Abstract
The methylation of specific lysine residues in histone H3 is integral to transcription regulation; however, little is known about how combinations of methylated lysine residues act in concert to regulate genome-wide transcription. We have systematically mutated methylated histone lysine residues in yeast and found that the triple mutation of H3K4, H3K36, and H3K79 to arginine (H3 K4,36,79R) is lethal. The histone H3 K4,36,79R mutant causes a mitotic cell cycle delay and a progressive transcription defect that initiates in telomere regions and then spreads into the chromosome. This effect is mediated by the silent information regulator (SIR) silencing complex, as we observe increased binding of the SIR complex to genomic regions adjacent to yeast telomeres in the H3 K4,36,79R mutant and deletion of SIR2, SIR3, or SIR4 rescues the lethal phenotype. Curiously, a yeast strain in which the histone methyltransferase genes are simultaneously deleted is viable. Indeed, deletion of the histone methyltransferase genes can suppress the H3 K4,36,79R lethal phenotype. These and other data suggest that the cause of lethality may in part be due to the association of histone methyltransferase enzymes with a histone substrate that cannot be methylated.
Eukaryotic chromosomes are organized into distinct domains of transcriptionally active euchromatin and repressive heterochromatin. These distinct chromatin domains profoundly influence the transcription, replication, repair, and segregation of their concomitant chromosomal sequences. Histone modifications, particularly histone lysine methylation, have important roles in initiating and maintaining these distinct chromatin domains. For example, methylation of histone H3 lysine-4 (H3K4), lysine-36 (H3K36), and lysine-79 (H3K79) directs the formation of euchromatin (reviewed in reference 21).
In the yeast Saccharomyces cerevisiae, the euchromatin-specific lysine residues are methylated by three histone lysine methyltransferases: Set1 (H3K4), Set2 (H3K36), and Dot1 (H3K79). These methyltransferases are recruited by the transcription elongation machinery, and their subsequent methylation of histone H3 is associated with actively transcribed genes (reviewed in references 31 and 33). However, while H3K4, H3K36, and H3K79 methylation is intricately coupled with transcription elongation, mutations in these residues or their cognate histone methyltransferases have relatively little effect on cell viability and gene expression. Deletion of SET1 results in decreased mRNA levels for a large number of genes (2, 27), in accordance with its proposed role in transcription elongation and euchromatin formation, but the magnitude of these transcriptional defects is relatively minor. In contrast, Set2-catalyzed methylation appears to primarily repress transcription by recruiting the Rpd3 histone deacetylase complex (3, 9, 11).
These studies suggest that histone H3 methylation plays only a modest role in transcription elongation and euchromatin formation in S. cerevisiae. We have investigated this possibility by systematically mutating the methylated lysine residues in histone H3. Surprisingly, we find that the triple mutation of H3K4, H3K36, and H3K79 to arginine (H3 K4,36,79R) is lethal in S. cerevisiae. Our data indicate that this lethal phenotype is not due to a defect in histone H3 stability or its incorporation in the nucleosome but is instead due to a progressive gene expression defect that initiates in telomeric heterochromatin and then spreads into the chromosome. We show that the H3 K4,36,79R mutation elicits the binding of the heterochromatin-associated silent information regulator (SIR) silencing complex (6) into regions of euchromatin adjacent to yeast telomeres. In addition, mutations in the SIR complex rescue this lethal phenotype. Curiously, mutations in the histone methyltransferase genes (e.g., SET1) also rescue the lethal H3 K4,36,79R mutant. These and other data suggest that the cause of lethality may in part be due to the association of histone methyltransferase enzymes with a histone substrate that cannot be methylated.
MATERIALS AND METHODS
Yeast strains and growth conditions.
A complete list of yeast strains used in this study is listed in Table S1 in the supplemental material (also see Y. Jin, A. M. Rodriguez, J. D. Stanton, A. A. Kitazono, and J. J. Wyrick, http://wyrick.sbs.wsu.edu/HistoneMethylation). Details of strain construction are available upon request. In general, yeast strains were propagated according to standard procedures in either rich medium (YPD) or in the appropriate selective medium (SC). The experimental procedures for yeast spotting, growth, viability, and cell cycle analysis are described in the supplemental material.
Plasmid construction and site-directed mutagenesis.
Details of plasmid construction are available upon request. Each histone H3 mutant was generated from plasmid pJW028 by site-directed mutagenesis (QuikChange kit, Stratagene). The set1-N1016Q and set1-C1068A mutants were generated from plasmid pJW048 (5) by site-directed mutagenesis. All mutants were confirmed by DNA sequencing. The complete list of mutagenic primer sequences is available at http://wyrick.sbs.wsu.edu/HistoneMethylation.
Genome-wide expression profiling.
Total RNA was isolated from each yeast culture. Equal amounts of five exogenous poly(A) control RNAs were added to each total RNA sample (8). The total RNA samples were used to prepare cDNA and biotin-cRNA, as previously described (20). The cRNA was then hybridized to a single S98 genome oligonucleotide array and scanned following standard protocols (Affymetrix). Intensities were captured using GeneChip software (Affymetrix), and a single raw expression level for each gene was determined. Complete data sets are available at http://wyrick.sbs.wsu.edu/HistoneMethylation/.
Data analysis.
The data from each chip were normalized using a normalization factor calculated from the signal intensities of five exogenous poly(A) controls, as previously described (8). A change in mRNA levels was deemed significant based on the following criteria: (i) the average change up or down was greater than twofold, (ii) the change (up or down) in each replicate experiment was greater than 1.5-fold, and (iii) the absolute intensity change was above background levels. See the report by Martin et al. (20) for more details.
Telomere-proximal gene analysis and statistics.
