a dilution of 1:3,000, washed with TBS + 0.1% Tween-20 (Sigma-Aldrich), and treated with a 1:10,000 dilution of goat anti–mouse HRP secondary antibody.
Online supplemental material
Fig. S1 shows the effects of DTDST siRNA treatment on mRNA expression levels of the other sulfate transporters. Fig. S2 (A and B) shows knockdown of individual proteoglycan mRNAs and reduced protein levels with specific siRNA treatments, and Fig. S2 C shows that syndecan-2 mRNA levels, but not levels of other proteoglycan mRNAs, are affected by syndecan-2 siRNA treatment. Fig. S3 shows increased cell surface sulfate levels with DTDST overexpression and no effect of DTDST knockdown or overexpression on syndecan-2 protein levels. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200707150/DC1.
Acknowledgments
We thank Dr. Kui Huang for providing the impetus for this work by performing the microarray that showed differential regulation of DTDST. We also thank Dr. Anne Woods and Dr. Antonio Rossi for helpful advice, Dr. Alan Rapraeger for providing a proteoglycan immunoblot protocol, Dr. Lawrence P. Karniski for supplying DTDST cDNA, Dr. Caroline H. Damsky for the BIIG2 integrin α5 function-blocking antibody, and Joseph Goodhouse for confocal expertise.
This work was supported by National Institutes of Health grants R01 GM059383 and P01 CA041086 to J.E. Schwarzbauer and T32 GM007388 to L.L. Galante.
Abbreviations used in this paper: DOC, deoxycholate; DTDST, diastrophic dysplasia sulfate transporter; FN, fibronectin; GAG, glycosaminoglycan; UTR, untranslated region.
References
- Ali, I.U., V. Mautner, R. Lanza, and R.O. Hynes. 1977. Restoration of normal morphology, adhesion and cytoskeleton in transformed cells by addition of a transformation-sensitive surface protein. Cell. 11:115–126. [DOI] [PubMed] [Google Scholar]
- Almeida, M.J., C. Pereira, C. Milet, J. Haigle, M. Barbosa, and E. Lopez. 2001. Comparative effects of nacre water-soluble matrix and dexamethasone on the alkaline phosphatase activity of MRC-5 fibroblasts. J. Biomed. Mater. Res. 57:306–312. [DOI] [PubMed] [Google Scholar]
- Aszodi, A., E.B. Hunziker, C. Brakebusch, and R. Fassler. 2003. Beta1 integrins regulate chondrocyte rotation, G1 progression, and cytokinesis. Genes Dev. 17:2465–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidart, J.M., C. Mian, V. Lazar, D. Russo, S. Filetti, B. Caillou, and M. Schlumberger. 2000. Expression of pendrin and the Pendred syndrome (PDS) gene in human thyroid tissues. J. Clin. Endocrinol. Metab. 85:2028–2033. [DOI] [PubMed] [Google Scholar]
- Brenner, K.A., S.A. Corbett, and J.E. Schwarzbauer. 2000. Regulation of fibronectin matrix assembly by activated Ras in transformed cells. Oncogene. 19:3156–3163. [DOI] [PubMed] [Google Scholar]
- Busby, T.F., W.S. Argraves, S.A. Brew, I. Pechik, G.L. Gilliland, and K.C. Ingham. 1995. Heparin binding by fibronectin module III-13 involves six discontinuous basic residues brought together to form a cationic cradle. J. Biol. Chem. 270:18558–18562. [DOI] [PubMed] [Google Scholar]
- Chang, J., H. Nakajima, and C.A. Poole. 1997. Structural colocalisation of type VI collagen and fibronectin in agarose cultured chondrocytes and isolated chondrons extracted from adult canine tibial cartilage. J. Anat. 190:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., and D.F. Mosher. 1996. Formation of sodium dodecyl sulfate-stable fibronectin multimers. J. Biol. Chem. 271:9084–9089. [DOI] [PubMed] [Google Scholar]
- Chung, C.Y., and H.P. Erickson. 1997. Glycosaminoglycans modulate fibronectin matrix assembly and are essential for matrix incorporation of tenascin-C. J. Cell Sci. 110:1413–1419. [DOI] [PubMed] [Google Scholar]
- Dessau, W., J. Sasse, R. Timpl, F. Jilek, and K. von der Mark. 1978. Synthesis and extracellular deposition of fibronectin in chondrocyte cultures. Response to the removal of extracellular cartilage matrix. J. Cell Biol. 79:342–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessau, W., H. von der Mark, K. von der Mark, and S. Fischer. 1980. Changes in the patterns of collagens and fibronectin during limb-bud chondrogenesis. J. Embryol. Exp. Morphol. 57:51–60. [PubMed] [Google Scholar]
