Abstract
Type II par operons harness polymerization of the dynamically unstable actin-like protein ParM to segregate low-copy plasmids in rod-shaped bacteria. In this study, we use time-lapse fluorescence microscopy to follow plasmid dynamics and ParM assembly in Escherichia coli. Plasmids lacking a par operon undergo confined diffusion with a diffusion constant of 5 × 10−5 μm2/s and a confinement radius of 0.28 μm. Single par-containing plasmids also move diffusively but with a larger diffusion constant (4 × 10−4 μm2/s) and confinement radius (0.42 μm). ParM filaments are dynamically unstable in vivo and form spindles that link pairs of par-containing plasmids and drive them rapidly (3.1 μm/min) toward opposite poles of the cell. After reaching the poles, ParM filaments rapidly and completely depolymerize. After ParM disassembly, segregated plasmids resume diffusive motion, often encountering each other many times and undergoing multiple rounds of ParM-dependent segregation in a single cell cycle. We propose that in addition to driving segregation, the par operon enables plasmids to search space and find sister plasmids more effectively.
Introduction
Transmission of genetic information from one generation to the next requires efficient partitioning of DNA molecules between daughter cells. Eukaryotic DNA segregation relies on the construction of a complex mitotic spindle that finds and aligns sister chromatids and then moves them to opposite sides of the cell division plane (Inoue, 1953). This process is conserved across eukaryotic phyla, and the molecular mechanisms driving it have been extensively studied. In contrast, the mechanisms driving prokaryotic chromosome segregation are still poorly understood.
The best understood example of bacterial DNA segregation is the partitioning of low-copy plasmids. Plasmids are circular pieces of extrachromosomal DNA that often contribute to virulence and antibiotic resistance in pathogenic bacteria. Large low-copy plasmids encode simple segregation systems to ensure that at least one copy of the plasmid ends up on each side of the cell division plane and is inherited by each daughter. The majority of these segregation systems fall into one of two classes: type I or type II. The segregation operons of both classes are composed of three components: (1) a centromeric DNA sequence, (2) genes encoding a DNA-binding protein, and (3) genes encoding an ATPase. The classes are distinguished primarily by the structure of the ATPase component (for review see Ebersbach and Gerdes, 2005). Type I segregation operons encode a deviant Walker-box ATPase (ParA), whereas type II segregation operons encode an actin-like ATPase (ParM).
The most thoroughly characterized plasmid segregation system is the type II mechanism that drives segregation of the R1 multidrug resistance plasmid isolated from Escherichia coli (Gerdes and Molin, 1986). The R1 par operon encodes a ParM that forms actin-like filaments in the presence of ATP (Moller-Jensen et al., 2002) and a DNA-binding protein, ParR, that binds a series of 10 direct sequence repeats in the centromeric parC DNA (Jensen et al., 1998). By total internal fluorescence microscopy, purified ParM forms filaments that are dynamically unstable and polymerize bidirectionally (Garner et al., 2004). The ParR–parC protein–DNA complex forms a structure analogous to a eukaryotic kinetochore that binds to either end of a ParM filament and suppresses dynamic instability. Insertional polymerization at the plasmid/filament interface generates force capable of segregating parC-coated beads. Based on these data, it has been proposed that two plasmids can capture opposite ends of the same ParM filament and that polymerization pushes the plasmids apart (Garner et al., 2007).
ParM-dependent DNA segregation has been reconstituted and studied in vitro, but the dynamics of plasmid segregation in vivo are unknown. To date, microscopic studies of type II plasmid segregation in bacteria have been based on either fixed cells or static images of live cells (Jensen and Gerdes, 1999; Weitao et al., 2000; Moller-Jensen et al., 2003). Time-lapse light microscopy of living cells has provided key insights into the structure and function of the mitotic spindle in eukaryotes (Inoue, 1953; Rieder et al., 1986). In this study, we use time-lapse imaging of fluorescently labeled ParM filaments and plasmids to follow the in vivo dynamics of ParM filament assembly and plasmid segregation. We find that segregation driven by the par operon is a highly dynamic process. Individual par-containing plasmids undergo diffusive motions that are considerably faster than those of control plasmids lacking the par operon. Pairs of plasmids are rapidly pushed toward the poles of the cell by elongating ParM filaments. Once they reach the poles, the ParM filaments disassemble, and the plasmids resume diffusive motion. Remarkably, multiple plasmids in the same cell can interact multiple times in a single cell cycle and undergo multiple rounds of segregation.