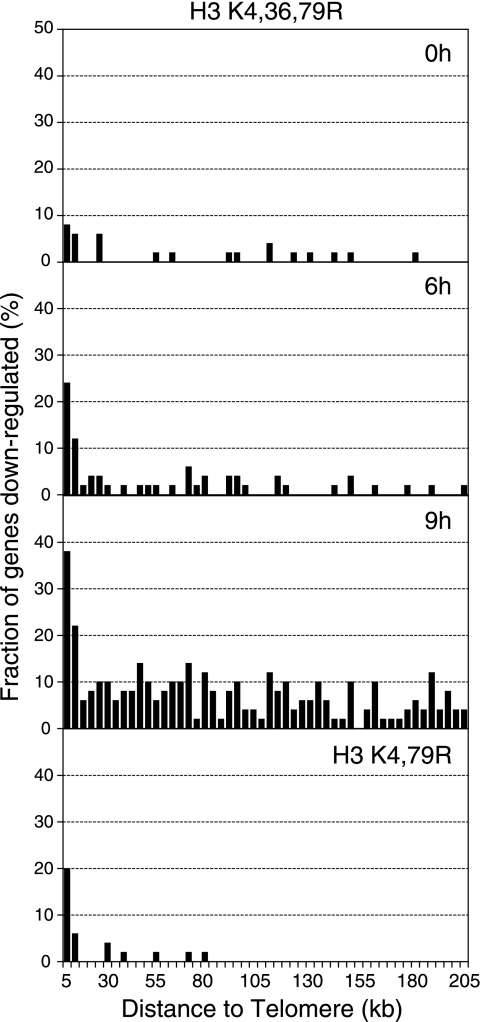
For the chromosome plots (see Fig. 3), all yeast genes were ordered according to their distance from a telomere and divided into consecutive 50-gene bins. The fraction of genes down-regulated in the mutant strain and the average distance of the genes from the telomere were plotted for each bin. Only those genes within ∼210 kb of a telomere were plotted. Statistical analysis of enrichment for telomere-proximal genes (0 to 10 kb from a telomere) was calculated using a hypergeometric probability distribution (20).
FIG. 3.
Effects of the histone H3 K4,36,79R mutant on genome-wide expression levels. Chromosome plots of the microarray data from the H3 K4,36,79R mutant at different time points following the depletion of wild-type histone H3. For each time point, a histogram of the fraction of genes whose mRNA levels are down-regulated in H3 mutants is plotted as a function of their distance from a chromosome end. The microarray data for the H3 K4,79R mutant are also shown for comparison.
Micrococcal nuclease digestion assay.
Micrococcal nuclease (MNase) digestion assays were performed as described previously (10). Briefly, yeast cells were grown and harvested under conditions similar to those used for genome-wide expression experiments. Cells were spheroplasted with Zymolyase (Zymo Research) and treated with doubling concentrations of MNase as indicated. After digestion, DNA was purified and resolved on a 1.5% agarose Tris-borate-EDTA (TBE) gel. The gels were stained with ethidium bromide and photographed.
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (17), with slight modifications. Briefly, yeast cells were grown under conditions similar to those for genome-wide expression experiments and cross-linked with 1% formaldehyde. Cells were lysed by glass beads and sonicated to shear the chromatin to fragment sizes of 150 to 400 bp. Cross-linked chromatin fragments were immunoprecipitated with anti-Myc antibody (catalog number AH00052; Biosource) or anti-Sir2 antibody (catalog number sc-6666; Santa Cruz Biotechnology) bound to magnetic beads (Dynal Biotech). After the cross-links were reversed, the DNA fragments were extracted for PCR. Taq DNA polymerase (New England Biolabs) and appropriate primer pairs were used in the PCR amplification reactions. PCR products were resolved on 2% agarose TBE gels stained with ethidium bromide and quantified using a GelDoc EQ imager with Quantity One software (Bio-Rad). The complete list of ChIP primer sequences is available at http://wyrick.sbs.wsu.edu/HistoneMethylation/.
Microarray data accession numbers.
The data discussed in this report have been deposited in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession numbers GSE6319, GSE6326, GSE6327, GSE6328, and GSE6331.
RESULTS
Systematic mutagenesis of histone H3 methylated lysine residues.
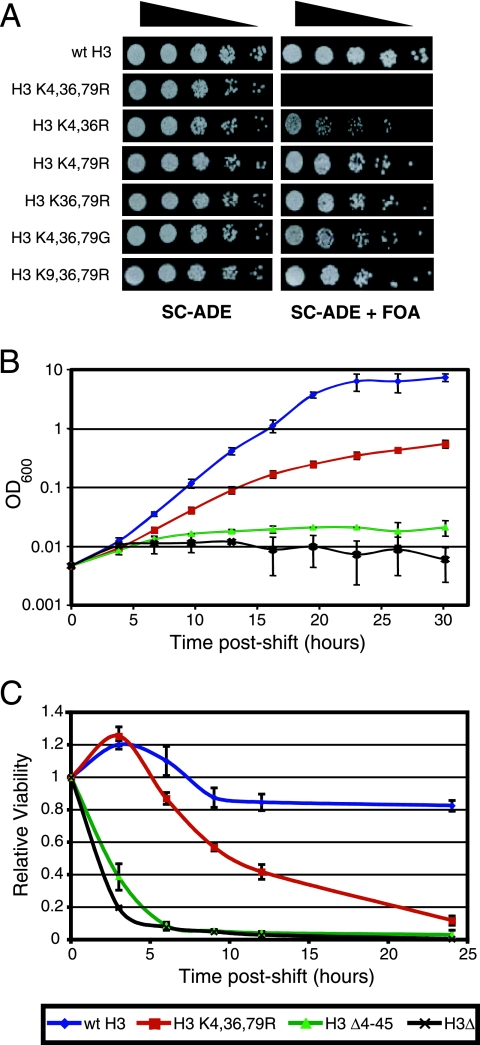
To investigate whether histone H3 methylated lysine residues have redundant functions in transcription elongation and chromatin organization, we systematically mutated H3K4, H3K36, and H3K79, both individually and in all possible combinations. The phenotype of each histone H3 mutant was tested by transforming a yeast strain (WY121) with a plasmid bearing the mutant H3 allele and subsequently removing the endogenous plasmid carrying the sole wild-type H3 allele by negative selection, using 5-fluoroorotic acid (5-FOA). The results showed that each of the H3 double-mutant strains (K4,36R, K4,79R, and K36,79R) was viable, though some mutants exhibited a slow-growth phenotype (Fig. 1A). However, the triple-mutant strain, in which all three methylated lysine residues were mutated to arginine (H3 K4,36,79R), was lethal (Fig. 1A).
FIG. 1.
The histone H3 K4,36,79R mutant strain is inviable. (A) The growth phenotypes of double and triple mutants in histone H3 methylated lysine residues were examined. Fivefold serial dilutions of each yeast strain (from about 104 cells) were spotted on SC-ADE medium or SC-ADE with 5-FOA as indicated. A wild-type (wt) histone H3 allele was present in each strain on SC-ADE plates but was lost from SC-ADE plates with 5-FOA. (B) Growth rates of histone mutant strains. The cell density of strains containing the H3 K4,36,79R, H3 Δ4-45, H3Δ, or wild-type alleles of histone H3 was monitored by measuring the optical density at different time points following the depletion of wild-type histone H3. (C) The viability for each of the mutant strains shown in panel B was determined by plating equal amounts of cells on galactose plates at different time points following the depletion of wild-type histone H3. The colonies formed on plates were scored after 72 h of incubation. Relative viability was calculated as the ratio of the colony number at certain time point to the 0-h time point. Each error bar represents the standard deviation of three independent experiments.
