Abstract
The ras proto-oncogenes, of which there are four isoforms, are molecular switches that function in signal transduction pathways to control cell differentiation, proliferation, and survival. How the Ras isoforms orchestrate cellular processes that affect behavior is poorly understood. Further, why cells express two or more Ras isoforms is unknown. Here, using a genetically defined system, we show that the presence of both wild-type KRas and NRas isoforms is required for transformation because they perform distinct nonoverlapping functions: wild-type NRas regulates adhesion, and KRas coordinates motility. Remarkably, we find that Ras isoforms achieve functional specificity by engaging different signaling pathways to affect the same cellular processes, thereby coordinating cellular outcome. Although we find that signaling from both isoforms intersects in actin and microtubule cytoskeletons, our results suggest that KRas signals through Akt and Cdc42 while NRas signals through Raf and RhoA. Our analyses suggest a previously unappreciated convergence of different Ras isoforms on the dynamics of the processes involved in transformation.
Mammalian cells express three highly homologous ras proto-oncogenes that encode HRas, KRas4a, KRas4b, and NRas that affect cell proliferation, differentiation, and survival (31, 33). The different Ras isoforms are ubiquitously expressed, although the ratio between isoforms varies from tissue to tissue. Ras proteins affect signaling by toggling between an active GTP-bound and an inactive GDP-bound state largely in response to extracellular cues. Among the best-characterized signaling pathways affected by Ras are the Raf/MEK/mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K)/Akt pathways (32). Ras has also been shown to affect signaling by Rho family GTPases RhoA, Rac, and Cdc42 (34, 46, 48, 53). Despite the high degree of homology between the Ras isoforms, differences at their C termini have been shown to result in differential subcellular localization, with consequent differential signaling (22, 33).
Activating mutations in the Ras isoforms that render them constitutively active are observed in about a third of all human cancers. Depending on tissue type, there appears to be a preference for a particular Ras isoform that is mutated (30). For example, activating mutations in KRas are observed in greater than 90% of pancreatic cancers (5). Though there is some evidence that the different Ras isoforms possess differing oncogenic potential as a function of cell type, the reason for tissue-specific preference for activating mutation in a particular Ras isoform is unknown.
In contrast to our knowledge about the requirement for oncogenic Ras in neoplastic transformation, relatively little is known about the biological function of the wild-type Ras isoforms. Further, why cells express two or more Ras isoforms is unknown. Efforts to bridge this gap have been initiated through the development of mice lacking different ras alleles. Mice nullizygous for Hras or Nras, or both genes, are developmentally normal (12, 52). By contrast, Kras−/− mice die midgestation with evidence of a defect in fetal liver development (25, 26). Characterization of mouse embryo fibroblasts (MEFs) lacking Kras or Nras has revealed differences in steady-state signaling (29, 56). Like their mutant, activated counterparts, there is evidence that wild-type Ras also contributes to transformation (42). Loss of Kras or Nras has been shown to suppress chemically induced lung tumors or lymphomas, respectively (11, 58), and in tumor-prone mice heterozygous for the retinoblastoma gene, Rb, loss of Nras or Kras has been shown to suppress, promote, or have no impact on tumor progression as a function of cell type (50, 51). However, the cellular processes affected by wild-type ras deficiency to elicit an impact on tumorigenesis are unknown.
Here we present a functional and biological dissection of the contribution of Kras and Nras to transformation of MEFs using a genetically defined system. We find that Kras and Nras are uniquely required for transformation. NRas regulates cell adhesion, whereas KRas coordinates motility. Although signaling from both Ras isoforms intersects in the actin and microtubule cytoskeletons, our findings suggest that KRas signals through Akt and Cdc42, while NRas signals through Raf and RhoA to effect cellular outcome. Collectively, our findings suggest the convergence of signaling by different Ras isoforms on the dynamics of the processes that contribute to cellular transformation.
MATERIALS AND METHODS
Tissue culture.
MEFs were prepared as described elsewhere (49) and maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 20% fetal bovine serum (FBS) and antibiotics. MCF-7 cells were grown in DMEM supplemented with 10% FBS and antibiotics. All MEFs were transduced at passage 3. The genotype sets were from the same littermate set(s). Cell growth curves were generated by plating 5,000 cells in triplicate in 24-well plates; at the indicated time points cells were counted using a Z1 Coulter particle counter (Beckman-Coulter). For the soft agar assay, cells (21,000/ml) suspended in DMEM containing 20% FBS were seeded in triplicate wells in 0.4% agar over an underlayer of 0.6% agar in six-well plates and fed every 2 days with DMEM containing 10% FBS. Cells were incubated for 3 weeks before scoring the colonies. Micrographs were taken on a Nikon XLS microscope. Where short hairpin RNAs (shRNAs) were employed, the degree of knockdown was assessed at the start and finish of each experiment, and no significant differences were noted. Soft agar assays were performed with at least seven independent isolates of MEFs. When pharmacological inhibition was used, cells starting on day 2 were fed every 2 days thereafter with fresh medium supplemented with U0126 (10 μM; Cell Signaling) or LY294002 (20 μM; Calbiochem) with 500 μl of medium, left on the top of the agar until the next feeding, while toxicity of the inhibitors was monitored with long-term clonogenicity assays. Briefly, single-cell suspensions were seeded in six-well plates and incubated at 37°C for 21 days in the presence or absence of inhibitors. Cells were then fixed with methanol, stained with 0.1% crystal violet solution, and photographed on a Nikon XLS microscope.
Vectors and infections.
Retroviral vectors encoding large T antigen (TAg; pBabe-Neo-TAg; gift from J. DeCaprio), NRas (pBabe-N-Ras [49]), and KRas4b (pBabe-K-Ras [50]) were used to infect MEFs. A palmitoylation-defective mutant of NRas, C 181 to S, NRas181S, was generated in pBabe-GFP (bicistronic; gift from J. Boehm) by site-directed mutagenesis (Stratagene) according to the manufacturer's recommendations and confirmed by sequencing. Wild-type PTEN was excised from pWZL-PTEN (gift from L. Garraway) and subcloned into pBabe-GFP, a bicistronic vector. Retroviruses were generated as previously described (49). Infections were performed serially by using drug selection or fluorescence-activated cell sorting to purify cell populations 48 h after infection. The drug concentrations used were as follows: neomycin (G418), 400 μg/ml; puromycin, 3 μg/ml. All shRNAs, cloned into the lentiviral vector pLKO-puro, were obtained from the TRC Consortium (Broad Institute, Cambridge, MA), and the sequences are available upon request. To avoid off-target effects, infection with five different specific shRNAs for each gene was performed with consistent results. Virus directing the expression of shRNA to green fluorescent protein (GFP) was included in all experiments employing shRNAs as control and gave the same result as vector-infected cells; in some figures these data are not shown. Lentiviruses were generated (4) and used to infect MEFs.
Migration and invasion assays.
Migration and invasion assays were performed by using BD Biocoat migration and invasion chambers (Corning Costar Corp.) according to the manufacturer's guidelines. FBS (10%) was used as a chemoattractant in the lower chamber. Cells (21,000/ml) were suspended in the top chamber in serum-free medium containing 0.1% bovine serum albumin. After 48 h, the cells that had invaded the Matrigel or migrated through the control inserts were stained with Giemsa and counted under the microscope from a total of 10 regions on the filter, and the cell number/cm2 was calculated.
Immunoblotting.
Cells were lysed in RIPA buffer (50 mM Tris HCl, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 0.2 mM phenylmethylsulfonyl fluoride, and 2 mM sodium orthovanadate) supplemented with protease inhibitors cocktail (Roche). Total cell lysates (40 μg) were resolved by SDS-polyacrylamide gel electrophoresis, transferred to Immobilon membranes, and immunoblotted with the following primary antibodies: anti-simian virus 40 (SV40) TAg (Pab 101), anti-KRas (F234), anti-HRas (F235), anti-NRas (F155), anti-Cdc42 (B-8), anti-RhoA (26C4), and anti-phospho-cofilin (sc-21867-R) were from Santa Cruz Biotechnology; anti-c-Raf (9422), anti-phospho-Akt (Ser473; 9271), anti-Akt (9272), anti-phospho-p44/42 MAPK (ERK1/2) (9101), anti-p44/42 MAPK (ERK1/2) (9102), anti-phospho-MEK1/2 (MEK1 and -2; 9121), anti-MEK1/2 (9122), anti-phosho-GSK-3β (9336S), anti-FAK (3285), and anti-PTEN (9552) were from Cell Signaling; anti-pan-Ras (Calbiochem), anti-Rac1 (clone 23A8; Upstate/Millipore), anti-GSK-3β (clone 4G; Upstate/Millipore), anti-FAK-Y397 (44-624G; BioSource), anti-Glu tubulin (Chemicon), and antitubulin (Sigma) were also used.