Results
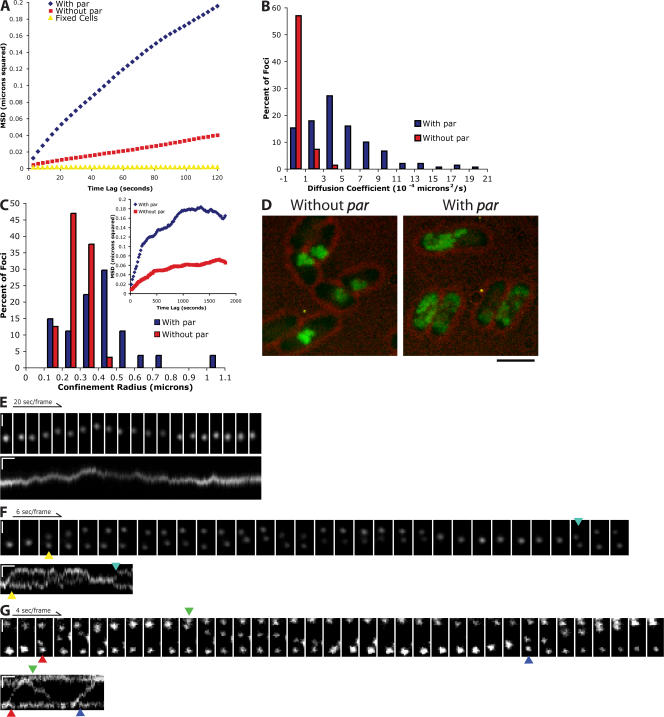
We used time-lapse fluorescence microscopy to compare plasmid dynamics in the absence and presence of the par operon. To visualize plasmids, we used a system first developed by Straight et al. (1996) in which LacI-GFP is bound to a tandem array of lacO sites integrated into the plasmid. We cotransformed E. coli cells with a low-copy mini–F plasmid containing the par operon from plasmid R1 and a series of 256 lacO repeats together with a plasmid expressing LacI-GFP. Cotransformed cells contained bright fluorescent foci. Control cells transformed with one plasmid or the other by itself contained no fluorescent foci, indicating that both LacI-GFP and the lacO repeats are necessary to generate fluorescent foci. We determined plasmid positions at every time point and plotted mean squared displacement (MSD) as a function of time. As a control, we looked at plasmids in fixed cells to show that noise and stage drift do not substantially affect our MSD measurements (Fig. 1 A). When analyzed on short time scales (<60 s), plasmids lacking the par operon undergo very slow, diffusive motions, with an average diffusion coefficient of 5 × 10−5 μm2/s (Fig. 1, A and B). The rate is considerably less than that for GFP alone (8 μm2/s; Elowitz et al., 1999) or GFP-labeled mRNA (10−3–10−2 μm2/s; Golding and Cox, 2006). Additionally, at time intervals <200 s, the MSD increases linearly with time, meaning that the movements are neither directed nor constrained. At longer time scales (>1,000 s), the MSD of the labeled plasmids departs from linearity and eventually plateaus, indicating that the diffusive motions of the plasmids are confined within a small volume. The average confinement radius is 275 nm, smaller than the dimensions of the bacterium (Fig. 1 C), implying that the plasmids are confined to subcellular compartments in which they are free to diffuse but from which they rarely escape. These cytoplasmic pockets can be viewed directly by projecting the maximum intensities of all of the pixels on all of the images of a time-lapse sequence onto a single image (Fig. 1 D).
Figure 1.
Par-containing plasmids undergo both rapid, directional and slow, diffusive movements. Arrows point to the initiation of segregation events. The colors of the arrows are coordinated between image montages and kymographs. (A) MSD versus time lag for plasmids with the par operon (pRBJ460; average of 151 foci) and those that do not contain the par operon (pRBJ461; average of 99 foci). (B) Distribution of diffusion coefficients for plasmids with and without the par operon. The diffusion coefficient was measured by taking the slope of individual MSD versus time lag traces and dividing by four (for the 2° of freedom). (C) Distribution of confinement radii for plasmids with (n = 27) and without (n = 32) the par operon. The plateau of each MSD was estimated by averaging the 1,600–1,800-s time points. The square root of the plateau value was then taken to obtain the confinement radius for each trace. (inset) Representative traces of MSD versus time lag for the longer time intervals used to estimate confinement sizes. (D) Maximum intensity projection of plasmids with and without the par operon over the course of a 2,000-s time series. Bar, 1.8 μm. (E) Cell with a single par-containing plasmid focus exhibiting diffusive movements. (F) Cell with two par-containing plasmid foci displaying mostly diffusive movements with occasional periods of rapid segregation. (G) Example of the rapid pole to pole movements seen in cells with three par-containing plasmid foci. Vertical bars, 1 μm; horizontal bars, 20 s. All images were contrast adjusted for clarity and rotated for ease of presentation.
The maximum number of foci observed in a given cell over a time course ranged from zero to four (Table S2, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1). This is in agreement with estimates by Collins and Pritchard (1973) that the F plasmid is present at approximately one copy per chromosome equivalent. We next analyzed the motion of plasmids containing the par operon. In cells with only a single fluorescent spot, the plasmid also undergoes apparently diffusive motion (Fig. 1 E and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1) but with a markedly higher diffusion coefficient (4 × 10−4 μm2/s) and confinement radius (420 nm), indicating that the par operon increases the mobility of individual plasmids (Fig. 1, A–C; and Video 1).