- Dessau, W., B.M. Vertel, H. von der Mark, and K. von der Mark. 1981. Extracellular matrix formation by chondrocytes in monolayer culture. J. Cell Biol. 90:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko, J.D. 1991. Genetic analysis of proteoglycan structure, function and metabolism. Curr. Opin. Cell Biol. 3:805–816. [DOI] [PubMed] [Google Scholar]
- Esko, J.D., T.E. Stewart, and W.H. Taylor. 1985. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. USA. 82:3197–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlino, A., R. Piazza, C. Tiveron, S. Della Torre, L. Tatangelo, L. Bonafe, B. Gualeni, A. Romano, F. Pecora, A. Superti-Furga, et al. 2005. A diastrophic dysplasia sulfate transporter (SLC26A2) mutant mouse: morphological and biochemical characterization of the resulting chondrodysplasia phenotype. Hum. Mol. Genet. 14:859–871. [DOI] [PubMed] [Google Scholar]
- Fritz, T.A., and J.D. Esko. 2001. Xyloside priming of glycosaminoglycan biosynthesis and inhibition of proteoglycan assembly. Methods Mol. Biol. 171:317–323. [DOI] [PubMed] [Google Scholar]
- Haila, S., J. Hastbacka, T. Bohling, M.L. Karjalainen-Lindsberg, J. Kere, and U. Saarialho-Kere. 2001. SLC26A2 (diastrophic dysplasia sulfate transporter) is expressed in developing and mature cartilage but also in other tissues and cell types. J. Histochem. Cytochem. 49:973–982. [DOI] [PubMed] [Google Scholar]
- Hastbacka, J., A. delaChapelle, M.M. Mahtani, G. Clines, M.P. Reeve-Daly, M. Daly, B.A. Hamilton, K. Kusumi, B. Trivedi, A. Weaver, et al. 1994. The diastrophic dysplasia gene encodes a novel sulfate transporter: positional cloning by fine-structure linkage disequilibrium mapping. Cell. 78:1073–1087. [DOI] [PubMed] [Google Scholar]
- Hedman, K., K. Alitalo, S. Lehtinen, R. Timpl, and A. Vaheri. 1982. Deposition of an intermediate form of procollagen type III (pN-collagen) into fibrils in the matrix of amniotic epithelial cells. EMBO J. 1:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes, R.O. 1990. Fibronectins. Springer-Verlag, New York. 546 pp.
- Itano, N., K. Oguri, H. Nakanishi, and M. Okayama. 1993. Membrane-intercalated proteoglycan of a stroma-inducing clone from Lewis lung carcinoma binds to fibronectin via its heparan sulfate chains. J. Biochem. (Tokyo). 114:862–873. [DOI] [PubMed] [Google Scholar]
- Itano, N., K. Oguri, Y. Nagayasu, Y. Kusano, H. Nakanishi, G. David, and M. Okayama. 1996. Phosphorylation of a membrane-intercalated proteoglycan, syndecan-2, expressed in a stroma-inducing clone from mouse Lewis lung carcinoma. Biochem. J. 315:925–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson, M., and S. Bjornsson. 2001. Quantitation of proteoglycans in biological fluids using Alcian blue. Methods Mol. Biol. 171:159–173. [DOI] [PubMed] [Google Scholar]
- Karniski, L.P. 2001. Mutations in the diastrophic dysplasia sulfate transporter (DTDST) gene: correlation between sulfate transport activity and chondrodysplasia phenotype. Hum. Mol. Genet. 10:1485–1490. [DOI] [PubMed] [Google Scholar]
- Kato, M., S. Saunders, H. Nguyen, and M. Bernfield. 1995. Loss of cell surface syndecan-1 causes epithelia to transform into anchorage-independent mesenchyme-like cells. Mol. Biol. Cell. 6:559–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, K.M., P.R. Brauer, and J.M. Keller. 1989. Modulation of cell surface heparan sulfate structure by growth of cells in the presence of chlorate. Biochemistry. 28:8100–8107. [DOI] [PubMed] [Google Scholar]
- Klaassen, C.D., and J.W. Boles. 1997. The importance of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) in the regulation of sulfation. FASEB J. 11:404–418. [DOI] [PubMed] [Google Scholar]
- Klass, C.M., J.R. Couchman, and A. Woods. 2000. Control of extracellular matrix assembly by syndecan-2 proteoglycan. J. Cell Sci. 113:493–506. [DOI] [PubMed] [Google Scholar]
- Lee, R.W.H., and W.B. Huttner. 1983. Tyrosine-0-sulfated proteins of PC12 pheochromocytoma cells and their sulfation by a tyrosylprotein sulfotransferase. J. Biol. Chem. 258:11326–11334. [PubMed] [Google Scholar]