This lethal phenotype was observed only when all three methylated lysine residues were mutated to arginine. For example, the triple glycine mutant (H3 K4,36,79G) was viable (Fig. 1A). The arginine mutation is thought to mimic the hypomethylated lysine side chain, while the glycine mutation eliminates the side chain entirely, suggesting that the lethal phenotype may be due to the replacement of the methylated lysine residue with a hypomethylated mimic. Similarly, the H3 K9,36,79R mutant, in which two methylated lysine residues and one acetylated lysine residue (H3K9) were mutated to arginine, was viable (Fig. 1A). This result suggests that the lethal phenotype was likely to be specific to methylated H3 lysine residues.
To further investigate this lethal phenotype, we employed a histone depletion strain (WY151) in which the sole copies of the histone H3 and H4 genes were regulated by the GAL1-10 promoter (19). Plasmids bearing the histone H3 mutant alleles (and wild-type histone H4) under the control of their endogenous promoters were transformed into the histone depletion strain, and the resulting growth defects were examined by shifting the cells to glucose medium. The glucose shutdown represses transcription of the wild-type histone H3 gene, leaving the H3 mutant gene as the sole source of histone H3 in the cell.
The growth curve of the histone H3 K4,36,79R mutant is shown in Fig. 1B. The H3 K4,36,79R mutant displays a growth defect at 6 h postshift compared to the wild type, and this growth defect is exacerbated over time, until the cells gradually cease growing by 25 to 30 h postshift. This phenotype is in sharp contrast with the rapid growth arrest observed for the H3 Δ4-45 mutant, which deletes part of the histone H3 core domain (19), and for the histone H3 (and H4) depletion strain (H3Δ; Fig. 1B). We also measured the loss of viability of the histone H3 mutants by returning the glucose-treated cells to the permissive galactose medium at various time points postshift. Figure 1C shows that the H3 K4,36,79R mutant shows a gradual decrease in viability beginning at 6 to 9 h postshift and is almost completely inviable by 24 h. In contrast, the H3 Δ4-45 and H3 depletion mutants show a rapid loss of cell viability at 3 h postshift and are almost completely inviable by 6 h postshift. Taken together, these results argue that the lethality of the H3 K4,36,79R mutant is not due to the loss or instability of histone H3 protein, as is the case with the H3 Δ4-45 and H3 depletion mutants, as the severity and kinetics of the H3 K4,36,79R mutant growth defect are strikingly different from these other mutants.
The H3 K4,36,79R mutant is stable and competent for nucleosome assembly.
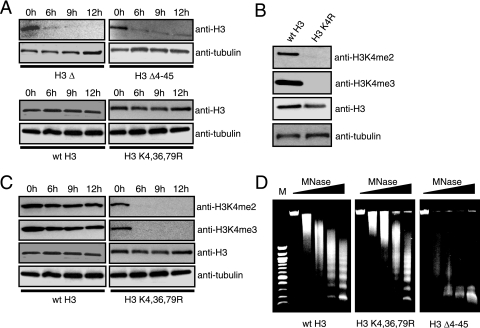
To directly test whether the H3 K4,36,79R mutant destabilizes the histone H3 protein, we measured H3 protein levels at consecutive time points following glucose shutdown of wild-type H3 expression. Western blotting analysis of yeast cell extracts indicated that histone H3 protein levels were relatively constant over the time course in the wild-type and H3 K4,36,79R mutant strains (Fig. 2A). In contrast, the H3 Δ4-45 and H3 depletion mutants showed a rapid loss in histone H3 protein. These data suggest that the H3 K4,36,79R mutant protein is stable in vivo; however, an alternative explanation for these data is that the wild-type H3 protein may continue to be expressed in the H3 K4,36,79R mutant strain, despite the presence of glucose in the medium.
FIG. 2.
The histone H3 K4,36,79R mutant protein is stable and competent for nucleosome assembly. (A) The protein stability of different histone mutants was measured by Western blotting analysis. Cell extracts from each of the mutants at four time points following the depletion of wild-type histone H3 were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed with anti-H3 antibody. As an internal loading control, anti-α-tubulin antibody was used. (B) Antibodies against di- and trimethylated histone H3K4 do not detect the histone H3 K4R mutant while antibody against histone H3 detects both wild-type histone H3 and the H3 K4R mutant. Cell extracts from wild-type and histone H3 K4R mutant strains were analyzed with antibodies as indicated. The level of the α-tubulin subunit was used as an internal loading control. (C) Loss of histone H3K4 methylation at different time points following the depletion of wild-type histone H3. Cell extracts from wild-type histone H3 and the H3 K4,36,79R mutant at different time points following the depletion of wild-type histone H3 were examined by Western blotting analysis with appropriate antibodies, as described in the panel B legend. (D) Genomic chromatin structure of the different histone mutants and wild-type strains was examined by MNase digestion assays. MNase digestion profiles for cells harvested at the 6-h time point following the depletion of wild-type histone H3 are shown as representative patterns. Subsequent lanes had doubling concentration of MNase from 0 to 37.5 units/ml. M, 100-bp DNA molecular size ladder.
To rule out this possibility, we measured the levels of H3K4 methylation in this strain over the glucose shutdown time course. As shown in Fig. 2B, the anti-H3K4 dimethylation (me2) and trimethylation (me3) antibodies readily detected methylation of wild-type histone H3, but no signal was detected in the H3 K4R mutant strain, as expected. Both the wild-type and the K4R mutant histone H3 proteins were detected by the anti-H3 antibody (Fig. 2B). Western blotting analysis of yeast cell extracts showed that while histone H3 is present at constant levels in the H3 K4,36,79R mutant over the glucose shutdown time course, H3K4 monomethylation, dimethylation, and trimethylation were almost completely absent by the 6-h time point (Fig. 2C and data not shown). We interpret this result to mean that the H3 K4,36,79R mutant protein had almost completely replaced the wild-type histone H3 by 6 h postshift. While it remains a formal possibility that the H3 wild-type histone is present but unmethylated at K4, this possibility seems unlikely, particularly since the wild-type H3 strain has normal H3K4 di- and trimethylation levels throughout the glucose shutdown time course (Fig. 2C).