In cells containing multiple foci, the labeled plasmids frequently converge to form clusters, as observed in previous studies (Pogliano et al., 2001; Li and Austin, 2002; Ebersbach et al., 2005). Therefore, the number of fluorescent foci in an individual image does not necessarily reflect the total number of plasmids present in a cell. For this reason, we classify cells based on the maximum number of foci observed over the entire acquisition time.
Cells with two foci exhibit a much different behavior from those containing only a single focus (Fig. 1 F and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1). Both foci move randomly and independently until they come into close proximity and merge into a single fluorescent spot. The plasmids then move together as a single unit for a few seconds until they split and move rapidly in opposite directions, one bright spot moving to each pole of the cell. After remaining immobilized at the poles for several seconds, the plasmids begin to move diffusively again and slowly migrate around the cell until they encounter each other once more and repeat the entire process.
In cells with three foci, the plasmids often develop a stable oscillatory pattern in which one fluorescent spot is maintained at each pole while a third spot undergoes repeated pole to pole movements (Fig. 1 G and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1). We observe this ping-pong behavior only in cells with three plasmid foci. The pole to pole movements take 10–30 s and always alternate direction. This oscillatory behavior maintains at least one plasmid at each end at all times. Plasmids undergoing rapid pole to pole motion occasionally pause near the center of the cell before continuing to the pole (Fig. 1 G, second translocation). The time between pole to pole movements is highly variable, ranging from 0 to 150 s (59 ± 37 s; n = 30). 0-s dwell times indicate that plasmids switched directions before traveling all the way to the other pole. After merging, a cluster of two foci often moves slightly away from the pole immediately before segregation (Fig. 1, F [first segregation] and G [first and third segregations]). Cells with four foci are rare, and the frequent clustering and unclustering of the plasmids make generalizations about their behavior difficult (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1). Although the dynamics in this case are complicated, one plasmid is almost always located near each pole.
For plasmid inheritance, the most important DNA segregation events are those that occur immediately before cell division. For this reason, we wanted to see whether plasmid behavior changed during septation. In actively dividing cells containing three plasmid foci, plasmids continue to oscillate from pole to pole. We observed cells in which a plasmid moved through a narrowing septum and then attempted to move back but failed to pass the site of septation, which was presumably blocked by septum closure (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1).
By immunofluorescence, Moller-Jensen et al. (2002, 2003) observed ParM filaments in ∼40% of cells containing plasmids with the R1 par operon. These filaments often ran from pole to pole and were associated with fluorescently labeled plasmids. To better understand the mechanism behind the observed plasmid dynamics, we used time-lapse fluorescence microscopy to study the assembly of ParM filaments in vivo. To monitor ParM dynamics, we used a GFP-ParM fusion protein expressed in cells with a par-containing plasmid. By having the unlabeled ParM expressed from the wild-type operon present, we minimize any negative effects of the GFP fusion. This technique has been successful for visualizing actin and other actin-related proteins (Westphal et al., 1997; Gitai et al., 2004). Overexpression of GFP-ParM produced bright, stable filaments in all cells observed (unpublished data). Filaments appeared regardless of the presence of ParR and parC. When we turned off the expression of GFP-ParM and diluted the fluorescent protein by allowing cells to multiply for four to six generations, we began to see dynamic filaments (Fig. 2). In contrast to the bundles seen when GFP-ParM was overexpressed, we observed these filaments only in the presence of the additional plasmid containing the par operon. These structures, which we call spindles by analogy with the eukaryotic DNA segregation machinery, elongate for up to 1 min before switching from elongation to rapid shortening. This behavior is very similar to the dynamic instability seen in vitro with purified ParM (Garner et al., 2004). The spindles never last longer than 3 min, indicating that stabilization against catastrophe by the ParR–parC complex is limited in vivo. Consistent with our observations of plasmid movement, elongation of ParM spindles sometimes slows when one end nears the center of the cell (Fig. 2, A and B; magenta arrows; and Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1). More rarely, we observe pauses in shortening (Fig. 2, A and B; yellow arrows). We never observed a switch from shortening to elongation.
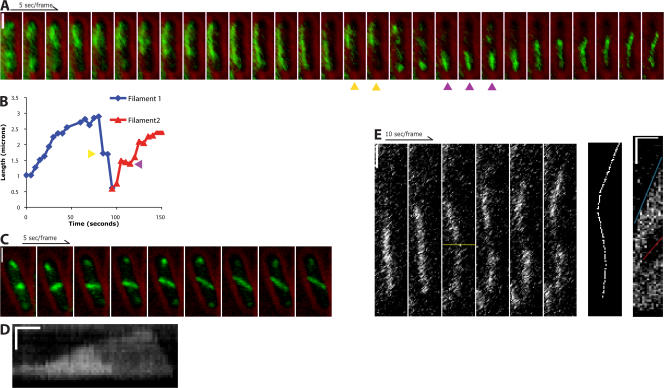
Figure 2.