Indirect immunofluorescence microscopy.
Cells were grown on eight-well slides, fixed with 3.7% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min, and then permeabilized with 0.5% Triton X-100 for 10 min, washed three times in PBS, and blocked with 1% FBS followed by staining with the primary antibody. Focal adhesions were stained with antivinculin antibody (hVIN-1; Sigma) and detected with Alexa Fluor 488 anti-mouse immunoglobulin G (Molecular Probes). Filamentous actin was visualized with fluorescein isothiocyanate (FITC)-phalloidin (Sigma). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). Micrographs were taken on a Nikon XLS microscope equipped with a charged-coupled camera and analyzed using Adobe Photoshop.
Rac, Cdc42, and RhoA activation assays.
Activation of Rac (Rac-GTP) and Cdc42 (Cdc42-GTP) was analyzed by glutathione S-transferase (GST)-PAK1-binding domain (GST-PBD) pull-down assays, essentially as described elsewhere (3). Briefly, cells were serum starved for 48 h and treated with epidermal growth factor (50 ng/ml) for 15 min, and then cell monolayers were washed twice with PBS and lysed for 5 min in GTPase activation buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% [vol/vol] Triton X-100, 10 mM MgCl2, 10% [vol/vol] glycerol, Complete protease inhibitor cocktail [Roche], 5 mM sodium fluoride, and 1 mM sodium orthovanadate). Approximately 20 μg of GST-PBD fusion protein (bound to glutathione beads; GE Healthcare/Amersham Biosciences) was added to the lysate, and the mixture was rotated for 1 h at 4°C. The beads were washed three times and eluted with SDS sample buffer. Bound proteins were resolved by SDS-polyacrylamide gel electrophoresis, transferred to an Immobilon membrane, and probed with antibodies to Rac (clone 23A8) or Cdc42 (polyclonal) from Upstate/Millipore. Equivalent amounts of each lysate were removed prior to GST-PBD addition and analyzed by immunoblotting to normalize total Rac or Cdc42 levels. A similar assay, using the Rho binding domain (RBD) of Rhotekin (43) as a GST fusion protein (Upstate/Millipore), was used to measure RhoA activation (RhoA-GTP). RhoA that bound the GST-RBD beads was detected as described above by immunoblotting with an antibody against RhoA (26C4; Santa Cruz).
Statistical analysis.
All P values were calculated using Student's t test (unpaired, two-tailed).
RESULTS
Wild-type Nras and Kras are required during transformation in MEFs.
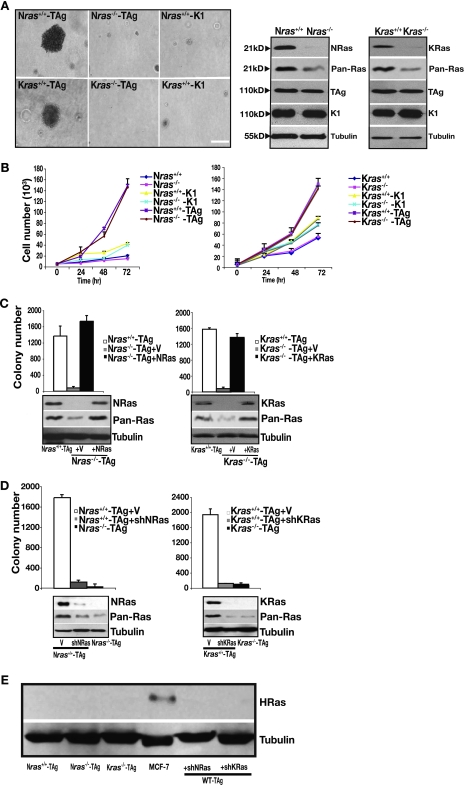
To assess the requirement of wild-type ras isoforms for transformation, we used early-passage primary MEFs derived from wild-type, Nras, and Kras knockout mice (25, 52). SV40 TAg was used as a driving oncogene to inactivate the pRb and p53 tumor suppressor pathways (1). Mutant versions of TAg (1), capable of only inactivating p53 (K1 mutant) or pRb (Δ434-444 mutant) (data not shown) served as negative controls. Wild-type MEFs were able to grow in an anchorage-independent manner as measured by soft agar assay following TAg transduction (Fig. 1A). By contrast, nullizygosity for either Kras or Nras prevented anchorage-independent growth. This occurred despite comparable levels of TAg expression in Nras−/−, Kras−/−, and wild-type MEFs (Fig. 1A), no signs of cell proliferation defects (Fig. 1B), and the presence of active GTP-bound Ras (data not shown). To rule out events secondary to loss of ras, we tested whether reconstitution of ras-deficient MEFs with their missing Ras isoform restored growth in soft agar. Reconstitution of wild-type NRas at physiological levels—in order to avoid upsetting the delicate balance between the steady-state signaling pathways—enabled Nras-deficient MEFs to form colonies in soft agar (Fig. 1C). Similarly, reconstitution of KRas4b (hereafter KRas) at physiological levels restored anchorage-independent growth in Kras-deficient cells. Further, acute and stable loss of either KRas or NRas by using shRNAs—five different gene-specific hairpins were used each time in order to exclude off-target effects—in wild-type MEFs inhibited their growth in soft agar (Fig. 1D), thereby recapitulating the outcome seen with chronic loss of Ras in ras-deficient cells (Fig. 1A). We have not detected HRas in murine MEFs (Fig. 1E), consistent with previous observations (57). Together, these observations suggest the presence of both wild-type NRas and KRas as a prerequisite for transformation of MEFs.
FIG. 1.
Kras and Nras-deficient MEFs are resistant to SV40-induced transformation. (A) MEFs of the indicated genotype were transduced with TAg or the K1 mutant and subjected to a soft agar assay. Shown are representative phase-contrast micrographs of their growth in soft agar. Expression levels of Ras isoforms, total Ras (Pan-Ras), and wild-type and mutant TAg were assessed by immunoblotting, with tubulin as a loading control. Bar, 10 μm. (B) Cell proliferation of parental and TAg- or K1 mutant-transduced MEFs of the indicated genotype. For each cell line, 5,000 cells were plated at t = 0 and counted at the indicated times. Each time point is represented as the mean ± standard deviation (SD) of triplicate determinations. (C) Anchorage-independent growth of Nras-deficient cells reconstituted with NRas and of Kras-deficient cells reconstituted with KRas4b (indicated as KRas) or vector (V) infected. Results are means ± SDs of five independent experiments. Expression levels of each Ras isoform and total Ras were assessed by immunoblotting to confirm physiological levels, with tubulin serving as a loading control (bottom panel). (D) Same experiment as in panel C, except wild-type cells following knockdown of either NRas or KRas (shRNAs directed to the genes encoding NRas and KRas are indicated by shNRas and shKRas, respectively) or infected with vector (V) were analyzed. Results are means ± SDs of five independent experiments. Protein knockdown and total Ras levels were determined by immunoblotting, with tubulin serving as a loading control (bottom panel). (E) Immunoblot analysis for HRas in the indicated cells. MCF-7 cells served as a positive control for HRas expression, and tubulin served as a loading control.
Wild-type Ras isoforms perform unique functions during transformation in MEFs.