To test whether the H3 K4,36,79R mutant can be assembled into the nucleosome, we used an MNase digestion assay to characterize the nucleosome content in the H3 mutant strain. MNase digestions were performed with the histone H3 K4,36,79R mutant at 6 h following the shift to glucose medium, when most of the cellular population of histone H3 is composed of the H3 K4,36,79R mutant protein (Fig. 2C). Comparison of the MNase digestion patterns of the H3 K4,36,79R mutant and the wild-type strains showed no significant alterations in nucleosome content or bulk genomic chromatin structure (Fig. 2D). In contrast, the H3 Δ4-45 mutant strain showed a general loss of nucleosome content (Fig. 2D), presumably due to a defect in nucleosome assembly. In summary, these data strongly suggest that the histone H3 K4,36,79R mutant protein is stable and competent for proper nucleosome assembly. However, these results do not rule out the possibility that the H3 K4,36,79R mutant may have more subtle effects on nucleosome positioning and chromatin structure, which would not be detected by the genomic MNase digestion assay.
The H3 K4,36,79R mutant causes gene repression, beginning with genes located in telomeric heterochromatin.
Previous studies have shown that the methylation of H3K4, H3K36, and H3K79 plays an important role in transcriptional regulation (33). To characterize the effects of the H3 K4,36,79R mutant on genome-wide transcription, we isolated RNA from parallel cultures of wild-type and H3 K4,36,79R mutant strains at 0, 6, and 9 h following the shift to glucose medium. The resulting genome-wide changes in mRNA levels were profiled using Affymetrix oligonucleotide arrays and are summarized in Table 1. The data for the 0-h time point indicate that the expression of many genes was altered prior to the shift to glucose medium, when both the wild-type and the mutant histone H3 proteins were expressed. This result suggests that even the partial replacement of wild-type histone H3 with the histone H3 K4,36,79R mutant protein can affect gene expression. Subsequent time points show that a general trend of progressively greater numbers of genes are down-regulated in the H3 K4,36,79R mutant, culminating in 361 genes with decreased expression levels by the 9-h time point.
TABLE 1.
Genome-wide expression changes in the H3 K4,36,79R mutant time course
| Time point (h) | No. of genes up-regulated | No. of genes down-regulated |
|---|---|---|
| 0 | 13 | 33 |
| 6 | 5 | 74 |
| 9 | 30 | 361 |
Chromosome plots of the genome-wide expression data showed that at each time point, a large fraction of the genes that were down-regulated in the H3 K4,36,79R mutant were clustered near yeast telomeres (Fig. 3). For example, 32% of the genes located in telomere-proximal regions (defined as a location within 10 kb of a telomere) showed decreased mRNA levels in the H3 K4,36,79R strain at the 9-h time point, compared to a genome-wide average of 6% (P = 9.7 × 10−13). Significant enrichment of telomere-proximal genes was also observed for the sets of down-regulated genes at the 0-h (P = 1.4 × 10−7) and 6-h time points (P = 4.2 × 10−16).
These data indicate that the H3 K4,36,79R mutant enhances telomeric gene silencing. Previous studies have shown that H3K4 and H3K79 methylation regulates telomeric silencing in yeast (31, 36), suggesting that the mutation of these residues alone may be responsible for the telomeric silencing effects observed for the H3 K4,36,79R mutant. To test this hypothesis, we used oligonucleotide arrays to profile the gene expression changes in the H3 K4,79R mutant. Analysis of the triplicate samples revealed that the mRNA levels of 61 genes were up-regulated and that the mRNA levels of 26 genes were down-regulated in the H3 K4,79R mutant, compared to that of the wild type. Further analysis shows that a significant fraction of the genes down-regulated in the H3 K4,79R mutant were also down-regulated in the H3 K4,36,79R mutant at the 6- and 9-h time points (data not shown). Nearly half of the genes down-regulated in the H3 K4,79R mutant are adjacent to yeast telomeres, a significant enrichment (P = 7.1 × 10−17). Indeed, the H3 K4,79R and H3 K4,36,79R mutants show strikingly similar effects of telomere-proximal gene expression (Fig. 3). In contrast, the H3 K36R mutant has a modest effect on telomere-proximal gene expression (data not shown). This analysis indicates that the initial increase in telomere silencing observed for the H3 K4,36,79R mutant time course is likely a consequence of the mutation of H3K4 and H3K79 to arginine.
At the 0-h and 6-h time points, relatively few genes outside of the telomere region are down-regulated in the H3 K4,36,79R mutant; however, at the 9-h time point, many genes in euchromatin regions of the genome are down-regulated (Fig. 3). We also profiled the gene expression changes at the 12-h and 18-h time points, and found that many more genes in euchromatin regions of yeast chromosomes were down-regulated at these later time points (data not shown). However, we are hesitant to draw conclusions from these data for two reasons. First, there is a considerable loss in the viability of the H3 K4,36,79R mutant at later time points (Fig. 1C). Hence, the gene expression changes we observed could have beeen an indirect consequence of cell death. Second, there is considerable variability in the microarray data for these later time points between replicate experiments.
In summary, the genome-wide expression data show the following trend: at the initial time points, the H3 K4,36,79R mutant represses the transcription of genes near telomere regions, while at later time points, the H3 K4,36,79R mutant-mediated gene repression begins to spread in the euchromatin regions of the chromosome. In contrast, the H3 K4,79R mutant represses gene transcription predominately at telomere proximal regions and shows little effect on the expression of genes located elsewhere.
The H3 K4,36,79R mutant leads to aberrant cell cycle progression.
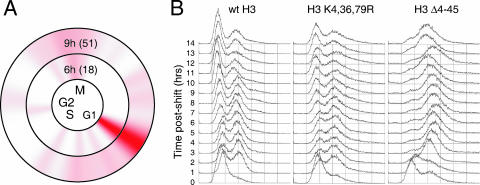
Cell cycle display of the H3 K4,36,79R mutant time course expression data revealed that a significant fraction of the genes down-regulated by the H3 K4,36,79R mutant are normally expressed during the G1 phase of the cell cycle (Fig. 4A). This result suggested the possibility that the H3 K4,36,79R mutant may have a cell cycle defect resulting in fewer cells cycling into G1 phase. We tested this hypothesis by performing fluorescence-activated cell sorter (FACS) analysis with propidium iodide-stained H3 K4,36,79R mutant cells at consecutive time points following the shift to glucose medium. The results indicated that there is a slight (∼10%) but reproducible decrease in the number of unbudded (G1-phase) cells in the H3 K4,36,79R mutant compared to that of the wild type (Fig. 4B), in accordance with the expression data. We also observed a broadening in the second (2N) peak for the H3 K4,36,79R mutant, which suggests slow progression through mitosis or cytokinesis in the mutant cells. The H3 Δ4-45 mutant, in contrast, shows an immediate G2/M arrest phenotype (Fig. 4B), which is indicative of nucleosome depletion (12). Similar results were obtained from FACS analysis of Sytox Green-stained cells (data not shown).