ParM forms dynamic filaments in E. coli. Time-lapse fluorescence microscopy of live cells expressing GFP-ParM and a plasmid containing the R1 par operon. (A and C) GFP-ParM (green) is superimposed over single brightfield images (red) taken directly before or after the time series. (A) A ParM spindle polymerizes from one pole to the other, stalls, and depolymerizes. A second spindle forms at the opposite pole and polymerizes in the other direction. Because of photobleaching, each frame in the series was contrast adjusted individually. (B) Length of the spindles in A measured over time. Pauses can be seen during the depolymerization of the first spindle (yellow arrows) and polymerization of the second spindle (magenta arrows). (C) Reorientation of a spindle upon contact with the sides of the cell. (D) Kymograph of a spindle that depolymerizes in two stages. Horizontal bar, 20 s of elapsed time. (E) Spindles elongate equally from each end. A polymerizing spindle was photobleached to create a fiducial mark (third frame, yellow line). The seventh frame is the line that was drawn to create the kymograph to the right. The red line is the rate of displacement of the photobleached spot by polymerization of the bottom of the spindle against the end of the bacterium (21 nm/s). The blue line is the rate of elongation of the entire spindle (44 nm/s). Horizontal bar in the kymograph, 50 s of elapsed time. Vertical bars, 1 μm. All images were contrast adjusted for clarity and rotated for ease of presentation.
How do ParM filaments find the long axis of the host cell? In our time-lapse videos, we note that ParM spindles do not always initially align with the long axis (Fig. 2 C and Video 6, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1). In cases in which spindles originally elongate orthogonal to the long axis, they quickly make contact with the sides of the cell. Continued elongation then proceeds together with alignment along the long axis. This result demonstrates that the initial spindle orientation is not predetermined by a cellular landmark and that alignment with the long axis is probably driven by elongation of the spindle itself.
In fixed cells, ParM spindles are frequently curved (Moller-Jensen et al., 2002). This could result from filament buckling caused by continued polymerization against both poles of the bacterium. However, spindles also frequently show curvature as they elongate across the cell (Fig. 2, A [last four frames] and E), suggesting that bending of the filaments often results from interaction with intracellular obstacles such as the nucleoid.
One important question is whether ParM spindles are composed of one filament or bundles of multiple filaments. Evidence that ParM spindles are filament bundles comes from the fact that spindles do not always depolymerize in a single step. In rare cases, spindles first show a considerable overall decrease in fluorescence before completely depolymerizing (Fig. 2 D, Fig. S1, and Video 7, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1). Kymograph analysis of these spindles shows that the decrease in fluorescence intensity is directional, moving from one end of the spindle to the other, and proceeds at the same rate as catastrophic depolymerization. The remaining spindle continues to elongate until it, too, depolymerizes. Although these data indicate that at least some spindles contain multiple filaments, we cannot rule out that these spindles are generated by clusters of linked plasmid rather than individual plasmid pairs.
To determine the directionality of polymerization, we photobleached a small section of a spindle to create a fiducial mark. Both ends of the spindle polymerize away from the mark at similar rates, indicating that, similar to in vitro reconstituted spindles (Garner et al., 2007), polymerization occurs simultaneously at both ends of a spindle. Also, when one end of the filament abuts the end of the cell, the bleached mark migrates in the opposite direction at a rate half that of the full spindle, indicating that polymerization against a barrier can drive the entire spindle through the cytoplasm (Fig. 2 E and Video 8, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1).
To determine the connection between plasmid movement and ParM dynamics, we expressed mCherry-ParM in cells containing GFP-labeled plasmids and imaged both labels. In cells that contain both ParM filaments and plasmid foci (n = 178), spindles always colocalized with the plasmid (Fig. 3). In all cases in which we observed pairs of plasmids moving rapidly in opposite directions, the plasmid pairs were separated by an elongating ParM spindle (Fig. 3 A and Video 9, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1). After plasmids reach the poles and stop moving, the associated spindles rapidly depolymerize. Plasmids move apart on elongating ParM spindles, but we never observed plasmids moving on shortening spindles.