- Levy, P., S. Emami, G. Cherqui, E. Chastre, C. Gespach, and J. Picard. 1990. Altered expression of proteoglycans in E1A-immortalized rat fetal intestinal epithelial cells in culture. Cancer Res. 50:6716–6722. [PubMed] [Google Scholar]
- Mackie, E.J., I. Thesleff, and R. Chiquet-Ehrismann. 1987. Tenascin is associated with chondrogenic and osteogenic differentiation in vivo and promotes chondrogenesis in vitro. J. Cell Biol. 105:2569–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, Y., and J.E. Schwarzbauer. 2005. Fibronectin fibrillogenesis, a cell- mediated matrix assembly process. Matrix Biol. 24:389–399. [DOI] [PubMed] [Google Scholar]
- Markovich, D. 2001. Physiological roles and regulation of mammalian sulfate transporters. Physiol. Rev. 81:1499–1533. [DOI] [PubMed] [Google Scholar]
- McKeown-Longo, P.J., and D.F. Mosher. 1983. Binding of plasma fibronectin to cell layers of human skin fibroblasts. J. Cell Biol. 97:466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown-Longo, P.J., and C.A. Etzler. 1987. Induction of fibronectin matrix assembly in human fibrosarcoma cells by dexamethasone. J. Cell Biol. 104:601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes de Aguiar, C.B., R.C. Garcez, M. Alvarez-Silva, and A.G. Trentin. 2002. Undersulfation of proteoglycans and proteins alter C6 glioma cells proliferation, adhesion and extracellular matrix organization. Int. J. Dev. Neurosci. 20:563–571. [DOI] [PubMed] [Google Scholar]
- Midwood, K.S., L.V. Valenick, H.C. Hsia, and J.E. Schwarzbauer. 2004. Coregulation of fibronectin signaling and matrix contraction by tenascin-C and syndecan-4. Mol. Biol. Cell. 15:5670–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranti, C.K., and J.S. Brugge. 2002. Sensing the environment: a historical perspective on integrin signal transduction. Nat. Cell Biol. 4:E83–90. [DOI] [PubMed] [Google Scholar]
- Moursi, A.M., C.H. Damsky, J. Lull, D. Zimmerman, S.B. Doty, S. Aota, and R.K. Globus. 1996. Fibronectin regulates calvarial osteoblast differentiation. J. Cell Sci. 109:1369–1380. [DOI] [PubMed] [Google Scholar]
- Nordahl, J., S. Mengarelli-Widholm, K. Hultenby, and F.P. Reinholt. 1995. Ultrastructural immunolocalization of fibronectin in epiphyseal and metaphyseal bone of young rats. Calcif. Tissue Int. 57:442–449. [DOI] [PubMed] [Google Scholar]
- Oliver, N., R.F. Newby, L.T. Furcht, and S. Bourgeois. 1983. Regulation of fibronectin biosynthesis by glucocorticoids in human fibrosarcoma cells and normal fibroblasts. Cell. 33:287–296. [DOI] [PubMed] [Google Scholar]
- Pasqualini, R., S. Bourdoulous, E. Koivunen, V.L. Woods, and E. Rouslahti. 1996. A polymeric form of fibronectin has antimetastatic effects against multiple tumor types. Nat. Med. 2:1197–1203. [DOI] [PubMed] [Google Scholar]
- Poole, C.A., S. Ayad, and R.T. Gilbert. 1992. Chondrons from articular cartilage. V. Immunohistochemical evaluation of type VI collagen organisation in isolated chondrons by light, confocal and electron microscopy. J. Cell Sci. 103:1101–1110. [DOI] [PubMed] [Google Scholar]
- Rapraeger, A.C., A. Krufka, and B.B. Olwin. 1991. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 252:1705–1708. [DOI] [PubMed] [Google Scholar]
- Romaris, M., J. Villena, A. Molist, A. Heredia, and A. Bassols. 1994. Ras transformation alters the composition of extracellular matrix proteoglycans in rat fibroblasts. Biochem. Biophys. Res. Commun. 200:925–932. [DOI] [PubMed] [Google Scholar]
- Rossi, A., J. Bonaventure, A.L. Delezoide, G. Cetta, and A. Superti-Furga. 1996. Undersulfation of proteoglycans synthesized by chondrocytes from a patient with achondrogenesis type 1B homozygous for an L483P substitution in the diastrophic dysplasia sulfate transporter. J. Biol. Chem. 271:18456–18464. [DOI] [PubMed] [Google Scholar]
- Rossi, A., I. Kaitila, W.R. Wilcox, D.L. Rimoin, B. Steinmann, G. Cetta, and A. Superti-Furga. 1998. Proteoglycan sulfation in cartilage and cell cultures from patients with sulfate transporter chondrodysplasias: relationship to clinical severity and indications on the role of intracellular sulfate production. Matrix Biol. 17:361–369. [DOI] [PubMed] [Google Scholar]
