Abstract
Gfi1 transcriptionally governs hematopoiesis, and its mutations produce neutropenia. In an effort to identify Gfi1-interacting proteins and also to generate new candidate genes causing neutropenia, we performed a yeast two-hybrid screen with Gfi1. Among other Gfi1-interacting proteins, we identified a previously uncharacterized member of the PR domain-containing family of tumor suppressors, PRDM5. PRDM5 has 16 zinc fingers, and we show that it acts as a sequence-specific, DNA binding transcription factor that targets hematopoiesis-associated protein-coding and microRNA genes, including many that are also targets of Gfi1. PRDM5 epigenetically regulates transcription similarly to Gfi1: it recruits the histone methyltransferase G9a and class I histone deacetylases to its target gene promoters and demonstrates repressor activity on synthetic reporters; on endogenous target genes, however, it functions as an activator, in addition to a repressor. Interestingly, genes that PRDM5 activates, as opposed to those it represses, are also targets of Gfi1, suggesting a competitive mechanism through which two repressors could cooperate in order to become transcriptional activators. In neutropenic patients, we identified PRDM5 protein sequence variants perturbing transcriptional function, suggesting a potentially important role in hematopoiesis.
Growth factor independent 1 (Gfi1) is a six-zinc-finger transcription factor originally identified through a mouse retroviral insertional mutagenesis screen for tumor progression to interleukin-2 (IL-2)-independent growth (19). Gfi1 plays a crucial role in hematopoiesis and inner ear development (14, 29). Gene-targeted mice deficient for Gfi1 display impaired blood cell formation characterized by a deficiency of neutrophil and lymphocyte numbers (“neutropenia” and “lymphopenia,” respectively) and the release from bone marrow of immature cells exhibiting features of both neutrophils and monocytes (26, 31). People with hereditary Gfi1 mutations manifest a phenotype similar to the mouse phenotype (50). More recently, mouse gene-targeting studies have identified Gfi1 as an intrinsic regulator of hematopoietic stem cell self-renewal (25, 62).
Emerging evidence indicates that Gfi1 participates in distinct pathways during hematopoiesis. Known Gfi1 targets in myeloid cells comprise a functionally heterogeneous collection of transcription factors, cell cycle regulators, and lineage-specific markers of terminal differentiation (16). During T-lymphocyte development, Gfi1 regulates TH2 cell proliferation through an IL-4- and STAT6-dependent pathway (63) and CD8+ T-cell survival through an IL-7-dependent pathway (49). Gfi1 also regulates STAT3-dependent dendritic cell differentiation and IL-6- and STAT3-mediated proliferative responses to antigenic stimulation (52, 53). One target of Gfi1 is the gene encoding neutrophil elastase, ELA2, whose mutations cause hereditary neutropenia (11, 27), a genetically heterogeneous disease in humans (15); consequently, the overexpression of ELA2 may contribute to neutropenia in both Gfi1-deficient mice and humans bearing GFI1 mutations (50).
Gfi1 demonstrates transcriptional-repressor activity (17, 50, 64), an effect largely mediated through its interaction with histone deacetylase enzymes (HDACs) and G9a methyltransferase (17, 41), which covalently modify histones in order to form epigenetically stable repressive chromatin. Loss of Gfi1, however, also reduces transcription from some of the genes whose promoters it occupies (17, 25, 62), suggesting that it can additionally function as a transcriptional activator in a gene- and/or cell-specific manner. The factors determining whether Gfi1 functions as a repressor or as an activator, however, are unknown.
To better understand Gfi1's molecular mechanism and to uncover new genes additionally responsible for hereditary neutropenia, we sought to discover Gfi1-interacting proteins using a yeast two-hybrid screen. Among other factors, we identified PRDM5, a previously uncharacterized zinc finger protein belonging to the PR domain-containing (PRD1-BF1 and RIZ homology) tumor suppressor protein family. We found that PRDM5 acts as a sequence-specific DNA binding transcription factor. Neutropenia-associated PRDM5 sequence variants interfere with its transcriptional activity. Large-scale target identification reveals that PRDM5 is capable of transcriptionally regulating many protein-coding and microRNA (miRNA) genes, including some involved in hematopoiesis. As with Gfi1, PRDM5 can function as a transcriptional repressor through the recruitment of histone-modifying enzymes to its genetic targets. Also similar to Gfi1, PRDM5 can activate some target genes, but only in the subset whose transcriptional regulation is under shared control by Gfi1, suggesting that when Gfi1 and PRDM5 interact they activate—rather than repress—transcription.
MATERIALS AND METHODS
Cell culture.
HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum. U937 cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% fetal bovine serum. Other cell lines and growth conditions are as previously described (17).
Plasmids.
The following plasmids were generous gifts: pCDNA3.1HA-G9a (K. L. Wright, University of South Florida), (Myc)3-SUV39H1 (T. Jenuwein, Research Institute of Molecular Pathology, Vienna, Austria), pCDNA3.1(−)-HDAC1 (K. Robertson, University of Florida), and pCMX-hHDAC1-, -2-, or -3-Flag (R. M. Evans, Salk Institute). pCF2-7 and pCF2-W3 were described previously (13). PRDM5 truncations were constructed by amplifying the corresponding regions of the human PRDM5 cDNA from pCF2-7 or pCF2-W3 and inserting them into the EcoRI/XhoI sites of the pCS2+Myc vector (59). Gfi1 truncations and plasmids containing sequential lysine substitutions of glutathione S-transferase-fused histone H3 were described previously (17). The luciferase reporter construct pGL3-promoter (containing two copies of the selected PRDMS binding site) was generated by inserting two head-to-tail copies of a PRDM5 binding sequence recovered from random sequence selection (data available on request) into the XhoI site of the vector pGL3-promoter (Promega). The plasmids pGL3-B-mi21-pro(1.2kb) and pGL3-B-mi196b-pro(1kb) were constructed by cloning the 1.2-kb hsa-mir-21 promoter and the 1-kb hsa-mir-196b promoter into the BglII/SacI and BglII/HindIII sites, respectively, of the vector pGL3-basic (Promega) (data available on request).
Yeast two-hybrid screen.
We used a Matchmaker two-hybrid system (Clontech) per the manufacturer's protocol. A construct containing only the zinc fingers of Gfi1 (17) was fused to the Gal4 DNA binding domain vector and screened at high stringency (in medium lacking adenine) against 106 clones from a human bone marrow cDNA library (Clontech) inserted into the Gal4 activation domain vector. Interacting plasmid clones were retransformed with the bait plasmid and a control bait encoding a LexA-lamin C fusion, and the identities of bait-dependent positive interactors were determined by DNA sequencing.
Random oligonucleotide selection and gel-shift assays.
An oligonucleotide containing 38 internal random nucleotides (data available on request) was incubated with Myc-PRDM5 and synthesized in vitro using a TnT transcription/translation system (Promega). The PRDM5-bound oligonucleotides were immunoprecipitated by Myc antibody (9E10; Roche) and amplified by PCR. After seven rounds of selection and amplification, the PCR products were cloned and sequenced. PRDM5 consensus binding motifs were determined by using MEME (http://meme.sdsc.edu/meme/intro.html). Gel-shift assays were carried out as previously described (16), with in vitro-synthesized Myc-tagged PRDM5.
Patient mutation screening.
For identification of mutations in neutropenic patients, DNA sample collection and DNA sequencing were performed as described previously (55).
Transient transfection reporter assay.
HeLa cells were transiently transfected using Lipofectamine Plus (Invitrogen). Luciferase activity assays (17) were performed using a dual-luciferase reporter assay system (Promega); each assay was completed in triplicate and repeated a total of three times. Error bars represent the standard errors of the means.
Immunostaining and protein immunoprecipitation and blotting.
For indirect immunofluorescence assay, HeLa and HEK293 cells were seeded onto coverslips the day before staining. Transient transfection employed Lipofectamine Plus. Cells were fixed with 1% paraformaldehyde for 10 min, permeabilized with 0.2% Triton X-100 for 5 min, and blocked with 5% normal goat serum (Jackson ImmunoResearch) and 1% bovine serum albumin in Tris-buffered saline for 1 h prior to incubation with 2.5 μg/ml PRDM5 rabbit polyclonal antibody (Life Span) or normal rabbit immunoglobulin G (IgG) (Santa Cruz) for 1 h at room temperature. Samples were incubated with 1:200 secondary rhodamine-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch) for 30 min and mounted using Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories). Phosphate-buffered saline was used for washing in between each step. Imaging employed a Zeiss 510 confocal laser scanning microscope (Scientific Imaging) (performed at Fred Hutchinson Cancer Research Center) with a 63× Plan-Apochromat objective and Zeiss LSM Image Browser software using the same magnification, vertical plane, and exposure condition for each image. Coimmunoprecipitation assays were performed as described previously (17). Western blotting assays were performed using ECL (Amersham) or Visualizer (Upstate) detection kits. The antibodies and their working concentrations are as follows: anti-PRDM5, 1:750; anti-Gfi1 (N20, Santa Cruz), 1:500; anti-Myc, 1:20,000; antihemagglutinin (anti-HA) (12CA5, Roche), 1:1,000; anti-G9a (Upstate), 1:500; and anti-HDAC1 (Affinity BioReagents); 1:2,000.