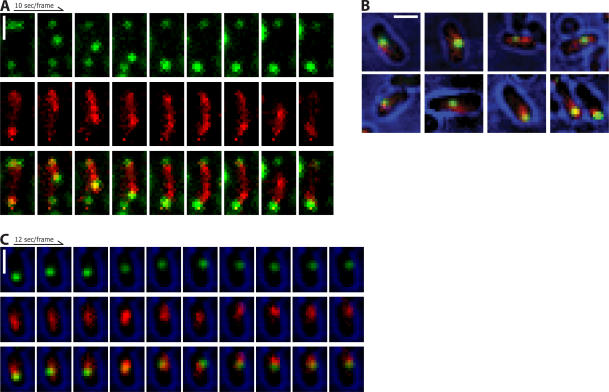
Figure 3.
Near-simultaneous visualization of ParM and plasmids. ParM, red; plasmids, green. (A) Plasmids colocalize with the ends of spindles as they polymerize across the cell. (B) Colocalization of ParM and plasmids in cells with only one plasmid focus. (C) Time-lapse series of a cell with a single plasmid focus. Brightfield (blue) is a single image taken directly after the time series and superimposed over the fluorescence images. Bars, 1 μm. All images were contrast adjusted for clarity and rotated for ease of presentation.
Single plasmid foci are also associated with increased mCherry fluorescence. The ParM fluorescence extends beyond the fluorescence of the plasmids, suggesting that short filaments extend from the plasmids (Fig. 3 B). These filaments are generally shorter, dimmer, and more dynamic than those that form the spindles associated with pairs of plasmid foci (Figs. 2 C and 3 C and Video 10, available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1). One end of these filaments appears to be bound and stabilized by the ParR–parC complex, whereas the other is presumably searching for additional plasmids. These structures are similar to ParM asters that form around isolated parC-coated beads in vitro (Garner et al., 2007) and probably explains the faster diffusive motions we observed for single plasmids containing the par operon. Thus, it appears that type II par operons evolved to enable plasmids to both search for partners and to be efficiently captured by other searching plasmids; that is, they play the role of both spindle pole and kinetochore.
We next measured the rates of spindle elongation and disassembly as well as the rate at which plasmids move apart from each other (Table I). The rates of spindle polymerization and plasmid segregation are nearly identical. This rate is also close to the previously measured rates of the polymerization of ParM in vitro (Garner et al., 2004, 2007). This result has two important implications. First, polymerization at both ends of ParM filaments powers plasmid segregation. Previous work has shown that ParM filaments polymerize equally from each end, both in isolation (Garner et al., 2004) and when associated with the ParR–parC complex (Garner et al., 2007). The in vitro rate in Table I is for both ends combined. Because plasmids move at this rate, they are being propelled by polymerization at both ends of the spindle, as confirmed by photobleaching experiments (Fig. 2 E). The second implication of these measurements is that the amount of monomer in the bacterium is at the steady state of 2.3 μM. Previous estimates for the total concentration of ParM protein expressed in E. coli are around 15 μM (Moller-Jensen et al., 2002). If this were the concentration of free, monomeric ParM, we would expect an initial rate of polymerization six to seven times greater than that observed. Therefore, we suggest that there is ParM polymer present in cells even when spindles are not visible. This result is in agreement with ParM having a very low nucleation barrier (Garner et al., 2004). Most likely, this polymer is not seen because it is in the form of short, rapidly diffusing filaments distributed throughout the cytoplasm. The simplest explanation for why the spindles can be seen over this background of dynamic polymer is that they are bundles of multiple filaments, making them brighter than single filaments. Alternatively, association with a 19-MD plasmid may decrease the diffusion of the filaments such that they are less likely to move during image acquisition.
Table I.
Comparison of ParM filament polymerization rates and plasmid segregation rates
| ParM in vitro | ParM in vivo | Plasmid segregation | |
|---|---|---|---|
| Polymerization rate (nm/s) |
58 ± 6 | 46 ± 17 | 52 ± 17 |
| Depolymerization rate (nm/s) |
157 ± 49 | 248 ± 73 | NA |
NA, not applicable. Averages ± SD. ParM rates in vitro are from Garner et al. (2004). ParM rates in vivo were measured from data such as those shown in Fig. 2. Plasmid segregation rates were measured from clear segregation events such as those seen in Fig. 1 (E and F). ParM polymerization, n = 22; ParM depolymerization, n = 9; plasmid segregation, n = 22.
The depolymerization rate of ParM spindles is variable but generally faster than that measured for single ParM filaments in vitro (Table I). This may be explained by the fact that the ParR–parC complex prevents catastrophe when filament polymerization stalls (Fig. 2 A, 50–80 s), allowing hydrolysis of more ATP in the filaments. When catastrophe eventually occurs, depolymerization proceeds more quickly because more of the polymer is in the unstable ADP-bound form.