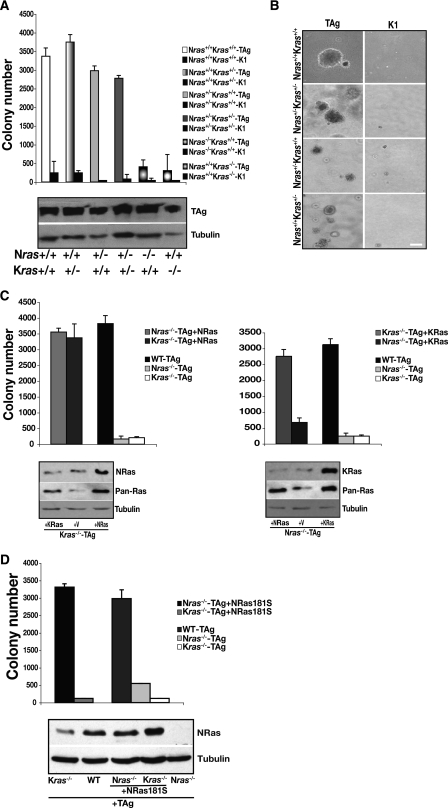
Requirement of both wild-type isoforms during transformation of MEFs could be attributable either to a threshold in ras gene dosage or to their functional uniqueness. To distinguish between these scenarios, we tested the minimum number of ras alleles that TAg-transduced heterozygous MEFs required in order to grow in soft agar. Heterozygous MEFs possessing any combination of three ras alleles were able to grow in an anchorage-independent manner (Fig. 2A), but that was not the minimum allele requirement: MEFs bearing only two ras alleles, one Nras and one Kras, were also fully competent for transformation (Fig. 2A and B). This genetic evidence suggests that both KRas and NRas isoforms perform distinct and essential biological functions during transformation.
FIG. 2.
Wild-type Kras and Nras perform unique functions during transformation. (A) MEFs of the indicated genotypes were transduced with TAg or the K1 mutant and subjected to soft agar assay. Results are means ± standard deviations (SDs) of five independent experiments. TAg expression was assessed by immunoblotting, with tubulin serving as a loading control (bottom panel). (B) Representative phase-contrast micrographs showing growth in soft agar of MEFs of the indicated genotypes transduced with TAg or the K1 mutant. (C) Anchorage-independent growth following expression of NRas in Nras- or Kras-deficient MEFs (left) and following expression of KRas4b (indicated as KRas) in Kras- or Nras-deficient cells (right). Results are means ± standard deviations (SDs) of five independent experiments. Expression of each Ras isoform and total Ras (Pan-Ras) in the indicated cells were assessed by immunoblotting, with tubulin as a loading control (bottom panels). (D) Same experiment as in panel C, except a palmitoylation-defective mutant of NRas, NRas181S, was expressed in Nras−/− and Kras−/− MEFs. Results are means ± SDs of three independent experiments. Expression of NRas181S in the indicated cells, with tubulin used as a loading control, was assessed by immunoblotting (bottom panel).
To explore the unique requirement for Nras and Kras during transformation further, we asked whether “cross-reconstitution” of ras-deficient MEFs restored transformation. Expression of KRas in Nras-deficient cells did not restore anchorage-independent growth, while expression of NRas in Kras−/− MEFs cells did allow these cells to grow in soft agar (Fig. 2C). Our genetic analysis (Fig. 2A) suggests that the ability of NRas to restore transformation in Kras-deficient cells was not due to gene dosage. We predicted, rather, that it might be due to subcellular compartmentalization of the different Ras isoforms. To address this possibility, we made use of the observations that while palmitoylation of NRas and HRas is required for their localization to the plasma membrane (23), a palmitolyation-defective mutant of HRas is biologically active, due to its ability to signal from the endoplasmic reticulum and Golgi apparatus (7). In contrast, KRas is thought to signal predominantly from the plasma membrane (8, 22). Expression of a palmitoylation-defective mutant of NRas, NRas181S, restored anchorage-independent growth in the Nras-deficient MEFs (Fig. 2D), suggesting that the ability of Ras to signal from the endoplasmic reticulum and Golgi complex participates in transformation. By contrast, expression of NRas181S did not enable Kras−/− MEFs to grow in soft agar, suggesting that signaling from the plasma membrane also contributes to transformation. These observations are consistent with the possibility that transformation of MEFs requires Ras isoform-dependent signaling events emanating from both the plasma membrane and endomembranes.
Wild-type Ras isoforms differentially affect cell migration and the actin cytoskeleton in transformed MEFs.
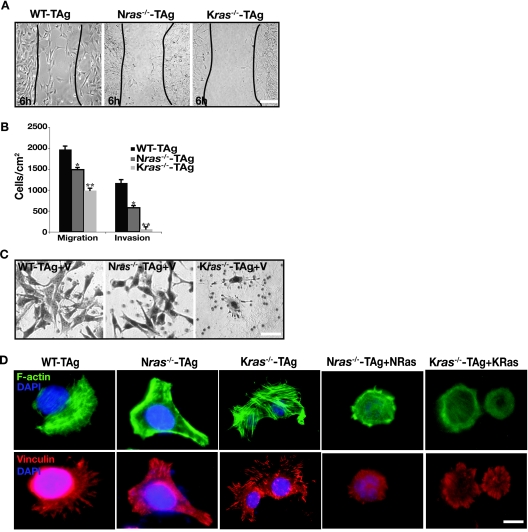
We next examined whether a deficiency in ras impinges on any other functional hallmarks of transformed cells in addition to anchorage-independent growth, such as cell motility and invasiveness (15, 21). We assessed cell motility of wild-type and ras-deficient cells in two-dimensional (2D) substrata by in vitro healing assay and in 3D substrata by using Boyden chambers. In the in vitro healing assay, wild-type and Nras-deficient cells started to heal the wound at 6 h postscratching and finished the healing at 12 h, whereas Kras-deficient cells started at 12 h postscratching and completed the process at 24 h (Fig. 3A). Further, Nras-deficient MEFs migrated through Boyden chambers 23% slower than their wild-type counterparts (Fig. 3B), and Kras−/− cells showed a 50% decrease in their migration speed. We also evaluated the invasive capacity of MEFs in Boyden chambers coated with a cell-derived extracellular matrix, Matrigel (Fig. 3B). When compared to wild-type cells, invasion of Nras-deficient cells through Matrigel was decreased by 50%, whereas the invasive capacity of Kras-deficient cells was severely impaired by 94% (Fig. 3B).
FIG. 3.
Ras deficiency affects cell migration and cytoskeletal dynamics. (A) Representative phase-contrast micrographs of transformed MEFs of the indicated genotypes 6 h after wounding. Similar results were obtained with five independent isolates of MEFs. (B) Migration and invasion assays of wild-type and ras isoform-deficient cells performed as described in Materials and Methods. Results are means ± standard deviations of three independent experiments performed in triplicate. Similar results were obtained with five independent isolates of MEFs. *, P < 0.05; **, P < 0.002. (C) Wild-type, Nras−/−, and Kras−/− cells show plasticity of migration. Representative micrographs are shown to demonstrate that wild-type and ras-deficient MEFs adopt different modes (elongated or rounded) of motility in 3D matrices, such as Matrigel. Similar results were obtained with five independent isolates of MEFs. Bar, 10 μm. (D) Immunofluorescence analysis of actin fibers and focal adhesions in wild-type and ras-deficient cells and in Nras−/− cells reconstituted with NRas and Kras−/− MEFs reconstituted with KRas4b. Shown is FITC-phalloidin staining (green) for filamentous actin, vinculin staining (red) for focal adhesions, and DAPI staining (blue) for nuclei in wild-type cells and ras-deficient cells. Similar results were obtained with five independent isolates of MEFs. Bar, 10 μm.
Our analysis of cell invasion also revealed the importance of Ras isoforms in controlling the plasticity of migration (55): wild-type and Nras-deficient cells invaded the 3D Matrigel by adopting an elongated morphology, whereas the very few invading Kras-deficient cells were of rounded morphology (Fig. 3C). Furthermore, this migratory behavior was reflected in the remodeling of actin cytoskeleton and redistribution of focal adhesion sites of integrin binding to the extracellular matrix (9, 16, 17). This was shown by indirect immunofluorescence to detect filamentous actin and vinculin-containing focal complexes. Wild-type cells displayed rounded morphology with their actin stress fibers distributed throughout the cell, as did Nras- and Kras-deficient MEFs reconstituted with NRas and KRas, respectively (Fig. 3D). Unlike wild-type cells, Nras-deficient cells exhibited polygonal morphology with intense cortical actin staining and lack of centrally located actin bundles, whereas Kras-deficient cells were roundish in shape with well-organized bundles of stress fibers throughout the cell (Fig. 3D). Despite similar amounts of cell surface β1 integrin expression (data not shown), the distribution of vinculin-containing focal complexes was also distinct. Wild-type cells had elongated peripheral focal adhesions. Nras-deficient MEFs possessed elongated focal adhesions toward, presumably, the leading edge of the cell and small intracellular focal contacts at the trailing cell edges (Fig. 3D). Kras-deficient cells lacked large peripheral vinculin-containing focal complexes; instead, they had small intracellular focal contacts distributed throughout the cell (Fig. 3D). Combined with the migration assay outcome, these results indicate that wild-type cells have low polarity, cell tension, and adhesiveness to the substratum, while Nras-deficient cells are more polarized with low adhesion strength and Kras-deficient cells are nonpolarized with increased tension and adhesion strength (18, 47). Our findings suggest that wild-type NRas and KRas differentially affect key features of transformation in MEFs, such as adhesion, cell migration, and the actin cytoskeleton.