ChIP assays.
HeLa and HEK293 cells were transiently transfected using Lipofectamine Plus. At 48 h after transfection, the cells were subjected to chromatin immunoprecipitation (ChIP) assays, as described previously (16), utilizing the following antibodies: 3 μg anti-Myc, 3 μg anti-HA, 10 μg anti-acetyl-H3 (Upstate), 10 μg anti-acetyl-H4 (Upstate), 2 μg anti-dimethyl-K9-H3 (Upstate), and 6 μg anti-PRDM5. Normal goat, mouse, and rabbit IgGs (Santa Cruz), as appropriate, were used as controls. Semiquantitative PCR or quantitative real-time PCR was performed with appropriate primer pairs (data available on request) based on the genomic sequences analyzed with PRIMER3 (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Each study was repeated a total of two times, for confirmation. Quantitative real-time reverse transcription-PCR (RT-PCR) was performed using an ABI 7300 real-time PCR system with a power SYBR green PCR kit (ABI). Each assay was performed in triplicate and verified by at least one separate repetition, though the results for only one assay are shown (because the quantification method of the ABI software has no easy means for the incorporation of error analysis for separate runs). Large-scale ChIP cloning was performed as previously described (60), where purified DNA fragments were end repaired, ligated to a HindIII linker (data available on request), and amplified by PCR. PCR products were cloned into the HindIII site of the pBluescript KS vector (Stratagene), and inserts ≥250 bp were DNA sequenced with an ABI BigDye Terminator cycle sequencing kit on an ABI 3100 capillary electrophoresis instrument. Their chromosomal location was annotated according to the UCSC genome browser (hg17, NCBI Build 35; http://genome.ucsc.edu).
ChIP-chip.
ChIP combined with high-density oligonucleotide array analysis (ChIP-chip) was carried out as described previously (36), using HEK293 cells that were transiently transfected with Myc-tagged PRDM5 expression vector by using Lipofectamine Plus. A total of 3.2 mg of chromatin, fragmented to ≤1 kb by sonication, was immunoprecipitated using 3 μg of anti-Myc antibody. After the reversal of cross-links, purified and enriched DNA, along with unenriched input DNA, was amplified by two-step ligation-mediated PCR. DNA samples from three independent transfection and ChIP experiments were pooled together and submitted to the NimbleGen ChIP-chip custom service. A custom DNA microarray comprising 50-mer probes tiled with a 12-bp overlap across nonrepeat masked regions of the chromosomal regions described in Results was manufactured. Each probe had three replicates. The DNA microarray hybridization, array signal scanning, data extraction, and data normalization were carried out by NimbleGen. DNA binding peaks were identified (3) by using the statistical model and methodology described at http://chipanalysis.genomecenter.ucdavis.edu/cgi-bin/tamalpais.cgi.
RNA interference (RNAi).
PRDM5 short hairpin RNA (shRNA) constructs V2HS_29874 and V2HS_29875, nonsilencing control vector RHS1703, and empty vector pSM2C were purchased from Open Biosystems. For virus production, pSM2C plasmids were transfected into LinX (Open Biosystems) packaging cells, and viral supernatants were harvested 48 h after transfection. Stable HEK293 and HL-60 cell lines were established by puromycin selection (2 μg/ml). For each construct, three to five independent cell pools were generated from different transduction experiments.
Gene expression assays.
Total RNA was prepared using an Absolutely RNA RT-PCR miniprep kit (Stratagene). An amount of 1 μg total RNA was used to produce cDNAs by using Superscript III reverse transcriptase with oligo(dT)12 primer (Invitrogen). TaqMan gene expression assays were purchased from Applied Bioystems (PRDM5, HS00218855_ml; Wnt10B, HS00559664_ml; EDNRA, HS00609865_ml; c-Myc, HS00509030_ml; LCN7, HS00223403_ml; CDH4, HS00242399_ml; SNF1LK, HS00545020_ml; human 18S rRNA, 4319413E; and human glyceraldehyde-3-phosphate dehydrogenase [GAPDH], 4326317E). miRNA expression profiling was performed by using TaqMan miRNA assays (ABI) and a mirVana miRNA detection kit (Ambion). Total RNA was prepared using TRIzol (Invitrogen). Quantitative real-time RT-PCR was performed on an ABI 7300 real-time PCR system, using the relative quantification method with triplicate samples, repeated a total of two times. The gene expression microarray was performed using a 60-mer NimbleGen high-density (eight probes per gene) human whole-genome expression array. Total RNA was extracted from three independent cell pools of each of the PRDM5 stable-knockdown cell lines and the control cell lines. Double-stranded cDNAs were made from each RNA sample by using a SuperScript double-stranded cDNA synthesis kit (Invitrogen). Three PRDM5 knockdown and control cDNA samples were each pooled. The cDNA samples were then submitted to NimbleGen's gene expression microarray service, which performed the expression hybridization, array signal scanning, data extraction, and data normalization.
HMTase assay.
In vitro histone methyltransferase (HMTase) assays were performed as described previously (17); 48 h posttransfection, HeLa cells were subjected to immunoprecipitation with anti-Myc (9E10) antibody or control IgG (0.8 μg per sample in a 1.5-ml volume) in radioimmunoprecipitation assay buffer. Purified G9a was obtained from Upstate.
Flow cytometry.
Cells were harvested, counted, fixed, and stained with 50 μg/ml propidium iodide for 3 h at 4°C. The DNA content was analyzed on a Becton Dickinson FACScan (by the University of Washington, Department of Immunology, Cell Analysis Facility) utilizing Tree Star FlowJo 8 software running in Apple Mac OS X. Each analysis was repeated a total of two times.
RESULTS
Identification of PRDM5 as a Gfi1-interacting protein.
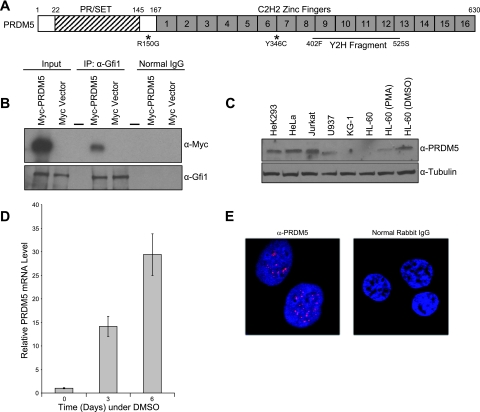
In order to further study Gfi1 function, and also move toward discovering candidate genes for cases of hereditary neutropenia in patients lacking mutations in known genes, we used a fragment of human Gfi1 comprised of only the zinc fingers as a bait for screening a yeast two-hybrid library of human bone marrow cDNA. A prior yeast two-hybrid screen of mouse full-length Gfi1 against a mouse embryonic library recovered just one interacting protein (53), the E3 SUMO ligase PIAS3 (38). Because zinc fingers can mediate protein-protein interactions (40), we reasoned that using only this portion as bait might uncover additional interactions otherwise sterically hindered in two-hybrid fusion constructs. The screening of 106 clones under high-stringency selection yielded only 56 interacting clones, corresponding to just 29 unique proteins (data available on request). We did not recover PIAS3, but most of the identified proteins are also involved in SUMO conjugation, including the closely related PIAS4 (39) and the E2 SUMO ligase, UBC9 (42). However, we were particularly intrigued by the interaction of Gfi1 with PRDM5, a protein containing 16 C2H2-type zinc fingers and belonging to the PR domain-containing family (30) of tumor suppressors (Fig. 1A). PRDM5 is transcriptionally silenced in some human cancers, but little else is known regarding its function (13). As a next step, we confirmed in vivo interaction between Gfi1 and PRDM5 by using bidirectional coimmunoprecipitation assays with epitope-tagged versions of both proteins expressed in transfected HeLa cells (Fig. 1B), although we could not demonstrate coimmunoprecipitation with endogenous proteins in HL-60 cells using native antibodies (data not shown). Deletion analysis shows that the 6 zinc fingers of Gfi1 (data available on request) and the first 10 zinc fingers of PRDM5 (data available on request) mediate their association.
FIG. 1.
PRDM5 structure, Gfi1 interaction, neutropenic sequence variants, and subcellular localization. (A) PRDM5 schematic representation. Asterisks denote variants found in neutropenic individuals. Zinc finger fragment recovered in yeast two-hybrid (Y2H) screen with zinc fingers of Gfi1 is indicated. Numbers on top of schematic correspond to protein sequence. (B) Interaction of PRDM5 with Gfi1 in cotransfected HeLa cells. Coimmunoprecipitation assays were performed using an antibody against Gfi1 or normal goat IgG as a negative control with HeLa cells cotransfected with the indicated plasmids. Dash indicates empty lane. (C) Western blot analysis of PRDM5 in human cell lines. PMA, phorbol myristate acetate; DMSO, dimethyl sulfoxide. (D) Up-regulation of PRDM5 during dimethyl sulfoxide (DMSO)-induced granulocytic differentiation of HL-60 cells, as measured by quantitative real-time RT-PCR with GAPDH as internal control. (E) Confocal microscopy of indirect immunofluorescent staining of HeLa cells with PRDM5 antibody (red) and control IgG. Nuclei counterstained with DAPI are blue. α, anti.