FIG. 4.
Cell cycle defects in the H3 K4,36,79R mutant strain. (A) Cell cycle display (32) of the genes down-regulated during the H3 K4,36,79R mutant time course. The circles represent the cell cycle expression patterns of 800 cell cycle-regulated genes (34). The intensity of red color indicates the number of genes that are normally expressed at that phase of the cell cycle which are down-regulated in the H3 K4,36,79R mutant strain. The total numbers of cell cycle genes that are down-regulated for each time point are listed in parentheses. (B) FACS analysis was used to measure the cell cycle profile of the H3 K4,36,79R mutant strain following depletion of wild-type (wt) histone H3.
At later time points (10 to 14 h postshift), there appeared to be significantly elevated numbers of rebudding cells in the H3 K4,36,79R mutant; we observed 11 to 16% multibudded cells in the H3 K4,36,79R mutant versus <2% multibudded cells in the wild type (see Fig. S1 in the supplemental material). We also observed more cells in anaphase in the H3 K4,36,79R mutant (see Fig. S1 in the supplemental material). These data suggest that the H3 K4,36,79R mutation causes mitotic defects, especially anaphase delay, in a small but significant subpopulation of the H3 K4,36,79R mutants.
The H3 K4,36,79R mutant enhances SIR complex binding to genomic regions adjacent to yeast telomeres.
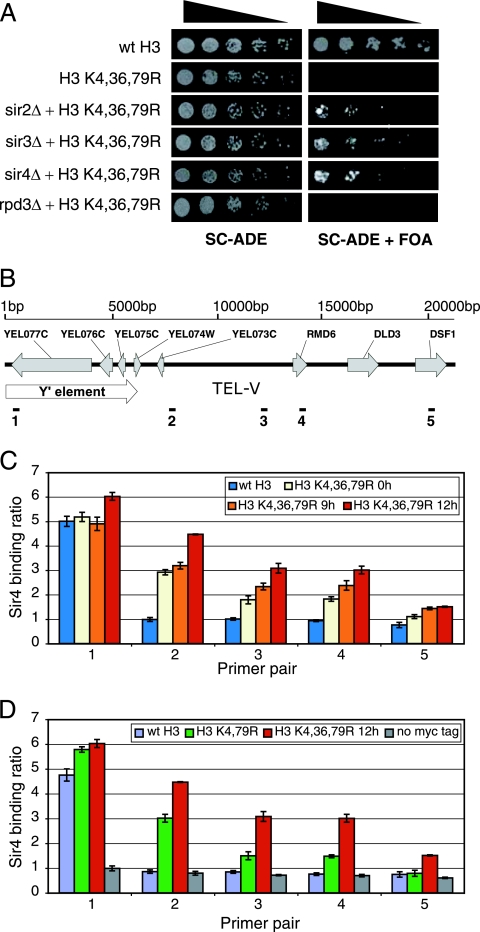
To gain insight into the mechanism underlying the H3 K4,36,79R lethality, we performed a genetic screen to identify extragenic mutations that suppressed the lethal phenotype. We isolated and cloned one of the suppressors, which partially rescued the lethal phenotype, and found that the suppressing mutation resided in the SIR3 gene. The suppressor mutant was a frameshift mutation caused by a single adenosine insertion into a stretch of 9 adenosines (nucleotides 1414 to 1422), resulting in the introduction of a premature stop codon in the Sir3 coding sequence following amino acid 485 (see Fig. S2 in the supplemental material). The addition of a wild-type allele of the SIR3 gene abolished suppression, demonstrating that the isolated sir3 suppressor mutant has a recessive, loss-of-function mutation (data not shown). The Sir3 protein is a component of the SIR complex, which silences gene expression at yeast telomeres and at the silent mating loci. We systematically deleted each subunit of the SIR complex (SIR2, SIR3, and SIR4) in the H3 K4,36,79R mutant strain and found that each of the deletions partially suppressed the H3 K4,36,79R lethal phenotype (Fig. 5A). In contrast, deletion of RPD3, which encodes a histone deacetylase whose association with chromatin is regulated by H3K36 methylation (3, 9, 11), did not suppress the H3 K4,36,79R lethal phenotype. In summary, these results indicate that the SIR complex is required for the lethality of the H3 K4,36,79R mutant.
FIG. 5.
The SIR silencing complex is required for the lethal phenotype of the histone H3 K4,36,79R mutant. (A) The growth of histone H3 K4,36,79R mutant strains lacking SIR2, SIR3, SIR4, or RPD3 was tested on SC-ADE plates with or without 5-FOA, as shown in Fig. 1A. (B) A schematic representation of the chromosome V-L telomere (TEL V-L) region. The location of the Y′ element, coding sequences, and ChIP primer pairs are depicted. (C and D) Increased binding by the SIR silencing complex near the chromosome V-L telomere region. The abundances of Sir4 at the TEL V-L region in wild-type strains and H3 mutants (and an untagged strain as control) were determined by ChIP analysis. The Sir4 binding ratio is shown as the ratio of each primer pair to the ACT1 primer pair. Each error bar represents the standard deviation of three independent experiments. Similar results were obtained with ChIP analysis of Sir2 (see Fig. S3 in the supplemental material).
This finding suggests that the transcriptional repression observed for the H3 K4,36,79R mutant may have been due, in part, to increased binding of the SIR silencing complex to genomic regions adjacent to telomeres. To test this possibility, we performed ChIP assays to measure the association of the Sir2 and Sir4 proteins with a series of DNA regions from 372 bp to 20,281 bp from telomere V-L (Fig. 5B) (16). For the Sir4 experiments, Sir4 was tagged with a 9-myc epitope, and anti-myc antibody was used for the ChIP assays (13); the Sir2 ChIP assays were performed using an anti-Sir2 antibody. ChIP assays were performed with the wild-type and the H3 K4,36,79R mutant strains at 0, 9, and 12 h following the shift to glucose medium. The results for the Sir4 ChIP experiments are shown in Fig. 5C. In the strain containing wild-type histone H3, strong Sir4 binding was detected at the most telomere-proximal DNA region (primer pair 1), but little if any binding was detected at DNA regions farther from the telomere (primer pairs 2 to 5). In contrast, in the H3 K4,36,79R mutant strain, considerable Sir4 binding is observed at more distant DNA regions (primer pairs 2 to 4). We observed substantial Sir4 binding even at the 0-h time point. This result is in accordance with the genome-wide expression data, which showed that a significant number of telomere-proximal genes are repressed in the H3 K4,36,79R mutant at the 0-h time point. The strength of the Sir4 interaction with these DNA regions increased at later time points. In addition, significant Sir4 binding was observed at the most distant DNA region assayed (primer pair 5) at the 9- and 12-h time points but not at the 0-h time point. Similar results were obtained with the Sir2 ChIP experiments (see Fig. S3 in the supplemental material). The increases in Sir2 and Sir4 occupancy at these distant DNA regions were not due to an increase in SIR protein, as Western blotting analysis indicates that Sir2 and Sir4 protein levels are constant throughout the time course (see Fig. S3C in the supplemental material).