Selective knockdown of Ras effectors rescues transformation of ras isoform-deficient MEFs.
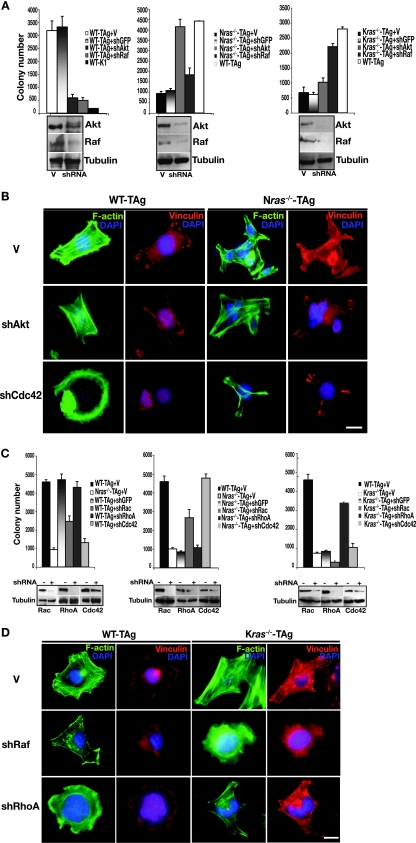
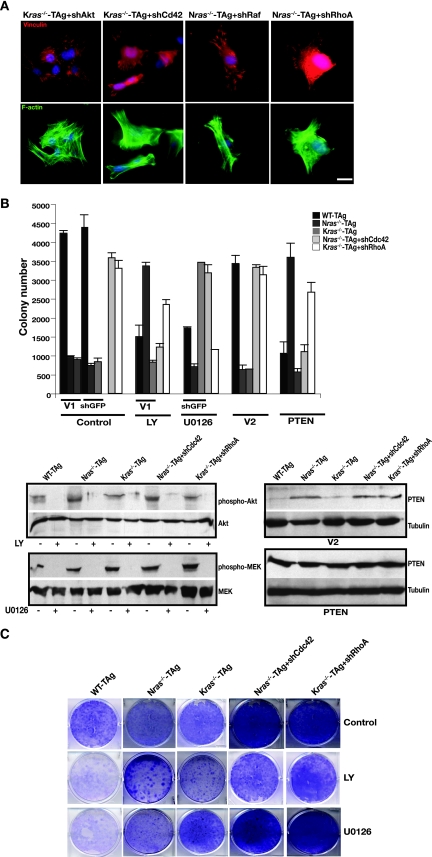
Next, we sought to unravel isoform-specific differences in inhibition of transformation seen in ras-deficient cells (Fig. 1 and 2) and, presumably, how Ras isoforms acquire biological specificity during transformation of wild-type cells. It has been suggested that the ability of Ras to signal from different subcellular locations may effect kinetically different outputs or allow activation of distinct effectors such as PI3K or Raf (41). We hypothesized that in the absence of a given Ras isoform that the remaining Ras isoform might impose a block to transformation by affecting specific signaling pathways. To address this possibility, we knocked down well-defined components of Ras effector pathways: Akt1 (which operates downstream of PI3K), and c-Raf by using gene-specific shRNAs. Depletion of either Akt1 or c-Raf (hereafter Akt and Raf, respectively) in wild-type cells inhibited their anchorage-independent growth (Fig. 4A). Similar treatment of MEFs bearing one Kras and one Nras allele (the minimum requirements to allow anchorage-independent growth in MEFs [Fig. 2]), also blocked their growth in soft agar (data not shown). However, inhibition of the same pathways had a drastically different outcome in ras-deficient cells: knockdown of Akt allowed anchorage-independent growth only in Nras-deficient cells, while depletion of Raf permitted soft agar growth only in Kras-deficient cells (Fig. 4A). In wild-type and ras-deficient MEFs knockdown of Akt resulted in attenuation of GSK-3β phosphorylation at serine 9, and depletion of Raf diminished ERK1 and ERK2 phosphorylation (Fig. 4E). These results reveal that in ras-deficient cells, wild-type Ras isoforms critically affect transformation through distinct and separable signal transduction pathways. Consequently, biological outcome appears to be dependent on the presence of specific Ras isoforms: our data suggest that in Nras-deficient cells the PI3K/Akt pathway inhibits transformation, and in Kras-deficient cells the Raf pathway blocks transformation, while in wild-type cells both the PI3K and Raf pathways are required for transformation.
FIG. 4.
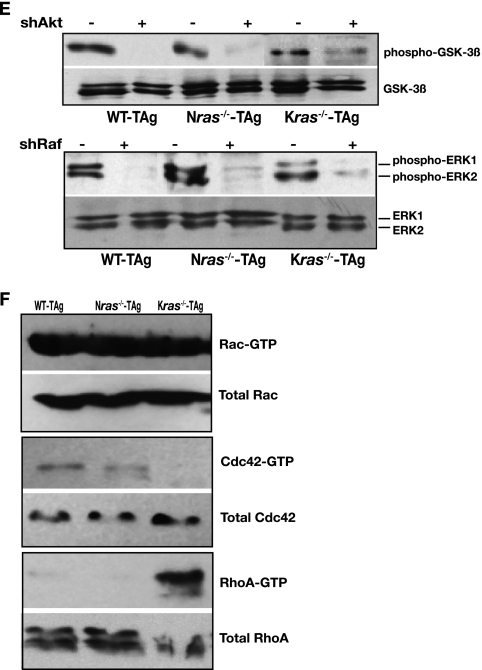
Effect of signaling components on the actin cytoskeleton, focal adhesions, and transformation. (A) Anchorage-independent growth of wild-type, Nras−/−, and Kras−/− MEFs following knockdown of Akt or Raf (shRNAs directed to the genes encoding Akt and Raf are indicated by shAkt and shRaf, respectively) or infected with empty vector (V) or vector directing the expression of an shRNA to green fluorescent protein (shGFP). Results are means ± standard deviations (SDs) of three independent experiments. Protein knockdown was confirmed by immunoblotting, with tubulin as a loading control (bottom panels). (B) Shown is FITC-phalloidin staining (green) for stress fibers, vinculin staining (red) for focal adhesions, and DAPI staining (blue) for nuclei in wild-type and Nras-deficient cells following knockdown of Akt or Cdc42. Bar, 10 μm. (C) Same experiment as in panel A, except cells of the indicated genotype following knockdown of small GTPases Rac, RhoA, or Cdc42 were analyzed (shRNAs directed to the genes encoding these Rho GTPases are denoted shRac, shRhoA, and shCdc42, respectively). Results are means ± SDs of three independent experiments. Depletion of proteins was confirmed by immunoblotting, with tubulin as a loading control (bottom panels). (D) Same experiment as in panel B, except wild-type and Kras-deficient cells following knockdown of Raf or RhoA were analyzed. (E) Immunoblotting of whole-cell lysates prepared from cells of the indicated genotype was performed with antibodies to phosphorylated GSK-3β and total GSK-3β, phosphorylated ERK1 and ERK2, and total ERK1 and ERK2. (F) In the indicated cells, the presence of activated, GTP-bound Rac, Cdc42, and RhoA was assayed as described in Materials and Methods. Whole-cell lysates were analyzed for total Rac, Cdc42, and RhoA as loading controls. Panels showing activated Rho GTPases are representative of three independent experiments, and loading controls are from the same lysates used for activation assays.