Expression and nuclear localization of PRDM5.
RT-PCR analysis of human tissues, as well as studies available from the Gene Expression Omnibus atlas (http://www.ncbi.nlm.nih.gov/geo/), indicates that PRDM5 is widely expressed (data not shown). Among human cell lines, Western blot analysis (Fig. 1C) shows that PRDM5 is expressed in HeLa and HEK293 epithelial cells and Jurkat T lymphocytes, but the protein is apparently less abundant in U937 monocytes and not detectable in early (KG-1) and late (HL-60) myeloblasts. Notably, the amounts of PRDM5 protein (Fig. 1C) and its transcript (Fig. 1D) increase with the granulocytic differentiation of HL-60 cells induced by dimethyl sulfoxide or all-trans-retinoic acid (not shown). Immunofluorescent localization reveals punctate nuclear staining of endogenous PRDM5 in HeLa cells (Fig. 1E).
PRDM5 variants associated with neutropenia.
A motive for these studies was to identify other genes whose mutation might cause unexplained cases of neutropenia. We determined the DNA sequence of the exons and intronic junctions of PRDM5 in 230 unrelated neutropenic patients lacking ELA2 or Gfi1 mutations and found two with different amino acid missense substitutions that were absent among more than 460 controls. The first individual is heterozygous for an R150G amino acid substitution at a phylogenetically conserved residue in between the SET domain and the first zinc finger; the second patient and three generations of other family members are heterozygous for amino acid substitution Y346C occurring two residues ahead of the first cysteine in the seventh zinc finger (marked in Fig. 1A; further clinical details available on request). The clinical and genetic data are insufficient to conclude that these variants are causative. Both variants had negligible effects on the PRDM5-Gfi1 interaction, as defined by coimmunoprecipitation (data available on request), but further analysis of their functional consequences may bear on their significance.
PRDM5 sequence-specific DNA binding and disruptive effect of neutropenia-associated variants.
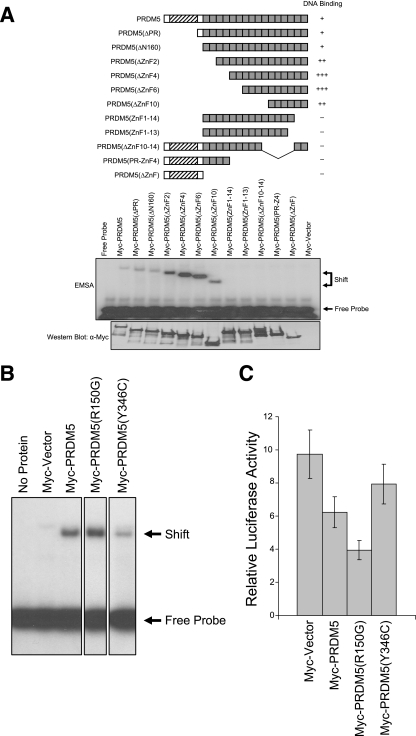
Zinc fingers can mediate protein-protein interactions, but they more commonly direct sequence-specific DNA binding (10). To determine if PRDM5 binds DNA, we performed random DNA sequence selection (28) using in vitro-translated protein and compiled a consensus sequence based on random DNA sequences to which PRDM5 bound (data available on request). The results of an electrophoretic mobility gel-shift assay confirm that PRDM5 binds this consensus DNA sequence (data available on request). Deletion analysis demonstrates that only the most-carboxyl six zinc fingers of PRDM5 are required for DNA binding (Fig. 2A), though others may contribute to specificity (data not shown). We evaluated whether neutropenic sequence variants affected PRDM5 DNA binding. The R150G substitution slightly increased the DNA affinity; the Y346C mutation markedly decreased binding, as might be expected with a substitution involving a cysteine in the spacer region between zinc fingers (Fig. 2B). (The variants had no effect on in vitro translational efficiency or protein stability [data not shown].)
FIG. 2.
DNA sequence-specific transcriptional repressor activity of PRDM5 and its disruption by neutropenia-associated sequence variants. (A) Deletion of PRDM5 to determine domains responsible for DNA binding via gel-shift assay. Hatching and shading are as indicated in Fig. 1A. EMSA, electrophoretic mobility shift assay. The number of plus signs indicates the relative strength of binding. −, no binding. (B) Effects of neutropenia-associated PRDM5 sequence variants on DNA binding in gel-shift assay. R150G shows slightly more DNA binding, Y346C less DNA binding. (An amount of 0.5 μl of 50 μl total in vitro TnT transcription/translation system product of each protein was used.) The figure shows a single gel image, where intervening lanes have been cropped for the purpose of clarity of presentation. α, anti. (C) PRDM5 transcriptional repressor activity disrupted by neutropenia-associated sequence variants. Reporter assay in HeLa cells transiently transfected with 100 ng of a construct containing a dimer of the PRDM5 DNA binding consensus sequence fused to a simian virus 40 promoter driving luciferase expression along with 50 ng of the indicated form of PRDM5 expression vector. Note that the R150G mutation increased repressor activity of PRDM5, while Y346C diminished activity, consistent with effects on DNA binding.
PRDM5 transcriptional repressor activity and disruptive effect of neutropenia-associated variants.
To determine if PRDM5 can function as a transcription factor, we constructed a luciferase reporter containing dimers of a sequence defined by random DNA selection, inserted upstream of the simian virus 40 promoter. Transient overexpression of PRDM5 in HeLa cells demonstrates that PRDM5 can, like Gfi1, function as a transcriptional repressor (Fig. 2C). We also evaluated the neutropenia-associated variants. Consistent with their effects on DNA binding, the variant (R150G) improving DNA binding increased repression, whereas the variant (Y346C) reducing DNA binding decreased repressor function (Fig. 2C), though the results for Y346C fell short of statistical significance.
PRDM5's protein-coding and miRNA target genes.
One means of characterizing the biological function of a novel transcription factor is to identify genes whose expression it regulates. State-of-the-art ChIP-chip approaches are generally cost prohibitive on a whole-genome scale. In order to most efficiently identify PRDM5 target genes, we utilized a discovery-based method in conjunction with a functionally oriented strategy. We designed a high-density chip array tiling four sets of sequences (Table 1). First, we performed ChIP in HeLa cells expressing epitope-tagged PRDM5 and cloned and sequenced the products, thus providing a list of 655 unique genomic sequences of ∼250 bp each as candidate PRDM5 binding sites. Second, because our goal is to determine how PRDM5 contributes to neutrophil differentiation, we included 3 kb of the promoter region from each of 92 genes participating in hematopoiesis (data available on request), as determined from the literature. Third, accumulating evidence indicates that miRNAs play important roles in both normal development and tumorigenesis (7, 8), and we reasoned that PRDM5, as a possible developmental factor and tumor suppressor, might function by regulating miRNA genes. Therefore, we included all (of the then) known human miRNAs. Fourth, we selected a 1.2-Mb contiguous region from chromosome 15q2, of interest because it contains the tumor suppressor gene PML, which is disrupted by translocations in promyelocytic leukemia (21). This region is also sufficiently large to permit surveying the density of PRDM5 binding sites among otherwise unselected genes and intergenic regions.
TABLE 1.
Candidate genes and region selected for PRDM5 binding analysis by ChIP-chip
| Candidate genes and region | Total no. | Size | No. binding PRDM |
|---|---|---|---|
| ChIP-cloned target candidates | 655 | 83 | |
| Candidates associated with hematopoiesis | 92 | 38 | |
| miRNA genes | 321 | 31 | |
| Human chromosome 15 region | 1.2 Mb | 22 |
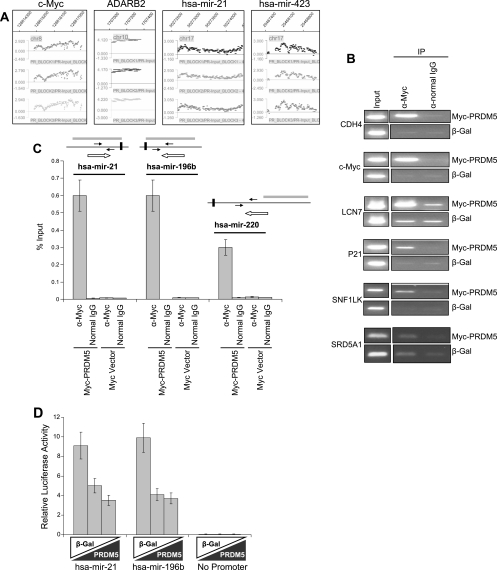
We then hybridized DNA fragments recovered by ChIP from HEK293 cells transfected with epitope-tagged PRDM5 to the array. (We reasoned that using a different cell line for ChIP in the hybridization phase of this experiment would reduce false positives among candidate targets derived from ChIP cloning in HeLa cells.) The results are summarized in Table 1; protein-coding and miRNA genes are available on request. Representative ChIP-chip hybridization signals are shown in Fig. 3A. Just 13% of the fragments obtained by ChIP cloning from HeLa cells were also bound by PRDM5 in HEK293 cells. In contrast, PRDM5 binding was detected in 41% of promoter sequences from hematopoiesis-associated candidate target genes. Notably, PRDM5 also occupied the proximal region (within 1 kb up- or downstream of the mature miRNA sequence) of 31 (10%) miRNAs. Of the miRNA genes targeted by PRDM5, the binding sites for 25 are intronic or intergenic; the remainder are contained within exons of protein-coding genes (data available on request). In the chromosome 15 contig, 22 sequences were bound by PRDM5.