Discussion
This study provides three basic insights into type II plasmid segregation. First, ParM filaments are dynamically unstable in vivo. When both plasmid-bound ends of a bipolar ParM spindle reach the poles, elongation stalls. After a few seconds, the spindle either breaks or dissociates from one of the plasmids, becomes unstable, and falls apart. We observed this directly by imaging fluorescent ParM spindles and indirectly by observing the behavior of the plasmids. After segregation, plasmids do not remain fixed at the poles, indicating that the spindle that pinned them to the poles has disassembled and that they are not anchored to polar landmarks in the host cell. Second, when only one plasmid is present, filaments are bound and stabilized at only one end. The other, unstable end is presumably competent to interact with a second plasmid and may mediate search and capture as observed in vitro (Garner et al., 2007). These monovalent attachments also drive plasmids in a diffusive random walk through the cytoplasm at rates greater than diffusion in the absence of the par operon. This may further increase the efficiency of the search for sister plasmids. Third, in vivo, ParM spindles can be composed of multiple filaments. Spindles reconstituted in vitro contain many filaments, but this is at least partially the result of having many copies of the parC DNA immobilized on a single bead (Garner et al., 2007). These results suggest that either a single ParR–parC complex may interact with multiple ParM filaments or multiple pairs of plasmids can cooperate to form a spindle.
The results from this study also confirm many conclusions based on in vitro experiments. Most notably, the behavior of filaments in cells is highly reminiscent of the dynamic instability of filaments observed in vitro (Garner et al., 2004). In both cases, filaments were short lived, and recovery from catastrophic depolymerization is not observed. Additionally, the binding of plasmids to each end of a spindle as it elongates is reminiscent of the in vitro reconstitution of the system using polystyrene beads (Garner et al., 2007). The rates of polymerization at steady state for single ParM filaments or ParR–parC-associated filament bundles in vitro were very similar to those measured in vivo. The similarities between the in vivo observations and the reconstitution with purified components strongly suggest that host factors are not required for segregation and that the conditions used for the in vitro experiments more or less mimic cellular conditions.
An important implication of the frequent directional movements of par-containing plasmids is that unlike previously observed mechanisms of DNA segregation (Mitchison and Salmon, 2001; Sherratt, 2003), type II plasmid segregation is a dynamic process that continues throughout the cell cycle. Plasmids are biased toward the ends of a cell and generally away from the plane of division rather than being immobilized at the poles. The apparent inefficiency of this system may reflect the small number of components used to construct the segregation machinery and the fact that it has not evolved to use host cell factors for anchoring to the poles. In fact, the broad host range of par-containing plasmids may make it impractical to rely on any specific host cell factor. The dynamics of segregation provide a potential mechanism for correcting mistakes. If one attempt at segregation fails and both plasmids end up on the same side of the division plane, the system can try again. Our observation that segregation is not coordinated with the cell cycle agrees with previous observations that R1 plasmid replication is uncoupled from chromosome replication (Gustafsson et al., 1978).
Our model for in vivo plasmid segregation (Fig. 4) highlights two important and unanswered questions regarding the interaction between ParM filaments and the ParR–parC complex. First, if the ParM-binding sites on the ParR–parC complex are occupied by the ends of searching filaments, how can they capture the ends of filaments attached to a different plasmid? We propose two possibilities: (1) free ends of monovalently attached filaments could anneal, creating a single stabilized filament, or (2) at a given time, only a fraction of filament-binding sites may be occupied by mono-attached filaments, leaving the rest available for capture. Another question concerning the filament–plasmid interaction is how a single complex binds and stabilizes the two structurally distinct filament ends. By electron microscopy, ParM filaments are composed of two parallel and polarized protofilaments, which create two completely different ends (van den Ent et al., 2002; Orlova et al., 2007). We previously showed that both ends of an individual ParM filament can bind to the ParR–parC complex (Garner et al., 2007). From a structural perspective, it is difficult to imagine the ParR–parC complex interacting with two different surfaces in the exact same manner. Either the ParR–parC complex contains two distinct ParM-binding sites, one specific for each end, or most of the molecular contacts are with the side of the filaments.
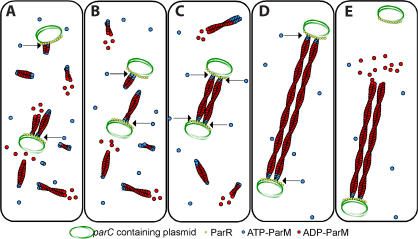
Figure 4.
Molecular model of plasmid segregation by the R1 par operon. (A) Nucleation of new filaments will happen throughout the cell. Filaments attached to one plasmid will search for a second plasmid. (B) Plasmids will diffuse around the cell until they get close enough to encounter each other. (C) When two plasmids come within close proximity, filaments will be bound at each end by a plasmid, forming a spindle. This will prevent the filaments from undergoing catastrophe. (D) As these stabilized filaments polymerize, the two plasmids will be forced to opposite poles. If the ends of a spindle run into the sides of the cell, it will be followed along the membrane to the ends of the cell. (E) After reaching a pole, pushing against both ends of the cell causes the filament to dissociate from the plasmid at one end and quickly depolymerize.