We also compared the levels of binding of the SIR complex in the H3 K4,79R and H3K4,36,79R mutants. As shown in Fig. 5D, the binding of Sir4 to DNA regions was considerably stronger in the H3 K4,36,79R mutant than in the H3 K4,79R mutant strain. This was particularly apparent at the more distant DNA regions (primer pairs 3 to 5). These data indicated that mutating all three methylated lysine residues in histone H3 causes considerably greater binding of the SIR complex to adjacent genomic regions than that observed when only K4 and K79 are mutated.
Deletion of the histone methyltransferase genes suppresses the H3 K4,36,79R lethal phenotype.
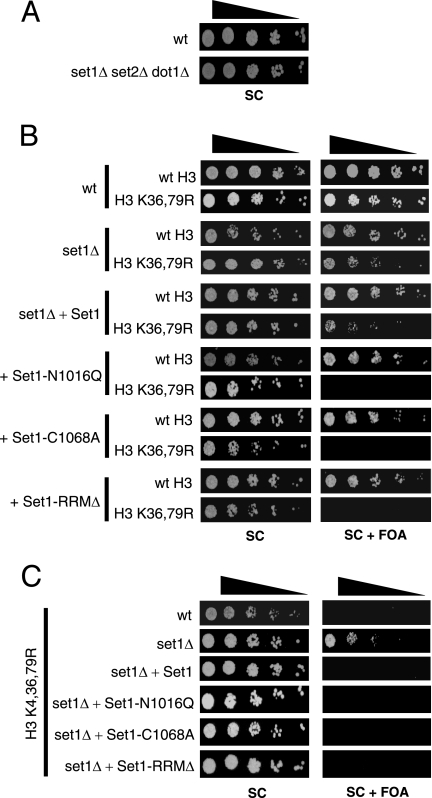
Because the arginine mutations are thought to mimic an unmethylated lysine side chain, we hypothesized that the H3 K4,36,79R mutant was lethal because it abolished histone H3 methylation. To test this hypothesis, we systematically deleted all three histone H3 lysine methyltransferases and characterized the phenotypes of the resulting set1Δ set2Δ dot1Δ mutant strain. Surprisingly, unlike the H3 K4,36,79R mutant, the triple methyltransferase mutant was viable (Fig. 6A). Phenotypic analysis of the set1Δ set2Δ dot1Δ mutant strain indicated that it grew normally under most growth conditions, though it showed caffeine sensitivity (see Fig. S4 in the supplemental material). This result calls into question whether the lethality of the H3 K4,36,79R mutant was due to the loss of lysine methylation.
FIG. 6.
Deletion of the histone methyltransferase genes suppresses the H3 K4,36,79R lethal phenotype. (A) Growth ability of the set1Δ set2Δ dot1Δ mutant strain. (B and C) Histone H3 mutant strains lacking Set1 methyltransferase were transformed with the wild-type SET1 or different set1 mutant alleles and then tested for viability, as shown in Fig. 1A.
To test this finding further, we constructed yeast strains containing combinations of methyltransferase deletions and histone H3 mutations. For example, deletion of SET1, which is required for H3K4 methylation, was not lethal in an H3 K36,79R mutant strain (Fig. 6B), as would be expected if the loss of H3 methylation were the cause of lethality in the H3 K4,36,79R mutant. Similar mutation combinations were constructed for the SET2 and DOT1 deletion strains, but none recapitulated the lethal phenotype of the H3 K4,36,79R mutant (data not shown).
In addition to the set1 deletion, we tested two different mutations in the Set1 active site (set1-N1016Q and set1-C1068A), which have been previously shown to render Set1 catalytically inactive (26, 28). Surprisingly, we found that both the set1-N1016Q and set1-C1068A mutations were lethal in combination with the H3 K36,79R mutation (Fig. 6B). The lethal phenotype of these set1 point mutants was specific to the H3 K36,79R mutant; the set1 point mutations were not lethal in combination with the H3 K4,79R or H3 K4,36R mutation or with the H3 K4,36,79G mutation (data not shown). We also tested the viability of strains containing the set1-RRMΔ mutant allele, in which the RNA recognition motif domain of Set1 (amino acids 230 to 335) had been deleted. Unlike the set1-N1016Q and set1-C1068A mutants, which eliminated all methylation of H3K4 (see Fig. S5A in the supplemental material), the set1-RRMΔ mutation only abolished H3K4 trimethylation and reduced H3K4 dimethylation, leaving H3K4 monomethylation intact (see Fig. S5B in the supplemental material), in accordance with previous studies (5, 29). The H3 K36,79R set1-RRMΔ mutant strain was also inviable (Fig. 6B). Again, the set1-RRMΔ mutation was not lethal in combination with the H3 K4,79R or H3 K4,36R mutation or with the H3 K4,36,79G mutation (data not shown).
Perhaps the simplest interpretation of these results is that the histone methyltransferase proteins (e.g., Set1) must be present in the cell for the loss of histone methylation to be lethal. If this hypothesis was correct, then we would expect that deleting the genes encoding the histone methyltransferase enzymes would rescue the H3 K4,36,79R lethal phenotype. To test this possibility, we measured the viability of the H3 K4,36,79R yeast strain in which the SET1 gene was deleted. As shown in Fig. 6C, deletion of SET1 rescued the lethal phenotype of the H3 K4,36,79R mutant, but this lethality was restored if a wild-type copy of the SET1 gene was added. Intriguingly, addition of any of the set1 mutants that abolished or disabled its methyltransferase activity (i.e., the set1-N1016Q, set1-C1068A, or set1-RRMΔ mutant) also restored lethality (Fig. 6C). This result indicates that the Set1 protein, but not its enzymatic activity, was required for the H3 K4,36,79R lethality. Deletion of the SET2 or DOT1 gene also rescued the lethal phenotype of the H3 K4,36,79R mutant, though the H3 K4,36,79R dot1Δ mutant strain was very slow growing (data not shown).