FIG. 3.
PRDM5 target gene identification. (A) Examples of ChIP-chip hybridization signals, with vertical axis representing log2 ratio of ChIP to input, showing results for three probe replicates as visualized using NimbleGen SignalMap software. (B) Confirmation of ChIP-chip results by semiquantitative PCR in transfected HEK293 cells. SRD5A1 (excluded as target by ChIP-chip) serves as negative control. Cells were transfected with either Myc-PRDM5 expression constructs or equivalent quantities of vector expressing β-gal. (C) Confirmation of ChIP-chip results for miRNA target genes via quantitative real-time PCR in transfected HEK293 cells. Paired arrows indicate location of PCR primers relative to mature RNA (horizontal black bars). Gray bars demonstrate enriched regions in ChIP-chip assays. Open arrows show transcription direction. (D) PRDM5 repression of hsa-mir-21 and hsa-mir-196b gene promoters, demonstrated by transient reporter assays in HeLa cells transfected with 0, 50, or 150 ng of PRDM5 expression vector (black triangles) and 100 ng of luciferase reporter construct. In each titration, the total quantity of expression vector was held constant at 150 ng with compensatory quantities of vector expressing β-gal (white triangles). α, anti.
As a test of the ChIP-chip reliability, we performed conventional ChIP followed by PCR on six protein-coding (Fig. 3B) and three miRNA (Fig. 3C) target genes, and corroborated each result independently. For two of the protein-coding genes (SNF1LK and CDH4), we additionally confirmed PRDM5 binding by gel-shift assay (data not shown). We also cloned the PRDM5 binding sites contained within the promoters (1-kb upstream region) of two miRNA genes, hsa-mir-21 and hsa-mir-196b, and fused each to a luciferase reporter; in transiently transfected HEK293 cells, each demonstrated strong promoter activity that could be repressed in the presence of PRDM5 (Fig. 3D).
Deregulation of the cell cycle following PRDM5 suppression.
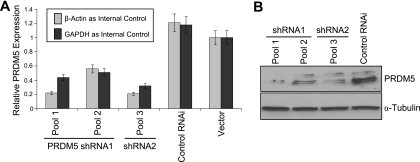
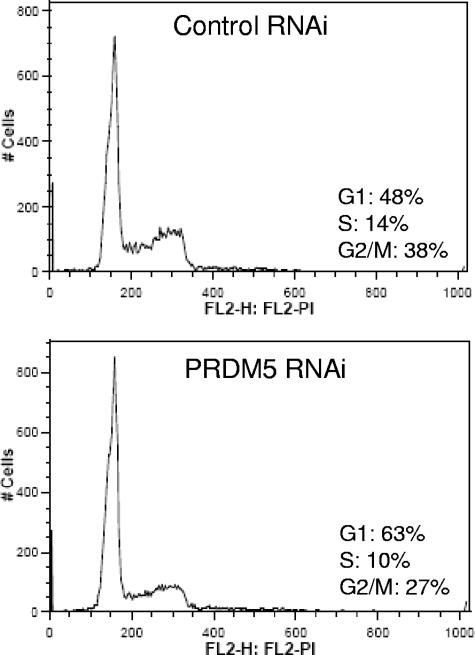
We used RNAi to investigate how reduced expression of PRDM5 influences the cell cycle. We employed two retrovirally expressed shRNAs. Each suppressed PRDM5 expression at both the mRNA (Fig. 4A) and protein (Fig. 4B) level in stably transduced HEK293 cells. Moreover, seven of eight probes tiled across the PRDM5 coding sequence demonstrate reduced abundance of the PRDM5 transcript when using microarray gene expression profiling to compare HEK293 cells transduced with either PRDM5 shRNA or control shRNA retrovirus (data available on request). The suppression of PRDM5 reduces the number of cells at the G2/M phase (Fig. 5), consistent with prior observation that restoring PRDM5 expression in cancer cells restores G2/M arrest (13).
FIG. 4.
Stable suppression of PRDM5 in HEK293 cells by RNAi. (A) Quantitative real-time RT-PCR assays demonstrating reduction of PRDM5 by two PRDM5 RNAi retroviral constructs, V2HS_29874 (shRNA1) and V2HS_29875 (shRNA2), compared to control vector RHS1703 and empty vector pSM2c. Pools correspond to different polyclonal populations selected for vector-conferred puromycin resistance. (B) Western blot assays demonstrating PRDM5 suppression at protein level. α, anti.
FIG. 5.
Fluorescence-activated cell sorter analysis of cell cycle profile following PRDM5 suppression in HEK293 cells compared to controls. FL2-H: FL2-PI corresponds to the height of the fluorescence signal for propidium iodide staining of DNA content. (Repetition of the experiment showed cells in G1, S, and G2/M stages with control RNAi and PRDM5 RNAi at 46%, 18%, and 36% and 63%, 13%, and 24%, respectively [data not shown].)
Deregulation of target genes following PRDM5 suppression.
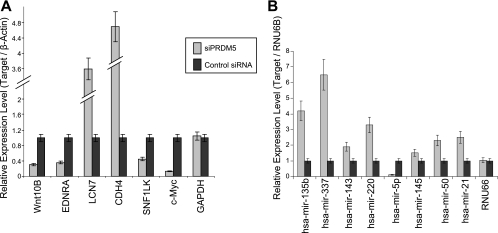
To determine how reduction of PRDM5 influences target gene expression in HEK293 cells stably suppressed for PRDM5 expression, we performed microarray profiling. Among putative protein-coding target genes (Table 2), 24 were found to be significantly up-regulated (≥1.5 times) after PRDM5 suppression (data available on request), consistent with PRDM5's apparent repressor activity. However, 55 protein-coding target genes demonstrated reduced (≤1.5 times) expression with PRDM5 suppression (data available on request). As a spot-check, we confirmed a subset of the microarray results by real-time RT-PCR (Fig. 6A).
TABLE 2.
Direct targets of PRDM5 as established by ChIP and deregulation following RNAi
| Primary gene function and gene name | Accession no.a |
|---|---|
| Signal transduction | |
| PIAS3 | NM_006099 |
| PPP1R12B | NM_032105 |
| MyD88 | NM_002468 |
| IL6R | NM_000565 |
| MyD118 | NM_015675 |
| GPR1 | NM_005279 |
| EDNRA | NM_001957 |
| NCOA7 | NM_181782 |
| WNT10B | NM_003394 |
| CKLF | NM_181641 |
| SOCS3 | NM_003955 |
| EVL | NM_016337 |
| CDC42EP4 | NM_012121 |
| Development | |
| ID-2 | NM_002166 |
| Myb | NM_005375 |
| PAX5 | NM_016734 |
| PAX6 | NM_000280 |
| ALX4 | NM_021926 |
| Eed | NM_003797 |
| RUNX1a | NM_001754 |
| Runx2c | NM_004348 |
| SOX2 | NM_003106 |
| EBF | NM_024007 |
| Oct-4 | NM_203289 |
| NAV2 | NM_145117 |
| OVOL1 | NM_004561 |
| RARa | NM_000964 |
| RXRb | NM_021976 |
| C5orf13 | NM_004772 |
| GABPa | NM_002040 |
| POU2F3 | NM_014352 |
| ENO2 | NM_001975 |
| Rae-28 | NM_004426 |
| KIAA1199 | NM_018689 |
| GOLGA6 | NM_001038640 |
| FOG1 | NM_153813 |
| PSG1 | NM_006905 |
| Cell cycle | |
| E2F3 | NM_001949 |
| Ets2 | NM_005239 |
| c-Myc | NM_002467 |
| ARID3B | NM_006465 |
| SNF1LK | NM_173354 |
| Tumor suppressor | |
| S100A2 | NM_005978 |
| P53 | NM_000546 |
| DKK3 | NM_001018057 |
| PML | NM_033246 |
| Cell-cell signaling | |
| Notch2 | NM_024408 |
| Notch1 | NM_017617 |
| Cell adhesion | |
| CLDN16 | NM_006580 |
| COL19A1 | NM_001858 |
| EMID1 | NM_133455 |
| CDH4 | NM_001794 |
| CD99 | NM_002414 |
| Protein modification | |
| FBXO33 | NM_203301 |
| UBL7 | NM_032907 |
| LCN7 | NM_022164 |
| CLK3 | NM_003992 |
| PPIL2 | NM_148175 |
| STK32C | NM_173575 |
| RNA processing | |
| ADARB2 | NM_018702 |
| C6orf151 | NM_152551 |
| APOBEC3B | NM_004900 |
| Calcium channel | |
| CACNA1C | NM_000719 |
| Metabolism | |
| GCK | NM_000162 |
| CYP3A7 | NM_000765 |
| ACSBG1 | NM_015162 |
| Muscle contraction | |
| MYOM2 | NM_003970 |
| Histones | |
| HST4H4 | NM_175054 |
| Unknown | |
| PALM2-AKAP2 | NM_007203 |
| CCDC33 | NM_182791 |
| NOD27 | NM_032206 |
| FLJ32499 | NM_144607 |
| FLJ34306 | NM_199340 |
| FLJ39713 | AK097032 |
| INTS3 | NM_023015 |
| FLJ45537 | NM_001001709 |
| LOC388135 | NM_001039614 |
| ISLR2 | NM_020851 |
| ISLR | NM_005545 |
| miRNA | |
| hsa-mir-135b | MI0000645 |
| hsa-mir-143 | MI0000459 |
| hsa-mir-145 | MI0000461 |
| hsa-mir-337 | MI0000806 |
| hsa-mir-299 | MI0000744 |
| hsa-mir-21 | MI0000077 |
| hsa-mir-150 | MI0000479 |
| hsa-mir-220 | MI0000297 |
Accession numbers for protein-coding genes are from GenBank (http://www.ncbi.nlm.nih.gov/) and for miRNA genes from miRBase (http://microrna.sanger.ac.uk/sequences/index.shtml).