Knockdown of Akt and Raf expression was also accompanied by cytoskeletal reorganization. In wild-type cells, depletion of either Akt or Raf resulted in loss of centrally located bundled actin stress fibers and vinculin-staining focal adhesions at the cell edges, compared to empty vector-infected cells (Fig. 4B and D). In contrast, downregulation of Akt in Nras-deficient cells induced a wild-type-like phenotype, with the appearance of more stress fibers in the central area of the cell and prominent elongated focal complexes (Fig. 4B). Inhibition of Raf produced marked changes in Kras-deficient cells, including reduction of actin bundles and small focal adhesions distributed throughout the cell surface, thereby more closely resembling wild-type cells (Fig. 4D). Further, the above changes in cytoskeletal organization were accompanied by inhibition of migration in wild-type cells and an increase in migratory capacity of ras-deficient cells (data not shown). Adaptation of a wild-type-like phenotype by ras-deficient cells following inhibition of Akt or Raf suggests that in wild-type cells a balance exists between NRas and KRas signaling that is essential for transformation. It appears that the presence of each wild-type Ras isoform influences the balance between sessile and migratory behavior via modulation of actin cytoskeleton.
Cross talk between wild-type Ras isoforms and small Rho GTPases.
To gain further insight into how each Ras isoform makes a distinct and specific contribution to transformation in ras-deficient cells, and by inference in wild-type cells, we determined the relative contribution of each Rho family small GTPase to anchorage-independent growth of MEFs. Rho family GTPases are key mediators of actin dynamics and organization (2, 6, 36, 44, 45) (as depicted in Fig. 3 and 4) and have been shown to drive various modes of cell migration (55) (Fig. 3C). Further, focal adhesions seen in Kras-deficient cells were similar to those mediated by activated RhoA in adherent stationary cells (35, 36), while focal complexes of Nras-deficient cells were similar to those mediated by Cdc42 and Rac1 in actively migrating cells (35, 36) (Fig. 3D). Consistent with these observations, the levels of activated Cdc42 were comparable and highest in Nras-deficient and wild-type cells, while activated RhoA was highest in Kras−/− MEFs (Fig. 4F).
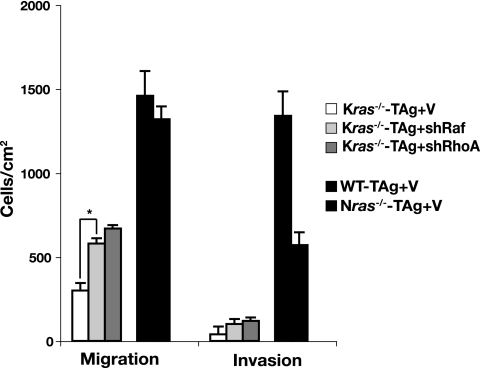
Knockdown of Cdc42 by using gene-specific shRNAs in wild-type cells blocked growth in soft agar and depletion of Rac inhibited transformation (∼50%), while RhoA depletion had no effect (Fig. 4C). Knockdown of Cdc42 was also accompanied by loss of stress fibers and disassembly of the focal complexes (Fig. 4B). Once again, Nras+/− Kras+/− MEFs behaved similar to wild-type cells following transduction with shRNAs targeting the genes encoding Cdc42, Rac, and RhoA (data not shown). By contrast, knockdown of Rho GTPases had opposite effects on ras-deficient MEFs, similar to the observed results with Akt and Raf: knockdown of Cdc42 permitted anchorage-independent growth in Nras-deficient cells and knockdown of Rac had a partial effect (Fig. 4C). Depletion of Cdc42 was accompanied by the appearance of bundled actin cables throughout the cell and elongated focal contacts at the cell edges (Fig. 4B). In Kras−/− cells, knockdown of RhoA was permissive for their growth in soft agar (Fig. 4D). These effects on anchorage-independent growth of Kras-deficient cells were paralleled by improved migration following knockdown of RhoA or Raf (Fig. 5). Increased invasion through Matrigel by Kras−/− MEFs correlated with a switch to an elongated morphology (data not shown), which coincided with a marked reduction in the centrally located bundles of stress fibers and small focal contacts (Fig. 4D). Altogether, our findings suggest that Akt, Raf, and Rho GTPases coordinately regulate motile behavior and adhesion to effect transformation, although their relative contribution to each process is dictated by the specific Ras isoform present.
FIG. 5.
Effect of knockdown of Raf or RhoA on the migration and invasion of Kras-deficient MEFs. Migration and invasion assays were performed with Kras−/− cells following knockdown of Raf or RhoA (shRNAs directed to genes encoding Raf and RhoA are indicated by shRaf and shRhoA, respectively) or infected with vector (V). Wild-type and Nras-deficient cells served as positive controls. Results are means ± standard deviations of two independent experiments performed in triplicate. *, P < 0.02.
We speculated that Akt and Cdc42 might operate through similar pathways and converge to remodel the actin cytoskeleton in Nras-deficient cells and likewise for Raf and RhoA in Kras-deficient MEFs. This was based on our observation that knockdown of Akt or Cdc42 not only preferably permitted transformation in Nras-deficient cells but also facilitated similar, specific changes in their actin cytoskeleton (Fig. 4 and 6A). Similarly, inhibition of Raf or RhoA allowed transformation only in Kras−/− cells by inducing specific actin remodeling accompanied by changes in cell motility (Fig. 5). To test this hypothesis, we inhibited the Akt and Raf pathways pharmacologically in Nras−/− MEFs depleted of Cdc42 and in Kras−/− MEFs depleted of RhoA. As predicted, the PI3K inhibitor LY294002 inhibited anchorage-independent growth of Nras-deficient cells in which Cdc42 was depleted, while having no impact on Kras−/− RhoA-depleted cells (Fig. 6B). Similarly, overexpression of PTEN, to antagonize the production of PIP3 by PI3K, blocked the growth of Nras−/− Cdc42-depleted cells in soft agar while having no effect on Kras-deficient cells in which RhoA was depleted (Fig. 6B). In contrast, inhibition of MEK with U0126 blocked anchorage-independent growth in Kras-deficient RhoA-depleted cells and had no effect on Nras−/− MEFs depleted of Cdc42 (Fig. 6B). Possible toxic effects of the inhibitors at the concentrations used were excluded by long-term clonogenicity assays (Fig. 6C). Together with the specific changes in the actin cytoskeleton noted above, these results can be reconciled by postulating that, in the presence of KRas, Akt and Cdc42 regulate polarity to affect cell migration, whereas in the presence of NRas alone, Raf and RhoA coordinate adhesion strength to promote motility.
FIG. 6.
Preferential contributions of downstream signaling effectors in ras isoform-deficient MEFs to the actin cytoskeleton, focal adhesions, and transformation. (A) Transformation of ras isoform-deficient cells correlates with specific actin remodeling. Shown is FITC-phalloidin staining (green) for actin, vinculin staining (red) for focal adhesions, and DAPI staining (blue) for nuclei in Kras-deficient MEFs following knockdown of Akt or Cdc42 (denoted shAkt and shCdc42, respectively) and in Nras-deficient cells depleted of Raf or RhoA (indicated by shRaf and shRhoA). Bar, 10 μm. (B) Effects of pharmacological inhibition of PI3K and Raf pathways and PTEN overexpression on the anchorage-independent growth of Nras-deficient Cdc42-depleted MEFs and Kras-deficient RhoA-depleted MEFs. Cells (genotype and expression of shRNA are indicated; V1 is vector) were subjected to a soft agar assay and subsequently treated with the PI3K inhibitor LY294002 (LY; 20 μM) and the MEK inhibitor U0126 (10 μM), as described in Materials and Methods. For PTEN overexpression, cells were infected with virus directing the expression of PTEN or vector (V2; pBabe-GFP), sorted, and subjected to a soft agar assay. Results are means ± standard deviations of two independent experiments. Total cell lysates were subjected to immunoblotting to verify the effectiveness of the pharmacological inhibitors (bottom left panel) and PTEN overexpression (bottom right panel). (C) Same analysis as in panel B, except long-term clonogenicity assays were performed as described in Materials and Methods.
Wild-type Ras isoforms coordinately regulate microtubule stability and actin polymerization during transformation.