Materials and methods
Construction of bacterial strains and plasmids
Splicing by overlapping extension PCR was used to make a GFP fusion to the ParM gene from plasmid R1-19 and was inserted into the NDE1 and BamH1 sites of pET11a (New England Biolabs, Inc.) to create pCC110. pCC121 was made by using splicing by overlapping extension PCR to fuse mCherry to ParM and inserting into the Xba1 and Sal1 sites of the CRIM vector pTB97 (containing a phage HK022 att site). This was then inserted into the E. coli strain TB20 (MG1655 Δlac) and checked for single integrants as previously described (Haldimann and Wanner, 2001) to create the strain CC1.
Growth and expression
All cultures were grown in Luria-Bertani (LB) media with the appropriate antibiotics at the following concentrations: 100 ng/ml ampicillin, 20 ng/ml kanamycin, and 68 ng/ml chloramphenicol. For expression of GFP-ParM, pCC110 was transformed into BL21 cells with pLysS (Studier et al., 1990) and pRBJ460 (Jensen and Gerdes, 1999). pLysS is required to turn off the expression of GFP-ParM. Cells were grown to an OD600 of 0.5 and induced with 3 mM IPTG for 1.5 h. Cells were then centrifuged, resuspended in the same volume of fresh LB, and diluted 1:100 in LB with kanamycin and chloramphenicol. After 2 h of outgrowth, samples were removed for microscopy. For plasmid visualization, pRBJ460 or pRBJ461 (Jensen and Gerdes, 1999) were cotransformed with pJMJ178 (Moller-Jensen et al., 2003) into the strain MG1655. Plasmids pRBJ460 and pRBJ461 are mini–F plasmids to which 256 lacO repeats and a portion of the R1 plasmid have been added. pRBJ460 contains the R1 par operon, and pRBJ461 contains the hok/sok system. For dual visualization of plasmids and filaments, pRBJ460 and pJMJ178 were cotransformed into CC1. Cultures were grown overnight in chloramphenicol and kanamycin. For cell fixation, 2× fixing solution (60 mM phosphate, pH 7.0, 4.8% PFA, and 0.08% gluteraldehyde) was mixed 1:1 with a bacterial culture and incubated at room temperature for 10 min and then at 4°C for at least 1 h. For microscopy, 1.5 μl of cells were placed on top of 70 μl M9 media (plus 0.2% glucose) agar (1.5%) pads and sealed with a 1:1:1 (by weight) mixture of petroleum jelly (Vaseline), lanalin, and parafin.
Microscopy
Three microscopes were used in this study. The first was a Nikon TU300 equipped with a CCD camera (ORCA 2 ER; Hamamatsu) and has been described previously (Garner et al., 2007). The primary microscope that was used was a Nikon TE 2000 with a Perfect Focus module equipped with a CCD camera (Ixon; Andor) and an ORCA ER camera. An Apo total internal reflection fluorescence 100× 1.49 NA oil immersion objective (Nikon) was used. For some experiments, an objective heater (Bioptechs) and air stream stage incubator (Nevtek) were used to heat the samples to 37°C. Images were acquired with the open source μManager software version 1.0.60 (http://micro-manager.org). Photobleaching experiments were performed with a confocal microscope (LSM510; Carl Zeiss, Inc.).
Image analysis
ImageJ (National Institutes of Health) was used for length measurements, contrast adjustment, image rotation, and making kymographs. For diffusion rates, plasmid tracking was performed with MATLAB (MathWorks). Peaks in fluorescence were identified using the pkfnd module of E. Dufresne (Yale University, New Haven, CT). The addition of a Gaussian-fitting module to obtain subpixel resolution did not alter the results and was not included in the final build. To determine which foci were considered to be from the same plasmid from frame to frame, the track module by J.C. Crocker (University of Chicago, Chicago, IL) was implemented. MSDs for each track were calculated by averaging the squares of the displacement for all pairs of time points for each time interval.
Online supplemental material
Fig. S1 presents quantification of the intensity of filaments. Fig. S2 shows plasmid dynamics in a dividing cell. Fig. S3 shows plasmid dynamics in a cell containing four foci. Videos 1–10 correspond to Figs. 1 A, 1 E, 1 F, 1 G, 2 A, 2 C, 2 D, 2 E, 3 A, and 3 C. Table S1 presents quantification of plasmid loss rates, and Table S2 presents a distribution of the maximum number of foci per cell. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200708206/DC1.
Acknowledgments
We thank members of the Mullins laboratory for moral support and helpful advice; K. Gerdes, T. Bernhardt, and P. Chien for supplying plasmids and strains; and W. Marshall, L. Campbell, and the ever cromulent E. Garner for careful reading of the manuscript.
This work was supported by National Institutes of Health grants R01GM61010 and R01GM675287, the Sandler Family Supporting Foundation, the University of California San Fransisco/University of California Berkeley Nanomedicine Development Center, and a National Science Foundation Predoctoral Fellowship to C.S. Campbell.
