Abstract
Allelic forms of DRG1/AFG2 confer resistance to the drug diazaborine, an inhibitor of ribosome biogenesis in Saccharomyces cerevisiae. Our results show that the AAA-ATPase Drg1 is essential for 60S maturation and associates with 60S precursor particles in the cytoplasm. Functional inactivation of Drg1 leads to an increased cytoplasmic localization of shuttling pre-60S maturation factors like Rlp24, Arx1, and Tif6. Surprisingly, Nog1, a nuclear pre-60S factor, was also relocalized to the cytoplasm under these conditions, suggesting that it is a previously unsuspected shuttling preribosomal factor that is exported with the precursor particles and very rapidly reimported. Proteins that became cytoplasmic under drg1 mutant conditions were blocked on pre-60S particles at a step that precedes the association of Rei1, a later-acting preribosomal factor. A similar cytoplasmic accumulation of Nog1 and Rlp24 in pre-60S-bound form could be seen after overexpression of a dominant-negative Drg1 variant mutated in the D2 ATPase domain. We conclude that the ATPase activity of Drg1 is required for the release of shuttling proteins from the pre-60S particles shortly after their nuclear export. This early cytoplasmic release reaction defines a novel step in eukaryotic ribosome maturation.
Biogenesis of the ribosomal subunits in Saccharomyces cerevisiae starts with transcription of the 35S pre-rRNA, which is a common precursor for the individual rRNAs for both the small and large ribosomal subunits. This pre-rRNA assembles with ribosomal and nonribosomal proteins to form a 90S preribosomal particle (for recent reviews on ribosome biogenesis, see references 5, 6, and 32). During maturation, preribosomal particles undergo substantial changes in protein composition, which are accompanied by a series of pre-rRNA processing events (see reference 5). Separation of the biogenesis pathways for the 40S and 60S subunits occurs when the 32S precursor rRNA is cleaved into the 20S and 27SA2 pre-rRNAs, the precursors for the small- and large-subunit rRNAs, respectively.
The maturation of 60S subunits involves a large number of nonribosomal factors, which are loaded onto and removed from the pre-60S particle in a sequential manner (21, 28). On the way from the nucleolus through the nucleoplasm to the cytoplasm, this maturation process can be characterized by the isolation of different pre-60S particles in which both pre-rRNA species and protein composition change (21). Maturation of the pre-60S subunit within the nucleus requires a number of different GTPases and at least two AAA (ATPases associated with a variety of cellular activities) proteins, Rix7 and Rea1 (8, 22). Since AAA-ATPases are regarded as specific chaperones, significant restructuring seems to be required during 60S maturation or for the formation of export-competent pre-60S particles.
Export of the pre-60S subunit through the nuclear pore requires the export adaptor Nmd3, which is a substrate for the exportin Xpo1/Crm1 (9, 13). The final maturation steps of the 60S subunit take place in the cytoplasm, where the last few nonribosomal proteins that accompany the particle have to be released. These shuttling proteins include Nmd3, Rlp24, Tif6, Arx1, and Alb1 (13, 17, 21, 28, 29). The release and recycling of shuttling proteins in the cytoplasm are crucial for ongoing ribosome biogenesis, since recycling defects lead to the depletion of preribosomal proteins from their site of action, resulting in pre-rRNA processing and assembly defects (16, 17, 29). Up to now, only the release mechanisms of Nmd3 and Tif6 were investigated in detail and linked to the function of the GTPases Lsg1 and Efl1 (12, 29).