DISCUSSION
Understanding how methylated histone lysine residues act in concert to regulate gene expression requires the systematic genetic analysis of each lysine mutant combination. Here, we demonstrate that while single or double lysine-to-arginine mutations in H3K4, H3K36, and H3K79 have relatively modest effects on yeast cell viability, the H3 K4,36,76R triple mutant is inviable. This lethal phenotype does not appear to be the consequence of a structural defect in the histone H3 protein, as the H3 K4,36,79R mutant protein is stable and appears to be incorporated into nucleosomes at normal levels. Instead, we find that the H3 K4,36,79R mutant triggers a progressive gene transcription defect, which originates in yeast telomeric heterochromatin and then spreads into the euchromatin regions of the genome. The H3 K4,36,79R mutant also causes cell cycle delays that are characteristic of defects in mitosis and cytokinesis. We show that the lethal phenotype requires a functional SIR silencing complex and that the SIR complex displays increased binding to genomic regions adjacent to yeast telomeres in the H3 K4,36,79R mutant strain. Intriguingly, we also show that the presence of intact histone methyltransferase proteins is required for lethality of the H3 K4,36,79R mutant.
The phenotype of the histone H3 K4,36,79R mutant.
Genome-wide expression profiling of the H3 K4,36,79R mutant strain following depletion of wild-type histone H3 revealed an initial decline in the transcription of genes proximal to telomere regions. At later time points, even genes located in euchromatin regions of yeast chromosomes suffered a decline in transcription. These data support a model in which the decrease in the transcription of essential genes in the H3 K4,36,79R mutant strain causes cell lethality. The genome-wide expression data alone do not distinguish whether the decline in transcriptional activity is due to a defect in transcriptional activation or elongation or to an enhancement in transcriptional repression. The isolation of loss-of-function suppressor mutations in the SIR transcriptional silencing complex and our observation that the SIR complex binds to adjacent euchromatin regions in the mutant strain, however, indicate that the transcriptional defect is due at least in part to SIR-mediated transcriptional repression.
The H3 K4,36,79R mutant strain also displays defects in cell cycle progression, particularly in mitosis. While a fairly small subset of mutant cells (11 to 16%) show cell cycle irregularities during the time course, the observed defects are significantly enriched in the mutant strain compared to that of the wild type and could be a contributing factor to the H3 K4,36,79R lethal phenotype. Previous studies have shown that the Set1 methylation of Dam1 plays an important role in regulating chromosome segregation during mitosis (38). Our data indicate that Set1-catalyzed methylation of histone H3, in parallel with Set2- and Dot1-catalyzed methylation, is also vital for proper mitotic progression.
It is important to note that previous studies have linked the spread of the SIR complex with yeast growth and cell cycle defects. Deletion of the GCN5 and ELP3 histone acetyltransferase genes, which caused the spread of the SIR complex into subtelomeric heterochromatin, resulted in a yeast strain with multiple growth defects (14). A second study showed that overexpression of the SIR complex led to cell cycle defects in mitosis and a decrease in chromosome stability (7). Hence, it is possible that the cell cycle defects observed for the H3 K4,36,79R mutant may be a consequence of the aberrant spread of SIR-mediated heterochromatin.
Role of the SIR complex in the H3 K4,36,79R lethal phenotype.
We have isolated two distinct classes of mutants that suppress the H3 K4,36,79R lethal phenotype. These classes of suppressor mutants provide insight into the mechanism underlying the H3 K4,36,79R lethal phenotype. The first class of suppressor mutants occurs in components of the SIR silencing complex.
Previous studies have shown that the association of the SIR silencing complex with chromatin is regulated by the methylation of H3K4 and H3K79. Trimethylation of H3K4 and H3K79 blocks SIR association (22, 26), while hypomethylated H3K4 and H3K79 favor SIR association (22, 26). This model fits our data, as the lysine-to-arginine mutations in the H3 K4,36,79R mutant, which elicit SIR binding (Fig. 5), are thought to mimic hypomethylated lysine residues. In contrast, the lysine-to-glycine mutations in the H3 K4,36,79G mutant would be expected to disrupt SIR binding because they eliminate the hypomethylated lysine side chain. This would explain why the H3 K4,36,79G mutant is viable and why the H3 K4,36,79G mutant disrupts telomeric gene silencing instead of enhancing it (see Fig. S6 in the supplemental material). It is also possible, however, that the arginine mutants bind more strongly to the SIR complex than would hypomethylated lysine residues and thus cause an artificially high degree of SIR binding.
We find that the H3 K4,79R mutation alone causes only a modest increase in gene silencing, primarily confined to genes located within 10 kb of a telomere. Only when H3K36 is also mutated to arginine is the silencing of euchromatin genes observed. This conclusion is supported by the SIR binding data, which clearly demonstrate more extensive binding of the SIR complex farther into the chromosome in the H3 K4,36,79R mutant than in the H3 K4,79R mutant. These data suggest that H3K36 plays an important role in regulating telomeric silencing. Previous studies have shown that mutants in the Paf1 complex, which regulates H3K4 and H3K36 methylation, have silencing phenotypes (15, 23, 24). This finding is also in accordance with a recent study which showed that Set2 regulated SIR complex spreading (35). Our microarray data indicate that the H3 K36R mutation has a significant but modest effect on telomeric gene expression (data not shown).
Role of histone methyltransferase proteins in the H3 K4,36,79R lethal phenotype.
The second class of suppressor mutants we have identified are in the histone lysine methyltransferase genes themselves. Deletion of either SET1, SET2, or DOT1 rescued the H3 K4,36,79R lethal phenotype (though the dot1 suppressor mutant was very slow growing). In light of this finding, it is not surprising that the set1Δ set2Δ dot1Δ triple deletion also yields a viable yeast strain. However, Set1 mutations that eliminate or disable its methylation activity are lethal in combination with either the H3 K4,36,79R or the H3 K36,79R histone mutant. It is important to note that while the set1-N1016Q and set1-C1068A mutations disrupt Set1 catalytic activity, they do not affect the stability of the Set1 protein or the integrity of the Set1-associated COMPASS complex (28), unlike the SET1 deletion mutant (25).