FIG. 6.
Confirmation of target gene deregulation following PRDM5 silencing in HEK293 cells. (A) Spot check of PRDM5 targets shown to be deregulated by microarray confirmed here by results of quantitative real-time RT-PCR with GAPDH and β-actin as internal controls. (B) PRDM5 miRNA target gene expression profiling following PRDM5 silencing, measured by TaqMan assays with RNU6B as internal control and RNU66 as negative control.
We also evaluated the expression pattern of miRNA target genes using TaqMan miRNA assays. Of eight miRNAs tested, seven become up-regulated, and one (hsa-mir-299-5p) demonstrated reduced expression (Fig. 6B). We then compared the expression pattern of these miRNAs with that of their corresponding host gene and found that PRDM5 generally regulates them independently of one another, with opposite effects with respect to repression or activation in some cases (data available on request).
In summary, ChIP data, in conjunction with RNAi profiling, indicates that PRDM5 targets numerous functionally diverse protein-coding and miRNA genes (Table 2). Many contribute to development and hematopoiesis. Another conclusion is that PRDM5 might also, depending on the target, act as a transcriptional activator in vivo. This may not be surprising, given that Gfi1, functioning as a repressor in vitro, can also transcriptionally activate some target genes in vivo (16, 17).
Transcriptional activation of genes targeted in common by PRDM5 and Gfi1.
Given that PRDM5 and Gfi1 interact, we asked if they each regulate the same or different target genes. We therefore repeated the ChIP-chip experiment, except that we hybridized to the array DNA fragments recovered by ChIP from HEK293 cells transfected with epitope-tagged Gfi1. Among many Gfi1 target genes identified, we found 40 protein-coding and 6 miRNA genes common to both PRDM5 and Gfi1 (data available on request). Of the 24 protein-coding target genes repressed by PRDM5, just 1 (PAX6) is also a target for Gfi1, but, interestingly, among the 55 protein-coding target genes activated by PRDM5, 12 (NOTCH2, PIAS3, IL6R, PPP1R12B, ID2, MYD88, C6ORF151, E2F3, NCOA7, MYB, C-MYC, and PAX5) are also Gfi1 target genes. These results suggest that PRDM5 is a transcriptional repressor when acting alone, but could be transformed into a transcriptional activator in the presence of Gfi1.
PRDM5-associated H3-K9 methyltransferase activity and interaction with histone-modifying enzymes.
In assays employing idealized PRDM5 binding sites and for miRNA genes that it targets, PRDM5 predominately acts as a repressor. We therefore investigated the mechanism through which it functions as a repressor. PR domains are closely related to SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domains, which possess HMTase activity. A distinguishing feature is that an essential histidine residue is changed to a cysteine in the PR domain; nevertheless, one PRDM family member, PRDM2/RIZ1, demonstrates intrinsic HMTase activity on histone H3 at the K9 position (34), a modification associated with gene repression. However, we could not detect HMTase activity for PRDM5 on histone substrates (data available on request).
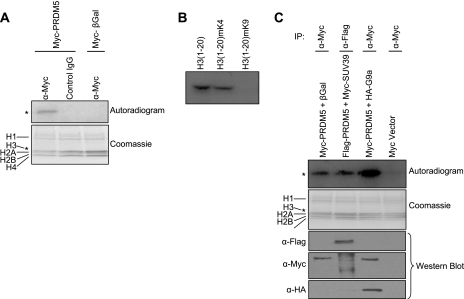
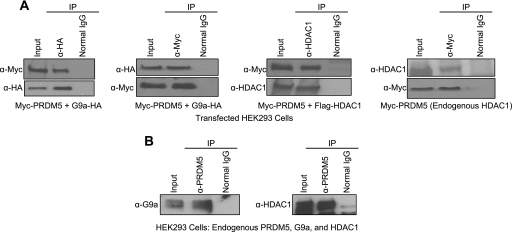
Some PR domain-containing proteins (9, 12, 22) also lack endogenous HMTase activity, but can recruit other histone-modifying enzymes. To address this possibility, we determined if PRDM5 might be present in a protein complex possessing HMTase activity. We transfected Myc epitope-tagged PRDM5 or a control plasmid (expressing β-galactosidase [β-gal]) into HeLa cells and subjected the cell lysates to immunoprecipitation with a Myc (9E10) antibody or a control IgG. Each of the immunoprecipitated complexes was then assayed for HMTase activity on free histones (including H1 and core histones) as substrates. Only the immunoprecipitated PRDM5 complex, but not ones derived from cells expressing the control plasmid or immunoprecipitated with control IgG, could incorporate tritiated S-adenosylmethionine into a band corresponding to histone H3 in autoradiograms (Fig. 7A). The HMTase activity recruited by PRDM5 is specific for the K9 position of H3 (Fig. 7B and data available on request). Moreover, HMTase activity was significantly increased upon cotransfection with G9a, but not with SUV39H1 (Fig. 7C), suggesting that, of the two, G9a is the responsible HMTase factor. Consequently, we determined if PRDM5 and G9a can associate. Cotransfected Myc-PRDM5 and HA-G9a can mutually immunoprecipitate one another in HEK293 cells (Fig. 8A). Immunoprecipitation of endogenous G9a also pulls down PRDM5 (Fig. 8B). Deletion mapping revealed that both the PR domain and zinc fingers 7 to 10 are sufficient for G9a interaction (data available on request).
FIG. 7.
PRDM5 recruitment of G9a HMTase. (A) Immunoprecipitated PRDM5 complex possesses HMTase activity. Vectors expressing Myc-PRDM5, or β-gal as a negative control, were transfected into HeLa cells, and cell extracts were immunoprecipitated with Myc or nonspecific antibody and then incubated with histones and S-[3H]adenosylmethionine and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis with Coomassie staining. Autoradiography demonstrates transfer of tritiated methyl to histone H3. (B) The PRDM5 complex methylates H3-K9, but not H3-K4. A synthetic peptide corresponding to the amino-terminal 20 residues of H3, as well as one in which the K4 was premethylated, are both methylated, but premethylation of K9 blocks its function as a substrate. (C) Coexpression of G9a, but not Suv39H1, in HeLa cells increases HMTase activity of immunoprecipitated PRDM5 complex. (Western blots were performed on cell extract to confirm expected expression.) Asterisks denote equivalent electrophoretic migration distance for H3 in different panels. α, anti.
FIG. 8.
In vivo association of PRDM5 with HDAC1 and G9a. (A) When coexpressing Myc-tagged PRDM5 with HA-tagged G9a (left two panels) and Flag-tagged HDAC1 (middle right panel) in transfected HEK293 cells, PRDM5 could be coimmunoprecipitated with G9a by antibodies against either HA epitope (left panel) or Myc epitope (middle left panel) and with HDAC1 by HDAC1 antibody (middle right panel), respectively. Transfected Myc-PRDM5 also coimmunoprecipitated with endogenous HDAC1 in HEK293 cells (right panel). (B) Endogenous PRDM5 forms an immunoprecipitable complex with endogenous G9a and HDAC1 in HEK293 cells. Antibody against PRDM5 coimmunoprecipitates G9a (left panel) and HDAC1 (right panel). α, anti.
Histone deacetylation is another epigenetic modification associated with gene repression, and PRDM5 similarly forms an immunoprecipitable complex with the class I HDAC enzyme HDAC1 (Fig. 8A and B), as well as HDAC2 and HDAC3 (data not shown). The same regions of PRDM5 (PR domain or zinc fingers 7 to 10) needed for G9a interaction are sufficient for HDAC1 binding (data available on request). These results parallel our earlier studies showing Gfi1 recruitment of G9a and HDAC1 to Gfi1 target promoters (17, 41), which is not surprising, given that both transcription factors can interact with each other and demonstrate some overlap in the target genes that they regulate.