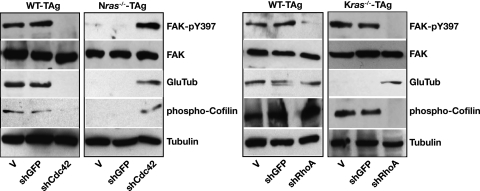
To elucidate further the downstream signaling events through which Ras isoforms afford specificity during transformation, we monitored the presence of phosphorylated FAK (FAK-pY397) (13, 54) upon focal adhesion assembly and integrin engagement, the presence of stabilized detyrosinated microtubules (38) (Glu MTs) for microtubule stability, and the phosphorylation of cofilin (24) for actin dynamics by immunoblotting. In wild-type cells, depletion of Cdc42 induced focal adhesion disassembly, destabilization of microtubules, and depolymerization of actin (Fig. 7A). In terms of RhoA, knockdown of this GTPase in wild-type cells caused focal adhesion disassembly but no changes in actin polymerization or microtubule stability (Fig. 7B). By contrast, Nras-deficient cells lacked phosphorylated FAK, Glu MTs, and phosphorylated cofilin. Knockdown of Cdc42 (or Akt [data not shown]), however, generated stabilized MTs coordinated with integrin engagement, focal adhesion assembly, and actin polymerization (Fig. 7A), similar to transformed wild-type cells. Glu MTs were also undetectable in Kras-deficient cells, but phosphorylated FAK and cofilin were detectable. Depletion of RhoA (or Raf [data not shown]) generated Glu MTs, focal adhesion disassembly, and reduced actin polymerization in Kras-deficient cells (Fig. 7B). Notably, microtubule stabilization was neither RhoA regulated nor integrin-FAK stimulated (13, 38, 54). Likely, it was either mDia stimulated (37) or due to other, yet-unidentified factors present in the serum of the growth medium (37, 38). Together these data suggest that, in the absence of a Ras isoform, KRas and NRas use distinct pathways to affect microtubule and actin cytoskeletons, while in wild-type cells they use a mechanistic interrelationship between these pathways to effect transformation.
FIG. 7.
Effect of knockdown of Cdc42 or RhoA on focal adhesion assembly, microtubule stability, and actin polymerization. (A) Immunoblotting of total cell lysates of wild-type and Nras-deficient cells following knockdown of Cdc42 (shCdc42) or GFP (shGFP) or infected with vector (V) with the indicated antibodies. (B) Same analysis as in panel A, except wild-type and Kras−/− cells were analyzed following knockdown of RhoA (shRhoA).
DISCUSSION
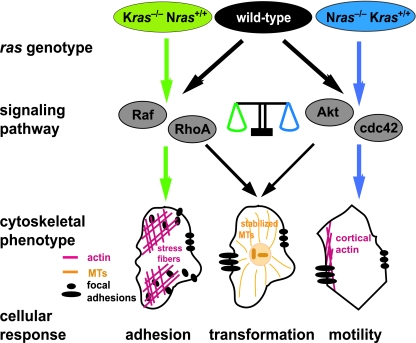
In order to understand how the Ras isoforms signal to effect biological outcome, we need to address the fundamental question of why mammalian cells express two or more isoforms. To approach this issue we asked whether Kras and Nras are required for cellular transformation and if so whether they perform unique functions. We have provided genetic evidence that both Kras and Nras perform essential and distinct functions during transformation. Our cell biological analyses suggest that NRas influences adhesion while KRas coordinates motility and that both isoforms regulate these cellular processes, in part, by affecting the actin and microtubule cytoskeletons. At the molecular level our findings suggest that KRas and NRas exert their influence on the cytoskeleton to effect migration, invasion, and anchorage-independent growth through distinct signaling pathways. KRas appears to signal through Akt and Cdc42, while NRas does so with Raf and RhoA. Together, the present evidence leads us to propose that in wild-type cells coordinate signaling from the different Ras isoforms through distinct effector pathways converges on the same cellular processes to effect biological outcome (Fig. 8).
FIG. 8.
Model illustrating a balance between Nras and Kras signaling necessary to effect transformation. In the absence of Kras, Nras signaling through Raf and RhoA effects excessive small focal adhesions and stress fiber formation (left; green arrows). In the absence of Nras, Kras signaling through Akt and Cdc42 effects cortical actin formation and more polarized focal contacts (right; blue arrows). In wild-type cells, a balance in signaling is achieved where both Nras and Kras affect the nature of the actin cytosketon and focal adhesions (center; black arrows). Similarly, neither Nras- nor Kras-deficient cells display stabilized microtubules; in wild-type cells a balance in signaling is achieved wherein Nras signaling through Raf and RhoA and Kras through Akt and Cdc42 effects the formation of stabilized microtubules, a scenario that correlates with transformation.
Though we have used an artificial system in which Kras and Nras are missing, we believe our results have bearing on the behavior and wiring of wild-type cells. An interpretation of our findings is that the behavior of Kras and Nras nullizygous cells represents extremes along a continuum where the Ras isoforms act together as a rheostat (Fig. 8). An alternative model to explain our findings might be that loss of Kras or Nras results in aberrant signaling and cellular behavior that are never encountered in a physiological or pathophysiological setting. We favor the former scenario for several reasons. The actin cytoskeleton and focal adhesions observed in Kras−/− and Nras−/− cells bear resemblance to those of wild-type cells in which activated forms of specific Rho GTPases have been introduced (Fig. 3D). Further, the disturbance in the balance in signaling resulting from loss of Kras or Nras can be restored, i.e., made more similar to wild-type cells, through knockdown of specific signaling components (Fig. 4). For example, knockdown of Cdc42 in Nras-deficient cells shifts the polarity of the actin cytoskeleton from high to low, thereby making these cells display an actin cytoskeleton more closely resembling that of wild-type cells. Similarly, Kras nullizygous cells display excess focal adhesion assembly, which imparts on these cells a block to invasion and reduced cell motility, consistent with the notion that KRas is the predominant Ras isoform that regulates cell migration (28). And, knockdown of RhoA in Kras−/− cells results in a shift to smaller focal contacts similar to that observed in wild-type cells. In each case the specific genetic manipulation also conferred upon ras isoform-deficient cells the ability to be transformed, a property of wild-type cells. Based on these observations and the specific knockdowns used to restore transformation, we suggest that in wild-type cells signaling by KRas through Akt and Cdc42 and signaling by NRas through Raf and RhoA represent pathways through which wild-type cells affect their actin and microtubule cytoskeletons and, in turn, various aspects of cellular transformation, such as adhesion, motility, and anchorage-independent growth.
Our finding that knockdown of RhoA or Raf can restore transformation in Kras-deficient cells may appear counterintuitive given that Raf is required for the transformation of wild-type cells. However, this observation and concept are not entirely unprecedented. Nobes and Hall reported that the ability of rat embryo fibroblasts to migrate could be inhibited by blocking all Ras signaling (using the Y13-259 neutralizing antibody), which affected focal adhesion and stress fiber turnover (35). And while inhibition of RhoA signaling also imposed a block to migration, Nobes and Hall observed that simultaneous inhibition of Ras and RhoA function restored migration (35). Our findings extend those of Nobes and Hall. We would suggest that it is the specific inhibition of KRas signaling that is responsible for the block to migration and that in this setting inhibition of RhoA or Raf is capable of partially reversing this defect (Fig. 5). How the block to migration occurs, whether it is due to the lack of KRas signaling per se or the presence of NRas signaling specifically in the absence of KRas signaling, remains to be determined.
It is known that both the actin and microtubule cytoskeletons are key participants in the migratory behavior of neoplastic cells. That the dynamics of the actin cytoskeleton and stabilization of microtubules are coordinately regulated is indicated by the observation that Rho GTPases directly affect both cytoskeletons (44). Our findings suggest an added layer of regulation, operating upstream of Rho GTPases, that coordinates the actin and microtubule cytoskeletons. Transformed wild-type cells display stabilized microtubules, whereas Kras−/− and Nras−/− cells do not, and these cells fail to transform. Further, specific genetic manipulations of ras-deficient cells can restore the appearance of stabilized microtubules, and this correlates with transformation (Fig. 4C, 7, and 8). It is the same signaling elements whose knockdown restores the stabilization of microtubules that also shift the polarity of the actin cytoskeleton toward that of wild-type cells in ras-deficient cells, e.g., aspects of the actin and microtubule cytoskeletons in Nras−/− cells depleted of Akt or Cdc42 resemble those of wild-type cells, and this correlates with transformation (Fig. 4 and 8). Whether the differential signaling in ras isoform-deficient cells is caused exclusively by the absence of direct regulatory mechanisms mediated by the missing ras alleles or by other compensatory mechanisms cannot be discerned from our findings. Nevertheless, it is tempting to speculate that in wild-type cells coordinate regulation of the actin and microtubule cytoskeletons is achieved with specific signaling by KRas through Akt and Cdc42 and by NRas through Raf and RhoA.