We have yet to test the phenotypes of catalytically inactive mutations in Set2 and Dot1, as inactivating mutations in these methyltransferases have not been as well as studied as those in Set1. However, we have found that the deletion of CTK1, which is required for Set2-catalyzed methylation of H3K36 (37), is lethal in combination with the H3 K4,79R mutation (data not shown). The CTK1 deletion disrupts Set2 methyltransferase activity but not the integrity of the Set2 protein; hence, this result mirrors the effects seen for the Set1 catalytically inactive mutants.
In summary, we have shown that the histone methyltransferases are required for the lethality of the H3 K4,36,79R mutant. Perhaps the simplest explanation for this observation is what we call the “stymied methyltransferase” model. This model asserts that the lethal phenotype arises, in part, due to the association of histone methyltransferase enzymes with a histone substrate that cannot be methylated. Thus, this model would predict that the histone H3 lysine-to-arginine mutants and the catalytically inactive methyltransferase mutants are lethal because they prevent lysine methylation without disrupting the association of the methyltransferase with its substrate. On the other hand, mutations that disrupted this association (e.g., methyltransferase deletion or histone lysine-to-glycine mutations) would not be expected to be lethal, in accordance with our results. While it is not clear why a “stymied methyltransferase” would necessarily be lethal, it is intriguing to speculate that the Set1 protein may have a direct inhibitory function, as is the case for the yeast Kss1 mitogen-activated protein kinase (1, 4, 18, 30). For example, Set1 binding to histone substrates in the absence of histone methylation could inhibit gene transcription. Alternatively, Set1 could exert its inhibitory effect through its interaction with other substrates, such as Dam1 (38).
A second possible explanation for these genetic data is that proteins associated with the histone methyltransferases may have additional functions that are required for lethality. Deletion of the histone methyltransferase would disrupt these functions and suppress the lethal phenotype. For example, it is possible that other components of the COMPASS complex have additional functions, which may be compromised by the deletion of SET1. In any case, future studies are needed to test these hypotheses.
Supplementary Material
Acknowledgments
We thank Bill Davis, Ray Reeves, and Michael Smerdon for helpful discussions and comments on the manuscript. We thank Michael Grunstein for the generous gift of yeast strains and plasmids. We thank Scott Briggs for the gift of the Set1 plasmids.
This work was supported by American Cancer Society grant RSG-03-181-01-GMC. A.R. was supported by NIH postdoctoral fellowship GM074541-01 from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 30 July 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bardwell, L., J. G. Cook, D. Voora, D. M. Baggott, A. R. Martinez, and J. Thorner. 1998. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 12:2887-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boa, S., C. Coert, and H. G. Patterton. 2003. Saccharomyces cerevisiae Set1p is a methyltransferase specific for lysine 4 of histone H3 and is required for efficient gene expression. Yeast 20:827-835. [DOI] [PubMed] [Google Scholar]
- 3.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581-592. [DOI] [PubMed] [Google Scholar]
- 4.Cook, J. G., L. Bardwell, and J. Thorner. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390:85-88. [DOI] [PubMed] [Google Scholar]
- 5.Fingerman, I. M., C. L. Wu, B. D. Wilson, and S. D. Briggs. 2005. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J. Biol. Chem. 280:28761-28765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarente, L. 1999. Diverse and dynamic functions of the Sir silencing complex. Nat. Genet. 23:281-285. [DOI] [PubMed] [Google Scholar]
- 7.Holmes, S. G., A. B. Rose, K. Steuerle, E. Saez, S. Sayegh, Y. M. Lee, and J. R. Broach. 1997. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics 145:605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 9.Joshi, A. A., and K. Struhl. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20:971-978. [DOI] [PubMed] [Google Scholar]
- 10.Kent, N. A., and J. Mellor. 1995. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 23:3786-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, N. J. Thompson, C. Boone, A. Emili, J. S. Weissman, T. R. Hughes, B. D. Strahl, M. Grunstein, J. F. Greenblatt, S. Buratowski, and N. J. Krogan. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593-605. [DOI] [PubMed] [Google Scholar]
- 12.Kim, U. J., M. Han, P. Kayne, and M. Grunstein. 1988. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 7:2211-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 14.Kristjuhan, A., B. O. Wittschieben, J. Walker, D. Roberts, B. R. Cairns, and J. Q. Svejstrup. 2003. Spreading of Sir3 protein in cells with severe histone H3 hypoacetylation. Proc. Natl. Acad. Sci. USA 100:7551-7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 16.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 17.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 18.Madhani, H. D., C. A. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673-684. [DOI] [PubMed] [Google Scholar]
- 19.Mann, R. K., and M. Grunstein. 1992. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 11:3297-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin, A. M., D. J. Pouchnik, J. L. Walker, and J. J. Wyrick. 2004. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics 167:1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6:838-849. [DOI] [PubMed] [Google Scholar]
- 22.Ng, H. H., D. N. Ciccone, K. B. Morshead, M. A. Oettinger, and K. Struhl. 2003. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. USA 100:1820-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng, H. H., S. Dole, and K. Struhl. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278:33625-33628. [DOI] [PubMed] [Google Scholar]
- 24.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 25.Roguev, A., D. Schaft, A. Shevchenko, W. W. Pijnappel, M. Wilm, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20:7137-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos-Rosa, H., A. J. Bannister, P. M. Dehe, V. Geli, and T. Kouzarides. 2004. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 279:47506-47512. [DOI] [PubMed] [Google Scholar]
- 27.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 28.Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon, C. Weise, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12:1325-1332. [DOI] [PubMed] [Google Scholar]
- 29.Schlichter, A., and B. R. Cairns. 2005. Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. EMBO J. 24:1222-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz, M. A., and H. D. Madhani. 2004. Principles of MAP kinase signaling specificity in Saccharomyces cerevisiae. Annu. Rev. Genet. 38:725-748. [DOI] [PubMed] [Google Scholar]
- 31.Shilatifard, A. 2006. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75:243-269. [DOI] [PubMed] [Google Scholar]
- 32.Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi, T. L. Volkert, J. J. Wyrick, J. Zeitlinger, D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106:697-708. [DOI] [PubMed] [Google Scholar]
- 33.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 34.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tompa, R., and H. D. Madhani. 2007. Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex. Genetics 175:585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Leeuwen, F., and D. E. Gottschling. 2002. Genome-wide histone modifications: gaining specificity by preventing promiscuity. Curr. Opin. Cell Biol. 14:756-762. [DOI] [PubMed] [Google Scholar]
- 37.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, K., W. Lin, J. A. Latham, G. M. Riefler, J. M. Schumacher, C. Chan, K. Tatchell, D. H. Hawke, R. Kobayashi, and S. Y. Dent. 2005. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell 122:723-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.