PRDM5-directed recruitment of G9a and HDAC1 to target miRNA promoters and resulting histone modifications.
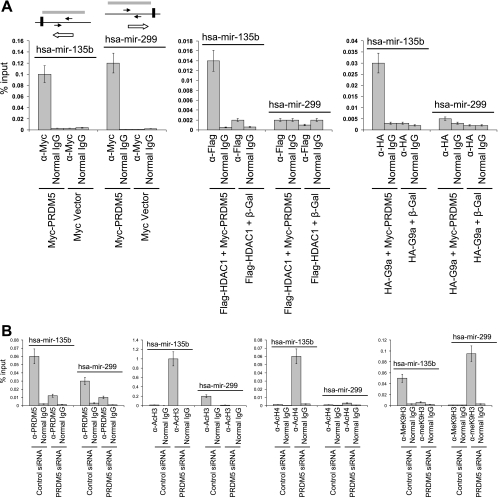
We determined if PRDM5 also recruits histone-modifying enzymes to its miRNA target genes. We focused on the promoters of two miRNA genes, hsa-mir-135b, which was strongly up-regulated after PRDM5 suppression, and hsa-mir-229-5p, which was markedly down-regulated with PRDM5 suppression. (Neither is a target for Gfi1.) We cotransfected HEK293 cells with vectors expressing either Myc-tagged PRDM5 or a β-gal control along with Flag-tagged HDAC1 or HA-tagged G9a and performed ChIP with antibodies to the Myc-, Flag-, or HA-tags, respectively. Flag-HDAC1 was recruited to the hsa-mir-135b promoter (which is repressed by PRDM5) only when Myc-PRDM5 was coexpressed and bound the promoter; similarly, recruitment of HA-G9a to the hsa-mir-135b promoter correlated with occupancy by PRDM5 (Fig. 9A). These results indicate that the recruitment of HA-G9a and Flag-HDAC1 to the hsa-mir-135b promoter is dependent on PRDM5. For hsa-mir-229-5p (which is activated by PRDM5), on the other hand (Fig. 9A), PRDM5 also bound to the promoter, but neither Flag-HDAC1 nor HA-G9a did so. Thus, PRDM5 represses the expression of hsa-mir-135b through a different mechanism than it uses to activate hsa-mir-229-5p.
FIG. 9.
PRDM5 recruitment of G9a and HDAC1 to its miRNA target promoters and histone modification pattern changes associated with its silencing in HEK293 cells. (A) HEK293 cells were cotransfected with the noted combinations of plasmids and subjected to ChIP with antibodies against the indicated epitope tags or control IgG followed by semiquantitative PCR. Left panel, occupancy of Myc-PRDM5 on the promoters of hsa-mir-135b and hsa-mir-299 genes. Middle panel, PRDM5 recruitment of HDAC1 to hsa-mir-135b promoter, but not hsa-mir-299 promoter. Right panel, PRDM5 recruitment of G9a to hsa-mir-135b promoter, but not hsa-mir-299 promoter. Paired arrows indicate location of PCR primers relative to mature RNA (horizontal black bars). Gray bars demonstrate enriched regions in ChIP-chip assays. Open arrows show transcription direction. (B) Histone modification pattern changes associated with silencing of PRDM5 in HEK293 cells. Left panel, presence of endogenous PRDM5 on promoters of hsa-mir-135b and hsa-mir-299, demonstrated by diminished binding with RNAi depletion. Left middle panel, PRDM5 depletion results in acetylated histone H3 becoming increased in hsa-mir-135b and decreased in hsa-mir-299 promoters. Right middle panel, PRDM5 depletion results in acetylated histone H4 becoming increased in hsa-mir-135b and not significantly changed on H4 in hsa-mir-299 promoters. Right panel, depletion of PRDM5 results in K9-methylated histone H3 becoming decreased in hsa-mir-135b and increased in hsa-mir-299 promoters. α, anti.
We examined the acetylation pattern of histones H3 and H4 and the methylation pattern of H3-K9 in these two miRNA promoters in PRDM5-reduced HEK293 cells. ChIP with antibody directed against PRDM5 confirmed that endogenous PRDM5 binds to the hsa-mir-135b and hsa-mir-229-5p promoters, and, when PRDM5 is suppressed, the binding of PRDM5 to each promoter becomes decreased (Fig. 9B, left panel). For hsa-mir-135b (Fig. 9B), ChIP reveals that PRDM5 suppression induces H3 and H4 acetylation while reducing H3-K9 methylation. In contrast, for hsa-mir-229-5p (Fig. 9B), PRDM5 suppression leads to the reduction of H3 acetylation and induction of H3-K9 methylation. These observations indicate that PRDM5 may form a repressive complex with histone modifiers on the hsa-mir-135b promoter, but that on the hsa-mir-229-5p promoter, PRDM5 actually offers protection against repressive histone modification.
DISCUSSION
Gfi1 regulates hematopoiesis, and its mutations in mice and humans cause hereditary forms of neutropenia (15). A yeast two-hybrid screen with its zinc fingers as bait recovered 29 interacting proteins, many of which are involved in sumoylation. Relevant to Gfi1's role as a transcriptional repressor, the sumoylation of histones opposes acetylation and is linked to transcriptional repression (43, 57). Nevertheless, evidence that Gfi1 is involved in sumoylation, or is itself sumoylated, is lacking. We therefore focused our studies, instead, on PRDM5, a tumor suppressor, about which little had been previously known (13). We confirmed the interaction of Gfi1 with PRDM5 by coimmunoprecipitation and demonstrated that their association requires the zinc fingers of Gfi1 and the first 10 zinc fingers of PRDM5.
PRDM5 additionally possesses a PR/SET domain and belongs to the PR domain-containing family of tumor suppressors. Seventeen family members are known, yet most remain uncharacterized. The most studied is PRDM1/Blimp1, involved in B-lymphocyte and macrophage (56), as well as germ cell (46), development. PRDM2/Riz is somatically mutated and inactivated in several different human cancers (4). PRDM3/Mds1/EVI1 and PRDM16 participate in leukemogenesis (44, 48, 58). PRDM6/PRISM promotes smooth muscle cell proliferation (12). Another family member, Meisetz, controls epigenetic events required for gamete formation during meiosis (24). PRDM5 is thought to have tumor suppressor activity, because its promoter contains a CpG island which becomes hypermethylated and leads to silencing of its expression in a variety of cancer cell lines (13). Moreover, PRDM5 resides on a region of human chromosome 4q26 often associated with loss of heterozygosity in breast, ovarian, liver, lung, colon, and other cancers (13).
The PR domain is a derivative of the SET domain, which serves as the catalytic center for HMTase activity. Some PR domain-containing proteins (i.e., PRDM2/RIZ (34) or H3-K4 HMTase (i.e., Meisetz [24]) possess intrinsic H3-K9 HMTase activity. Others, instead, recruit chromatin-modifying enzymes. PRDM1/Blimp1 associates with HDAC1, G9a (22), and Prmt5 (2). PRDM6/PRISM partners with class I HDACs, p300, and G9a (12). Schwann cell factor 1 (SC1) also contains a PR domain and interacts with class I HDACs 1, 2, and 3 (9). We could not detect intrinsic HMTase activity for PRDM5, but found that PRDM5 can bind G9a and class I HDACs and recruit them to the promoters of PRDM5 target genes, yielding H3-K9 methylation and H3 and H4 deacetylation, respectively.
Although PRDM5, as well as Gfi1 (64), functions as a transcriptional repressor in reporter assays employing synthetic constructs containing idealized binding sites, in vivo, each alone can either repress or activate the transcription of its target genes, as demonstrated here for PRDM5 by RNAi and previously for Gfi1 by RNAi (17) and mouse gene targeting (25, 62). Thus, whether PRDM5 or Gfi1 functions as a repressor or an activator is a function of both promoter and cell type. Recently, it was shown that Gfi1 could either activate or repress the MMP8 promoter, but that activation required that Gfi1 interact with C/EBPɛ, a binding site for which is also present on that promoter (33). Our analysis of microarray expression data on the effects of reduction of PRDM5 in HEK293 cells extends upon this observation. Given that the two transcription factors interact and demonstrate overlapping function with respect to the recruitment of histone-modifying enzymes, it comes as no surprise that PRDM5 and Gfi1 regulate an overlapping set of target genes. Unexpectedly, however, we noted that, with just one exception, all of the protein-coding and miRNA genes that were common targets for both PRDM5 and Gfi1 were activated, rather than repressed, by PRDM5. Thus, it appears that the occupancy of a promoter by both PRDM5 and Gfi1 leads to transcriptional activation, whereas each acting alone functions as a repressor. The fact that the same regions of PRDM5 that are involved in its interaction with Gfi1 (the proximal zinc fingers) are also required for its association with G9a and HDAC1 (and that the same is also true for Gfi1 [17]) suggests a potential mechanism: conceivably, PRDM5 and Gfi1 (or C/EBPɛ and Gfi1), when present together on a promoter, could associate, function as an activator, and prevent one another from interacting with repressive HMTases and deacetylases. Likewise, PRDM2/RIZ, also ordinarily a repressor, becomes an activator upon interaction with the estrogen receptor (5). Collectively, these findings may point to a general mechanism for how two interacting repressors may become an activator. In the case of PRDM5, however, Gfi1 may not be the only PRDM5-interacting protein capable of switching it from a repressor to an activator. Our observation that the promoter of hsa-mir-229-5p, which is activated by PRDM5 but does not appear to be a target of Gfi1, demonstrates increased H3-K9 methylation and reduced H3 acetylation when PRDM5 is suppressed, suggests that yet another factor can recruit repressive histone-modifying enzymes to PRDM5 targets, but only when PRDM5 is not present to block its association with them.