How KRas and NRas achieve their functional specificities remains to be determined. Recently, it has been demonstrated that Kras−/−, but not Nras−/−, fibroblasts are defective in growth factor-induced Akt activation and that this correlates with a defect in cell migration (28). In this context, our findings establish a causal relationship between Akt or Raf activation and cell migration. We would suggest that in the absence of Kras the ability of NRas to signal through Raf imposes a block to migration and this effect can be at least partially reversed by inhibiting Raf signaling (Fig. 5). Consistent with a functional relationship between NRas and Raf, it has been shown that NRas is unique among Ras isoforms in that it is constitutively bound to c-Raf (19). Collectively, observations such as these might suggest that the ability of different Ras isoforms to engage specific effectors, e.g., KRas activation of Akt, figures prominently in transformation. However, there are alternative explanations for our findings. In the case of Nras deficiency, for example, KRas might be capable of signaling through both Raf and Akt but in this setting only Akt activation imposes a block to transformation. Work from Wolfman's group, using a cell system similar to ours, is consistent with the former scenario (28). Further, those authors suggested that differential signaling could be due to KRas and NRas associating with different microdomains in the plasma membrane or differential localization to endomembranes (22, 27, 28, 33, 39-41). Given our findings suggesting that NRas and KRas coordinate the stabilization of microtubules (Fig. 7) and the observation that this event requires a specialized membrane domain (38), it is tempting to speculate that KRas and NRas might need to signal from discrete microdomains in order to coordinate the behavior of microtubules.
A priori, we might have imagined that the differential contributions of Kras and Nras to transformation be reflected in their participation in distinct cellular behaviors, for example, cell survival and growth without anchorage to a substratum. While our findings do not rule out this possibility, they do beg the question of why cells use two Ras isoforms to effect the same cellular processes. Based on our findings, we suggest that the usage of two or more Ras isoforms to effect a given cellular process provides different cell types with the necessary intrinsic plasticity to meet their needs, i.e., to effect cell-type-specific cellular outcomes. This is exemplified by the observation that Kras- and Nras-deficient cells display widely differing phenotypes in 3D matrices (Fig. 3C). Kras−/− cells display a rounded morphology, while Nras−/− cells use an elongated morphology to invade Matrigel. In Kras−/− cells, this extreme in cellular behavior can be shifted specifically with knockdown of RhoA such that the cells adopt an elongated morphology. This dynamic range in cellular behavior afforded by the convergence of two Ras isoforms may be utilized during normal physiological events, such as the migration of different cell types during embryogenesis (14). In pathological scenarios, it has been demonstrated that different tumor types use different modes of invasion during metastasis (10, 14, 15, 55). Perhaps accommodating the diversity in behavior exhibited by different cell types that make up the body necessitates the need for four Ras isoforms. Whether the coordinate signaling achieved with multiple Ras isoforms seen in cells of mesenchymal origin described here is also operating in epithelial cells is currently under investigation.
Our findings might have bearing on why particular tumor types display a preference to acquire activating mutations in a particular Ras isoform (see the introduction). If we assumed that oncogenic Ras does not display a gain of function but rather represents exacerbated signaling, then we would suggest that its contribution to transformation should not be studied in isolation but also needs to take into consideration the contribution to signaling by the wild-type Ras isoforms. In other words, the net output of Ras signaling achieved through oncogenic Ras superimposed upon wild-type Ras likely dictates biological outcome. Depending on how a given cell type is wired and the particular ratios in the levels of wild-type Ras isoform activation, this may render them more susceptible to transformation by one oncogenic Ras isoform than the others. Indeed, in support of this notion it has been demonstrated that oncogenic HRas requires wild-type NRas for its transformation (20). These observations and ideas have therapeutic implications. They suggest the possibility that in a given tumor type harboring activated Ras, targeting either the Ras isoform that is mutated or the wild-type Ras isoforms can be beneficial; it remains to be determined which strategy is best. Further, our findings suggest that in tumors that do not harbor an activating mutation in Ras, targeting specific Ras isoforms in this setting may yield therapeutic benefit.
Acknowledgments
We thank J. Boehm, J. DeCaprio, W. Hahn, L. Garraway, W. Sellers, and M. Brown for providing reagents, T. Jacks and R. Kucherlapati for providing mice, and A. Toker, J. S. Brugge, D. Pellman, H. Shih, and C. Das for critically reading the manuscript.
This work was supported by funding from the National Cancer Institute (M.E.E.).
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Ali, S. H., and J. A. DeCaprio. 2001. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin. Cancer Biol. 11:15-23. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Sagi, D., and A. Hall. 2000. Ras and Rho GTPases: a family reunion. Cell 103:227-238. [DOI] [PubMed] [Google Scholar]
- 3.Benard, V., and G. M. Bokoch. 2002. Assay of Cdc42, Rac, and Rho GTPase activation by affinity methods. Methods Enzymol. 345:349-359. [DOI] [PubMed] [Google Scholar]
- 4.Boehm, J. S., M. T. Hession, S. E. Bulmer, and W. C. Hahn. 2005. Transformation of human and murine fibroblasts without viral oncoproteins. Mol. Cell. Biol. 25:6464-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bos, J. L. 1989. ras oncogenes in human cancer: a review. Cancer Res. 49:4682-4689. [PubMed] [Google Scholar]
- 6.Burridge, K., and K. Wennerberg. 2004. Rho and Rac take center stage. Cell 116:167-179. [DOI] [PubMed] [Google Scholar]
- 7.Chiu, V. K., T. Bivona, A. Hach, J. B. Sajous, J. Silletti, H. Wiener, R. L. Jounson, A. D. Cox, and M. R. Philips. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4:343-350. [DOI] [PubMed] [Google Scholar]
- 8.Choy, E., V. K. Chiu, J. Silletti, M. Feoktistov, T. Morimoto, D. Michaelson, I. E. Ivanov, and M. R. Philips. 1999. Endomembrane trafficking of Ras: the CAAX motif targets proteins to the ER and Golgi. Cell 98:69-80. [DOI] [PubMed] [Google Scholar]
- 9.Chung, C. Y., G. Potikyan, and R. A. Firtel. 2001. Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol. Cell 7:937-947. [DOI] [PubMed] [Google Scholar]
- 10.De Wever, O., Q. D. Nguyen, L. Van Hoorde, M. Bracke, E. Bruyneel, C. Gespach, and M. Mareel. 2004. Tenascin-C and SF/HGF produced by myofibroblasts in vitro provide convergent proinvasive signals to human colon cancer cells through RhoA and Rac. FASEB J. 18:1016-1018. [DOI] [PubMed] [Google Scholar]
- 11.Diaz, R., D. Ahn, L. Lopez-Barcons, M. Malumbres, I. Perez de Castro, J. Lue, N. Ferrer-Miralles, R. Mangues, J. Tsong, R. Garcia, R. Perez-Soler, and A. Pellicer. 2002. The N-ras proto-oncogene can suppress the malignant phenotype in the presence or absence of its oncogene. Cancer Res. 62:4514-4518. [PubMed] [Google Scholar]
- 12.Esteban, L. M., C. Vicario-Abejon, P. Fernandez-Salguero, A. Fernandez-Medarde, N. Swaminathan, K. Yienger, E. Lopez, M. Malumbres, R. McKay, J. M. Ward, A. Pellicer, and E. Santos. 2001. Targeted genomic disruption of H-ras and N-ras, individually or in combination, reveals the dispensability of both loci for mouse growth and development. Mol. Cell. Biol. 21:1444-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ezratty, E. J., M. A. Partridge, and G. G. Gundersen. 2005. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 7:581-590. [DOI] [PubMed] [Google Scholar]