Abbreviation used in this paper: MSD, mean squared displacement.
References
- Collins, J., and R.H. Pritchard. 1973. Relationship between chromosome replication and F'lac episome replication in Escherichia coli. J. Mol. Biol. 78:143–155. [DOI] [PubMed] [Google Scholar]
- Ebersbach, G., and K. Gerdes. 2005. Plasmid segregation mechanisms. Annu. Rev. Genet. 39:453–479. [DOI] [PubMed] [Google Scholar]
- Ebersbach, G., D.J. Sherratt, and K. Gerdes. 2005. Partition-associated incompatibility caused by random assortment of pure plasmid clusters. Mol. Microbiol. 56:1430–1440. [DOI] [PubMed] [Google Scholar]
- Elowitz, M.B., M.G. Surette, P.E. Wolf, J.B. Stock, and S. Leibler. 1999. Protein mobility in the cytoplasm of Escherichia coli. J. Bacteriol. 181:197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner, E.C., C.S. Campbell, and R.D. Mullins. 2004. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 306:1021–1025. [DOI] [PubMed] [Google Scholar]
- Garner, E.C., C.S. Campbell, D.B. Weibel, and R.D. Mullins. 2007. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 315:1270–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, K., and S. Molin. 1986. Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J. Mol. Biol. 190:269–279. [DOI] [PubMed] [Google Scholar]
- Gitai, Z., N. Dye, and L. Shapiro. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. USA. 101:8643–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding, I., and E.C. Cox. 2006. Physical nature of bacterial cytoplasm. Phys. Rev. Lett. 96:098102. [DOI] [PubMed] [Google Scholar]
- Gustafsson, P., K. Nordstrom, and J.W. Perram. 1978. Selection and timing of replication of plasmids R1drd-19 and F'lac in Escherichia coli. Plasmid. 1:187–203. [DOI] [PubMed] [Google Scholar]
- Haldimann, A., and B.L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, S. 1953. Polarization optical studies of the mitotic spindle. I. The demonstration of spindle fibers in living cells. Chromosoma. 5:487–500. [DOI] [PubMed] [Google Scholar]
- Jensen, R.B., and K. Gerdes. 1999. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO J. 18:4076–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, R.B., R. Lurz, and K. Gerdes. 1998. Mechanism of DNA segregation in prokaryotes: replicon pairing by parC of plasmid R1. Proc. Natl. Acad. Sci. USA. 95:8550–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., and S. Austin. 2002. The P1 plasmid is segregated to daughter cells by a ‘capture and ejection’ mechanism coordinated with Escherichia coli cell division. Mol. Microbiol. 46:63–74. [DOI] [PubMed] [Google Scholar]
- Mitchison, T.J., and E.D. Salmon. 2001. Mitosis: a history of division. Nat. Cell Biol. 3:E17–E21. [DOI] [PubMed] [Google Scholar]
- Moller-Jensen, J., R.B. Jensen, J. Lowe, and K. Gerdes. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21:3119–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller-Jensen, J., J. Borch, M. Dam, R.B. Jensen, P. Roepstorff, and K. Gerdes. 2003. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol. Cell. 12:1477–1487. [DOI] [PubMed] [Google Scholar]
- Orlova, A., E.C. Garner, V.E. Galkin, J. Heuser, R.D. Mullins, and E.H. Egelman. 2007. The structure of bacterial ParM filaments. Nat. Struct. Mol. Biol. 14:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano, J., T.Q. Ho, Z. Zhong, and D.R. Helinski. 2001. Multicopy plasmids are clustered and localized in Escherichia coli. Proc. Natl. Acad. Sci. USA. 98:4486–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., E.A. Davison, L.C. Jensen, L. Cassimeris, and E.D. Salmon. 1986. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 103:581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt, D.J. 2003. Bacterial chromosome dynamics. Science. 301:780–785. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., A.S. Belmont, C.C. Robinett, and A.W. Murray. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599–1608. [DOI] [PubMed] [Google Scholar]
- Studier, F.W., A.H. Rosenberg, J.J. Dunn, and J.W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60–89. [DOI] [PubMed] [Google Scholar]
- van den Ent, F., J. Moller-Jensen, L.A. Amos, K. Gerdes, and J. Lowe. 2002. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 21:6935–6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitao, T., S. Dasgupta, and K. Nordstrom. 2000. Plasmid R1 is present as clusters in the cells of Escherichia coli. Plasmid. 43:200–204. [DOI] [PubMed] [Google Scholar]
- Westphal, M., A. Jungbluth, M. Heidecker, B. Muhlbauer, C. Heizer, J.M. Schwartz, G. Marriott, and G. Gerisch. 1997. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr. Biol. 7:176–183. [DOI] [PubMed] [Google Scholar]