miRNAs play important roles in development and tumorigenesis (37), and several recent findings have shown that they are regulated by transcription factors involved in controlling the expression of protein-coding genes. c-Myc activates the expression of the hsa-mir-17-92 polycistron by directly binding to this locus (45). NFI-A and C/EBPα regulate the expression of miRNA-223 during granulopoiesis (18). Myogenic factors regulate the expression of muscle-specific miRNA genes (51, 54). We observed that PRDM5 binds to a large number of intronic miRNA gene promoters and recruits histone-modifying enzymes to them. In general, the regulation of miRNA genes by PRDM5 was independent of its influence on host gene expression. Since many of these miRNA genes are expressed in hematopoietic lineages, it is possible that PRDM5's regulation of miRNA expression contributes to control of hematopoiesis. In particular, hsa-mir-196 inhibits the HOXB8-induced granulocytic differentiation of HL-60 cells (32), and hsa-mir-196b is located upstream from HOXB9 (23), also implicated in granulocytic differentiation of HL-60 cells (47).
PRDM5 suppression by RNAi influenced the expression of many genes, but in the absence of specific ChIP data, we presume that these are indirect effects, attributable to PRDM5's regulation of other transcription factors or miRNA genes. Another explanation is that potential PRDM5 binding sites could be separated from the coding sequence by great linear genomic distances—for example, in the case of estrogen receptor binding sites, as identified through genome-wide analysis (6)—and then brought into proximity with each other in chromatin by looping or other mechanisms. Thus, knowing how far from a gene to look for a transcription factor's potential binding sites can be problematic, though a promising approach predicts transcription-binding neighborhood blocks based on the distribution of binding sites for the “insulator” protein CTCF (CCCTC binding factor) (35, 61).
Finally, we identified two individuals, with otherwise genetically unexplained neutropenia, who harbor amino acid missense substitutions in PRDM5 affecting transcriptional repressor activity in reporter assays. Family data were insufficient to differentiate polymorphism from mutation, but the construction of a mouse model could offer further relevance. We should note, however, that we generated Prdm5 mutant mice by utilizing commercially available (Lexicon Genetics) gene-trapped embryonic stem cells (1). No obvious phenotype, hematopoietic or otherwise, was apparent, but, unfortunately, the gene trapping occurred intronically, with consequent “leaky” expression of Prdm5. Moreover, gene-targeted mouse models of neutropenia-associated ELA2 mutations also demonstrate normal hematopoiesis (20), indicating that mouse models may lack relevance for human hereditary neutropenia.
Acknowledgments
This work was supported by NIH grants DK58161 (to M.S.H.), HL79507 (to M.S.H.), HL079574 (to H.L.G.), and CA112405 (to H.L.G.).
We thank W. Noble and S. Gupta for help with consensus sequence algorithms, G. Priestley for technical help, and M. Mealiffe for critical reading of the manuscript.
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Abuin, A., G. M. Hansen, and B. Zambrowicz. 2007. Gene trap mutagenesis. Handb. Exp. Pharmacol. 178:129-147. [DOI] [PubMed] [Google Scholar]
- 2.Ancelin, K., U. C. Lange, P. Hajkova, R. Schneider, A. J. Bannister, T. Kouzarides, and M. A. Surani. 2006. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 8:623-630. [DOI] [PubMed] [Google Scholar]
- 3.Bieda, M., X. Xu, M. A. Singer, R. Green, and P. J. Farnham. 2006. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 16:595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canote, R., Y. Du, T. Carling, F. Tian, Z. Peng, and S. Huang. 2002. The tumor suppressor gene RIZ in cancer gene therapy (review). Oncol. Rep. 9:57-60. [PubMed] [Google Scholar]
- 5.Carling, T., K. C. Kim, X. H. Yang, J. Gu, X. K. Zhang, and S. Huang. 2004. A histone methyltransferase is required for maximal response to female sex hormones. Mol. Cell. Biol. 24:7032-7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38:1289-1297. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. Z. 2005. MicroRNAs as oncogenes and tumor suppressors. N. Engl. J. Med. 353:1768-1771. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Z., and H. F. Lodish. 2005. MicroRNAs as regulators of mammalian hematopoiesis. Semin. Immunol. 17:155-165. [DOI] [PubMed] [Google Scholar]
- 9.Chittka, A., J. C. Arevalo, M. Rodriguez-Guzman, P. Perez, M. V. Chao, and M. Sendtner. 2004. The p75NTR-interacting protein SC1 inhibits cell cycle progression by transcriptional repression of cyclin E. J. Cell Biol. 164:985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choo, Y., and A. Klug. 1997. Physical basis of a protein-DNA recognition code. Curr. Opin. Struct. Biol. 7:117-125. [DOI] [PubMed] [Google Scholar]
- 11.Dale, D. C., R. E. Person, A. A. Bolyard, A. G. Aprikyan, C. Bos, M. A. Bonilla, L. A. Boxer, G. Kannourakis, C. Zeidler, K. Welte, K. F. Benson, and M. Horwitz. 2000. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood 96:2317-2322. [PubMed] [Google Scholar]
- 12.Davis, C. A., M. Haberland, M. A. Arnold, L. B. Sutherland, O. G. McDonald, J. A. Richardson, G. Childs, S. Harris, G. K. Owens, and E. N. Olson. 2006. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol. Cell. Biol. 26:2626-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, Q., and S. Huang. 2004. PRDM5 is silenced in human cancers and has growth suppressive activities. Oncogene 23:4903-4910. [DOI] [PubMed] [Google Scholar]
- 14.Duan, Z., and M. Horwitz. 2003. Gfi-1 oncoproteins in hematopoiesis. Hematology 8:339-344. [DOI] [PubMed] [Google Scholar]
- 15.Duan, Z., and M. Horwitz. 2005. Gfi-1 takes center stage in hematopoietic stem cells. Trends Mol. Med. 11:49-52. [DOI] [PubMed] [Google Scholar]
- 16.Duan, Z., and M. Horwitz. 2003. Targets of the transcriptional repressor oncoprotein Gfi-1. Proc. Natl. Acad. Sci. USA 100:5932-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan, Z., A. Zarebski, D. Montoya-Durango, H. L. Grimes, and M. Horwitz. 2005. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol. Cell. Biol. 25:10338-10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fazi, F., A. Rosa, A. Fatica, V. Gelmetti, M. L. De Marchis, C. Nervi, and I. Bozzoni. 2005. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 123:819-831. [DOI] [PubMed] [Google Scholar]
- 19.Gilks, C. B., S. E. Bear, H. L. Grimes, and P. N. Tsichlis. 1993. Progression of interleukin-2 (IL-2)-dependent rat T-cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol. Cell. Biol. 13:1759-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grenda, D. S., S. E. Johnson, J. R. Mayer, M. L. McLemore, K. F. Benson, M. Horwitz, and D. C. Link. 2002. Mice expressing a neutrophil elastase mutation derived from patients with severe congenital neutropenia have normal granulopoiesis. Blood 100:3221-3228. [DOI] [PubMed] [Google Scholar]
- 21.Grimwade, D., and E. Solomon. 1997. Characterisation of the PML/RAR alpha rearrangement associated with t(15;17) acute promyelocytic leukaemia. Curr. Top. Microbiol. Immunol. 220:81-112. [DOI] [PubMed] [Google Scholar]
- 22.Gyory, I., J. Wu, G. Fejer, E. Seto, and K. L. Wright. 2004. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 5:299-308. [DOI] [PubMed] [Google Scholar]
- 23.Harfe, B. D. 2005. MicroRNAs in vertebrate development. Curr. Opin. Genet. Dev. 15:410-415. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, K., K. Yoshida, and Y. Matsui. 2005. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 438:374-378. [DOI] [PubMed] [Google Scholar]
- 25.Hock, H., M. J. Hamblen, H. M. Rooke, J. W. Schindler, S. Saleque, Y. Fujiwara, and S. H. Orkin. 2004. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431:1002-1007. [DOI] [PubMed] [Google Scholar]
- 26.Hock, H., M. J. Hamblen, H. M. Rooke, D. Traver, R. T. Bronson, S. Cameron, and S. H. Orkin. 2003. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18:109-120. [DOI] [PubMed] [Google Scholar]