- Saoncella, S., F. Echtermeyer, F. Denhez, J.K. Nowlen, D.F. Mosher, S.D. Robinson, R.O. Hynes, and P.F. Goetinck. 1999. Syndecan-4 signals cooperatively with integrins in a Rho-dependent manner in the assembly of focal adhesions and actin stress fibers. Proc. Natl. Acad. Sci. USA. 96:2805–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer, J.E. 1991. Identification of the fibronectin sequences required for assembly of a fibrillar matrix. J. Cell Biol. 113:1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer, J.E., and J.L. Sechler. 1999. Fibronectin fibrillogenesis: a paradigm for extracellular matrix assembly. Curr. Opin. Cell Biol. 11:622–627. [DOI] [PubMed] [Google Scholar]
- Schweinfest, C.W., K.W. Henderson, S. Suster, N. Kondoh, and T.S. Papas. 1993. Identification of a colon mucosa gene that is down-regulated in colon adenomas and adenocarcinomas. Proc. Natl. Acad. Sci. USA. 90:4166–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechler, J.L., Y. Takada, and J.E. Schwarzbauer. 1996. Altered rate of fibronectin matrix assembly by deletion of the first type III repeats. J. Cell Biol. 134:573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shworak, N.W. 2001. High-specific-activity 35S-labeled heparan sulfate prepared from cultured cells. Methods Mol. Biol. 171:79–89. [DOI] [PubMed] [Google Scholar]
- Sjoberg, I., and A. Malmstrom. 1982. Biosynthesis of dermatan sulphate in cultured fibroblasts. Characterization of newly synthesized glycans from cells and microsomes. Eur. J. Biochem. 128:29–34. [PubMed] [Google Scholar]
- Superti-Furga, A., A. Rossi, B. Steinmann, and R. Gizelmann. 1996. A chondrodysplasia family produced by mutations in the diastrophic dysplasia sulfate transporter gene: genotype/phenotype correlations. Am. J. Med. Genet. 63:144–147. [DOI] [PubMed] [Google Scholar]
- Tavella, S., G. Bellese, P. Castagnola, I. Martin, D. Piccini, R. Doliana, A. Colombatti, R. Cancedda, and C. Tacchetti. 1997. Regulated expression of fibronectin, laminin and related integrin receptors during the early chondrocyte differentiation. J. Cell Sci. 110:2261–2270. [DOI] [PubMed] [Google Scholar]
- Timar, J., and H. Paterson. 1990. Localization and production of proteoglycans by HT1080 cell lines with altered N-ras expression. Cancer Lett. 53:145–150. [DOI] [PubMed] [Google Scholar]
- Tumova, S., A. Woods, and J.R. Couchman. 2000. Heparan sulfate chains from glypican and syndecans bind the Hep II domain of fibronectin similarly despite minor structural differences. J. Biol. Chem. 275:9410–9417. [DOI] [PubMed] [Google Scholar]
- Velling, T., J. Risteli, K. Wennerberg, D.F. Mosher, and S. Johansson. 2002. Polymerization of type I and III collagens is dependent on fibronectin and enhanced by integrins alpha 11beta 1 and alpha 2beta 1. J. Biol. Chem. 277:37377–37381. [DOI] [PubMed] [Google Scholar]
- Wang, A., T. Miralem, and D.M. Templeton. 1999. Heparan sulfate chains with antimitogenic properties arise from mesangial cell-surface proteoglycans. Metabolism. 48:1220–1229. [DOI] [PubMed] [Google Scholar]
- Winterbourne, D.J., and P.T. Mora. 1981. Cells selected for high tumorigenicity or transformed by simian virus 40 synthesize heparan sulfate with reduced degree of sulfation. J. Biol. Chem. 256:4310–4320. [PubMed] [Google Scholar]
- Woods, A., and J.R. Couchman. 1994. Syndecan-4 heparan sulfate proteoglycan is a selectively enriched and widespread focal adhesion component. Mol. Biol. Cell. 5:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., J.S. Bauer, R.L. Juliano, and J.A. McDonald. 1993. The alpha 5 beta 1 integrin fibronectin receptor, but not the alpha 5 cytoplasmic domain, functions in an early and essential step in fibronectin matrix assembly. J. Biol. Chem. 268:21883–21888. [PubMed] [Google Scholar]
- Yamada, K.M., S.S. Yamada, and I. Pastan. 1976. Cell surface protein partially restores morphology, adhesiveness, and contact inhibition of movement to transformed fibroblasts. Proc. Natl. Acad. Sci. USA. 73:1217–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]