- 14.Friedl, P. 2004. Prespecification and plasticity: shifting mechanisms of cell migration. Curr. Opin. Cell Biol. 16:14-23. [DOI] [PubMed] [Google Scholar]
- 15.Friedl, P., and K. Wolf. 2003. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer 3:362-374. [DOI] [PubMed] [Google Scholar]
- 16.Gomes, E. R., S. Jani, and G. G. Gundersen. 2005. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 121:451-463. [DOI] [PubMed] [Google Scholar]
- 17.Guo, W., and F. G. Giancotti. 2004. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 5:816-826. [DOI] [PubMed] [Google Scholar]
- 18.Gupton, S. L., and C. M. Waterman-Storer. 2006. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell 125:1361-1374. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton, M., J. Liao, M. K. Cathcart, and A. Wolfman. 2001. Constitutive association of c-N-Ras with c-Raf-1 and protein kinase Cɛ in latent signaling modules. J. Biol. Chem. 276:29079-29090. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton, M., and A. Wolfman. 1998. Ha-ras and N-ras regulate MAPK activity by distinct mechanisms in vivo. Oncogene 16:1417-1428. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 22.Hancock, J. F. 2003. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 4:373-384. [DOI] [PubMed] [Google Scholar]
- 23.Hancock, J. F., H. Paterson, and C. J. Marshall. 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63:133-139. [DOI] [PubMed] [Google Scholar]
- 24.Huang, T. Y., C. DerMardirossian, and G. M. Bokoch. 2006. Cofilin phosphatases and regulation of actin dynamics. Curr. Opin. Cell Biol. 18:26-31. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, L., D. Greenbaum, K. Cichowski, K. Mercer, E. Murphy, E. Schmitt, R. T. Bronson, H. Umanoff, E. Windfried, R. Kucherlapati, and T. Jacks. 1997. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 11:2468-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koera, K., K. Nakamura, K. Nakao, J. Miyoshi, K. Toyoshima, T. Hatta, H. Otani, A. Aiba, and M. Katsuki. 1997. K-Ras is essential for the development of the mouse embryo. Oncogene 15:1151-1159. [DOI] [PubMed] [Google Scholar]
- 27.Kranenburg, O., I. Verlaan, and W. H. Moolenaar. 2001. Regulating c-Ras function: cholesterol depletion affects caveolin association, GTP loading, and signaling. Curr. Biol. 11:1880-1884. [DOI] [PubMed] [Google Scholar]
- 28.Liao, J., S. M. Planchon, J. C. Wolfman, and A. Wolfman. 2006. Growth factor-dependent AKT activation and cell migration requires the function of c-K(B)-Ras versus other cellular Ras isoforms. J. Biol. Chem. 281:29730-29738. [DOI] [PubMed] [Google Scholar]
- 29.Liao, J., J. C. Wolfman, and A. Wolfman. 2003. K-Ras regulates the steady-state expression of matrix metalloproteinase 2 in fibroblasts. J. Biol. Chem. 278:31871-31878. [DOI] [PubMed] [Google Scholar]
- 30.Maher, J., D. A. Baker, M. Manning, N. J. Dibb, and I. A. G. Roberts. 1995. Evidence for cell-specific differences in transformation by N-, H- and K-ras. Oncogene 11:1639-1647. [PubMed] [Google Scholar]
- 31.Malumbres, M., and M. Barbacid. 2003. RAS oncogenes: the first 30 years. Nat. Rev. Cancer 3:459-465. [DOI] [PubMed] [Google Scholar]
- 32.Malumbres, M., and A. Pellicer. 1998. Ras pathways to cell cycle control and cell transformation. Front. Biosci. 3:d887-d912. [DOI] [PubMed] [Google Scholar]
- 33.Mor, A., and M. R. Philips. 2006. Compartmentalized Ras/MAPK signaling. Annu. Rev. Immunol. 24:771-800. [DOI] [PubMed] [Google Scholar]
- 34.Nimnual, A. S., B. A. Yatsula, and D. Bar-Sagi. 1998. Coupling of Ras and Rac guanine triphosphatases through the Ras exchange factor. Science 279:560-563. [DOI] [PubMed] [Google Scholar]
- 35.Nobes, C. D., and A. Hall. 1999. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144:1235-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nobes, C. D., and A. Hall. 1995. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53-62. [DOI] [PubMed] [Google Scholar]
- 37.Palazzo, A. F., T. A. Cook, A. S. Alberts, and G. G. Gundersen. 2001. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat. Cell Biol. 3:723-729. [DOI] [PubMed] [Google Scholar]
- 38.Palazzo, A. F., C. H. Eng, D. D. Schlaepfer, E. E. Marcantonio, and G. G. Gundersen. 2004. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science 303:836-839. [DOI] [PubMed] [Google Scholar]
- 39.Plowman, S. J., C. Muncke, R. G. Parton, and J. F. Hancock. 2005. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 102:15500-15505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prior, I. A., C. Muncke, R. G. Parton, and J. F. Hancock. 2003. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quatela, S. E., and M. R. Philips. 2006. Ras signaling on the Golgi. Curr. Opin. Cell Biol. 18:162-167. [DOI] [PubMed] [Google Scholar]
- 42.Raptis, L., H. L. Brownell, M. J. Corbley, K. W. Wood, D. Wang, and T. Haliotis. 1997. Cellular ras gene activity is required for full neoplastic transformation by the large tumor antigen of SV40. Cell Growth Diff. 8:891-901. [PubMed] [Google Scholar]
- 43.Ren, X.-D., W. B. Kiosses, and M. A. Schwartz. 1999. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ridley, A. J. 2001. Rho GTPases and cell migration. J. Cell Sci. 114:2713-2722. [DOI] [PubMed] [Google Scholar]
- 45.Sahai, E., and C. J. Marshall. 2002. RHO-GTPases and cancer. Nat. Rev. Cancer 2:133-142. [DOI] [PubMed] [Google Scholar]
- 46.Sahai, E., M. F. Olson, and C. J. Marshall. 2001. Cross-talk between Ras and Rho signalling pathways in transformation favours proliferation and increased motility. EMBO J. 20:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz, M. A., and A. R. Horwitz. 2006. Integrating adhesion, protrusion, and contraction during cell migration. Cell 125:1223-1225. [DOI] [PubMed] [Google Scholar]
- 48.Scita, G., J. Nordstrom, R. Carbone, P. Tenca, G. Giardina, S. Gutkind, M. Bjarnegard, C. Betsholtz, and P. P. Di Fiore. 1999. EPS8 and E3B1 transduce signals from Ras to Rac. Nature 401:290-392. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi, C., R. T. Bronson, M. Socolovsky, B. Contreras, K. Y. Lee, T. Jacks, M. Noda, R. Kucherlapati, and M. E. Ewen. 2003. Rb and N-ras function together to control differentiation in the mouse. Mol. Cell. Biol. 23:5256-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi, C., B. Contreras, R. T. Bronson, M. Loda, and M. E. Ewen. 2004. Genetic interaction between Rb and K-ras in the control of differentiation and tumor suppression. Mol. Cell. Biol. 24:10406-10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi, C., B. Contreras, T. Iwanaga, Y. Takegami, A. Bakker, R. T. Bronson, M. Noda, M. Loda, J. L. Hunt, and M. E. Ewen. 2006. Nras loss induces metastatic conversion of Rb1-deficient neuroendocrine thyroid tumor. Nat. Genet. 38:118-123. [DOI] [PubMed] [Google Scholar]
- 52.Umanoff, H., W. Edelmann, A. Pellicer, and R. Kucherlapati. 1995. The murine N-ras gene is not essential for growth and development. Proc. Natl. Acad. Sci. USA 92:1709-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watnick, R. S., Y.-N. Cheng, A. Rangarajan, T. A. Ince, and R. A. Weinberg. 2003. Ras modulates Myc activity to repress thrombospondin-1 expression and increase tumor angiogenesis. Cancer Cell 3:219-231. [DOI] [PubMed] [Google Scholar]
- 54.Webb, D. J., K. Donais, L. A. Whitmore, S. M. Thomas, C. E. Turner, J. T. Parsons, and A. F. Horwitz. 2004. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6:154-161. [DOI] [PubMed] [Google Scholar]
- 55.Wilkinson, S., H. F. Paterson, and C. J. Marshall. 2005. Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat. Cell Biol. 7:255-261. [DOI] [PubMed] [Google Scholar]
- 56.Wolfman, J. C., T. Palmby, C. J. Der, and A. Wolfman. 2002. Cellular N-Ras promotes cell survival by downregulation of Jun N-terminal kinase and p38. Mol. Cell. Biol. 22:1589-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolfman, J. C., and A. Wolfman. 2000. Endogenous c-N-Ras provides a steady-state anti-apoptotic signal. J. Biol. Chem. 275:19315-19323. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Z., Y. Wang, H. G. Vikis, L. Johnson, G. Liu, J. Li, M. W. Anderson, R. C. Sills, H. L. Hong, T. R. Devereux, T. Jacks, K.-L. Guan, and M. You. 2001. Wild-type Kras2 can inhibit lung carcinogenesis in mice. Nat. Genet. 29:25-33. [DOI] [PubMed] [Google Scholar]