- 27.Horwitz, M., K. F. Benson, R. E. Person, A. G. Aprikyan, and D. C. Dale. 1999. Mutations in ELA2, encoding neutrophil elastase, define a 21-day biological clock in cyclic haematopoiesis. Nat. Genet. 23:433-436. [DOI] [PubMed] [Google Scholar]
- 28.Horwitz, M. S., and L. A. Loeb. 1986. Promoters selected from random DNA sequences. Proc. Natl. Acad. Sci. USA 83:7405-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jafar-Nejad, H., and H. J. Bellen. 2004. Gfi/Pag-3/senseless zinc finger proteins: a unifying theme? Mol. Cell. Biol. 24:8803-8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, G. L., and S. Huang. 2000. The yin-yang of PR-domain family genes in tumorigenesis. Histol. Histopathol. 15:109-117. [DOI] [PubMed] [Google Scholar]
- 31.Karsunky, H., H. Zeng, T. Schmidt, B. Zevnik, R. Kluge, K. W. Schmid, U. Duhrsen, and T. Moroy. 2002. Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30:295-300. [DOI] [PubMed] [Google Scholar]
- 32.Kawasaki, H., and K. Taira. 2004. MicroRNA-196 inhibits HOXB8 expression in myeloid differentiation of HL60 cells. Nucleic Acids Symp. Ser. 48:211-212. [DOI] [PubMed] [Google Scholar]
- 33.Khanna-Gupta, A., H. Sun, T. Zibello, H. M. Lee, R. Dahl, L. A. Boxer, and N. Berliner. 2007. Growth factor independence 1 (Gfi-1) plays a role in mediating specific granule deficiency (SGD) in a patient lacking a gene inactivating mutation in the C/EBP{epsilon} gene. Blood 109:4181-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, K. C., L. Geng, and S. Huang. 2003. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 63:7619-7623. [PubMed] [Google Scholar]
- 35.Kim, T. H., Z. K. Abdullaev, A. D. Smith, K. A. Ching, D. I. Loukinov, R. D. Green, M. Q. Zhang, V. V. Lobanenkov, and B. Ren. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128:1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim, T. H., L. O. Barrera, M. Zheng, C. Qu, M. A. Singer, T. A. Richmond, Y. Wu, R. D. Green, and B. Ren. 2005. A high-resolution map of active promoters in the human genome. Nature 436:876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kloosterman, W. P., and R. H. Plasterk. 2006. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 11:441-450. [DOI] [PubMed] [Google Scholar]
- 38.Lyst, M. J., X. Nan, and I. Stancheva. 2006. Regulation of MBD1-mediated transcriptional repression by SUMO and PIAS proteins. EMBO J. 25:5317-5328. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Matsuura, T., Y. Shimono, K. Kawai, H. Murakami, T. Urano, Y. Niwa, H. Goto, and M. Takahashi. 2005. PIAS proteins are involved in the SUMO-1 modification, intracellular translocation and transcriptional repressive activity of RET finger protein. Exp. Cell Res. 308:65-77. [DOI] [PubMed] [Google Scholar]
- 40.Matthews, J. M., and M. Sunde. 2002. Zinc fingers—folds for many occasions. IUBMB Life 54:351-355. [DOI] [PubMed] [Google Scholar]
- 41.McGhee, L., J. Bryan, L. Elliott, H. L. Grimes, A. Kazanjian, J. N. Davis, and S. Meyers. 2003. Gfi-1 attaches to the nuclear matrix, associates with ETO (MTG8) and histone deacetylase proteins, and represses transcription using a TSA-sensitive mechanism. J. Cell. Biochem. 89:1005-1018. [DOI] [PubMed] [Google Scholar]
- 42.Mo, Y. Y., and S. J. Moschos. 2005. Targeting Ubc9 for cancer therapy. Expert Opin. Ther. Targets 9:1203-1216. [DOI] [PubMed] [Google Scholar]
- 43.Nathan, D., K. Ingvarsdottir, D. E. Sterner, G. R. Bylebyl, M. Dokmanovic, J. A. Dorsey, K. A. Whelan, M. Krsmanovic, W. S. Lane, P. B. Meluh, E. S. Johnson, and S. L. Berger. 2006. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 20:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishikata, I., H. Sasaki, M. Iga, Y. Tateno, S. Imayoshi, N. Asou, T. Nakamura, and K. Morishita. 2003. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood 102:3323-3332. [DOI] [PubMed] [Google Scholar]
- 45.O'Donnell, K. A., E. A. Wentzel, K. I. Zeller, C. V. Dang, and J. T. Mendell. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435:839-843. [DOI] [PubMed] [Google Scholar]
- 46.Ohinata, Y., B. Payer, D. O'Carroll, K. Ancelin, Y. Ono, M. Sano, S. C. Barton, T. Obukhanych, M. Nussenzweig, A. Tarakhovsky, M. Saitou, and M. A. Surani. 2005. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436:207-213. [DOI] [PubMed] [Google Scholar]
- 47.Ohnishi, K., T. Tobita, K. Sinjo, A. Takeshita, and R. Ohno. 1998. Modulation of homeobox B6 and B9 genes expression in human leukemia cell lines during myelomonocytic differentiation. Leuk. Lymphoma 31:599-608. [DOI] [PubMed] [Google Scholar]
- 48.Ott, M. G., M. Schmidt, K. Schwarzwaelder, S. Stein, U. Siler, U. Koehl, H. Glimm, K. Kuhlcke, A. Schilz, H. Kunkel, S. Naundorf, A. Brinkmann, A. Deichmann, M. Fischer, C. Ball, I. Pilz, C. Dunbar, Y. Du, N. A. Jenkins, N. G. Copeland, U. Luthi, M. Hassan, A. J. Thrasher, D. Hoelzer, C. von Kalle, R. Seger, and M. Grez. 2006. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 12:401-409. [DOI] [PubMed] [Google Scholar]
- 49.Park, J. H., Q. Yu, B. Erman, J. S. Appelbaum, D. Montoya-Durango, H. L. Grimes, and A. Singer. 2004. Suppression of IL7Ralpha transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity 21:289-302. [DOI] [PubMed] [Google Scholar]
- 50.Person, R. E., F. Q. Li, Z. Duan, K. F. Benson, J. Wechsler, H. A. Papadaki, G. Eliopoulos, C. Kaufman, S. J. Bertolone, B. Nakamoto, T. Papayannopoulou, H. L. Grimes, and M. Horwitz. 2003. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat. Genet. 34:308-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao, P. K., R. M. Kumar, M. Farkhondeh, S. Baskerville, and H. F. Lodish. 2006. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. USA 103:8721-8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rathinam, C., R. Geffers, R. Yucel, J. Buer, K. Welte, T. Moroy, and C. Klein. 2005. The transcriptional repressor Gfi1 controls STAT3-dependent dendritic cell development and function. Immunity 22:717-728. [DOI] [PubMed] [Google Scholar]
- 53.Rodel, B., K. Tavassoli, H. Karsunky, T. Schmidt, M. Bachmann, F. Schaper, P. Heinrich, K. Shuai, H. P. Elsasser, and T. Moroy. 2000. The zinc finger protein Gfi-1 can enhance STAT3 signaling by interacting with the STAT3 inhibitor PIAS3. EMBO J. 19:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenberg, M. I., S. A. Georges, A. Asawachaicharn, E. Analau, and S. J. Tapscott. 2006. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J. Cell Biol. 175:77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salipante, S. J., K. F. Benson, J. Luty, V. Hadavi, R. Kariminejad, M. H. Kariminejad, N. Rezaei, and M. S. Horwitz. 2007. Double de novo mutations of ELA2 in cyclic and severe congenital neutropenia. Hum. Mutat. 28:874-881. [DOI] [PubMed] [Google Scholar]
- 56.Schebesta, M., B. Heavey, and M. Busslinger. 2002. Transcriptional control of B-cell development. Curr. Opin. Immunol. 14:216-223. [DOI] [PubMed] [Google Scholar]
- 57.Shiio, Y., and R. N. Eisenman. 2003. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 100:13225-13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stevens-Kroef, M. J., E. F. Schoenmakers, M. van Kraaij, E. Huys, S. Vermeulen, B. van der Reijden, and A. G. van Kessel. 2006. Identification of truncated RUNX1 and RUNX1-PRDM16 fusion transcripts in a case of t(1;21)(p36;q22)-positive therapy-related AML. Leukemia 20:1187-1189. [DOI] [PubMed] [Google Scholar]
- 59.Turner, D. L., and H. Weintraub. 1994. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8:1434-1447. [DOI] [PubMed] [Google Scholar]
- 60.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie, X., T. S. Mikkelsen, A. Gnirke, K. Lindblad-Toh, M. Kellis, and E. S. Lander. 2007. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. USA 104:7145-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng, H., R. Yucel, C. Kosan, L. Klein-Hitpass, and T. Moroy. 2004. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 23:4116-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, J., L. Guo, B. Min, C. J. Watson, J. Hu-Li, H. A. Young, P. N. Tsichlis, and W. E. Paul. 2002. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity 16:733-744. [DOI] [PubMed] [Google Scholar]
- 64.Zweidler-Mckay, P. A., H. L. Grimes, M. M. Flubacher, and P. N. Tsichlis. 1996. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 16:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]