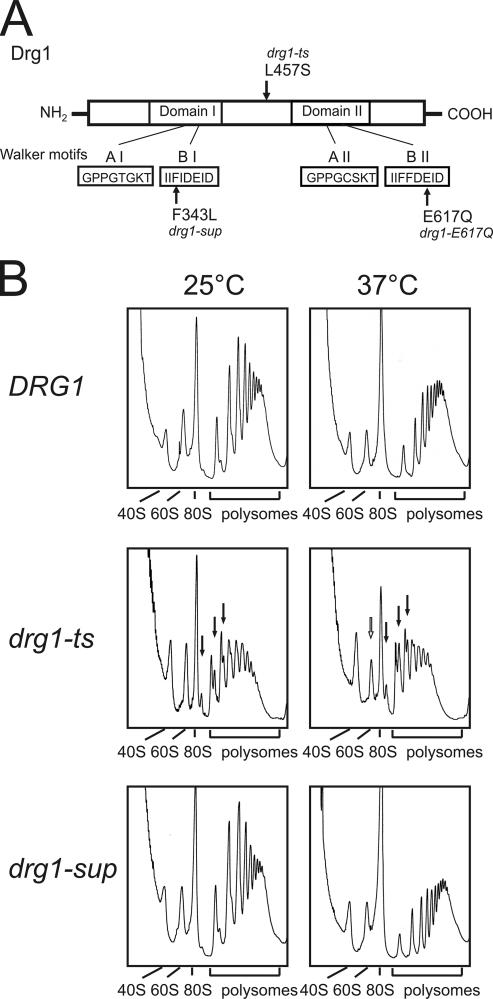
The S. cerevisiae DRG1 (diazaborine resistance gene 1) gene, alias AFG2 (ATPase family gene 2), encodes an essential AAA protein which is composed of two AAA domains (D1 and D2) (31, 35) (Fig. 1A). Drg1 is highly conserved between eukaryotes, sharing 48% identity with the human spermatogenesis-associated factor, SPAF (18). The purified Drg1 protein exhibits ATPase activity and forms hexamers in solution (37). A mutation which results in an L457S amino acid change causes the temperature-sensitive growth phenotype of the drg1-ts strain (afg2-18 allele) (23, 37). The amino acid change is located in the hinge region between the two AAA domains and affects the ATPase activity as well as the oligomeric structure of Drg1 (37). The thermosensitive growth phenotype of the drg1-ts strain can be suppressed by an additional mutation in the gene. This intragenic suppressor allele, designated drg1-sup, carries a second mutation resulting in the replacement of the conserved phenylalanine at position 343 in the Walker B box of D1 with leucine (37). Other allelic forms of DRG1 confer resistance to the drug diazaborine in yeast (35). Diazaborine inhibits maturation of the 60S ribosomal subunit by blocking 27SA2 pre-rRNA processing (24). This processing step is dependent on the nucleolar protein Nop4 (3, 30), which is relocalized from the nucleolus to the nuclear periphery upon diazaborine treatment. In the diazaborine-resistant DRG1-1 mutant, no inhibition of 60S biogenesis and no relocalization of Nop4 were observed (24).
FIG. 1.
The drg1-ts mutant shows ribosome half-mers in polysome profiles. (A) Schematic representation of protein Drg1. The amino acid changes leading to the thermosensitive phenotype, the suppressor phenotype, and the E617Q variant are marked by arrows. The Walker A and B motifs of the AAA domains D1 and D2 are indicated in the lower part of the diagram. (B) Polysome profiles of the wild-type strain W303, the drg1-ts mutant FWY111, and the drg1-sup suppressor mutant DTY4, incubated for 30 min at 37°C, are shown. Extracts were prepared, and 6.5 A260 units of each was loaded on 15 to 50% sucrose gradients as described in Materials and Methods. After ultracentrifugation for 2.5 h at 200,000 × g, polysome profiles were collected. Half-mer polysomes and reduced free 60S subunits observed in the drg1-ts mutant are indicated by filled and open arrows, respectively.
Here, we demonstrate that the AAA protein Drg1 is essential for pre-60S maturation, associates with cytoplasmic pre-60S particles, and is required for the release of several preribosome maturation factors at a very early cytoplasmic stage. Blocking this early maturation step allowed us to detect transient events, like the rapid shuttling of Nog1, that could not be seen under wild-type conditions. Our results directly connect an AAA protein involved in pre-60S formation with the release of shuttling proteins during structural remodeling of the nascent particles.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The yeast strains used in the present study are listed in Table 1. Chromosomal deletions or gene fusions were generated by homologous recombination using PCR products to transform the respective yeast strain. Alternatively, green fluorescent protein (GFP)- or tandem affinity purification (TAP)-tagged strains were obtained commercially, and the fusions were introduced into the desired strain background by classical yeast genetics. Strains were grown at different temperatures (25°C, 30°C, or 37°C) either in yeast extract-peptone-dextrose complex medium or, for metabolic labeling experiments or plasmid maintenance, in synthetic dextrose (SD) medium supplemented with the appropriate amino acids. Plasmids used in this study were pGZ252 (35), carrying the DRG1 gene in YEp351; pAZ7, carrying the glutathione S-transferase (GST)-Drg1 fusion in pYEX4-T1 (37); pAJ544 and pAJ368, overexpressing wild-type NMD3 or the dominant-negative nmd3-Δ100, respectively (a gift from A. W. Johnson); and pGFP-Nop4 (24) and pGFP-Drg1, which carry the respective wild-type genes in the centromeric GFP fusion vector pUG36 (a gift from J. H. Hegemann). The pGST-Drg1(E617Q) plasmid carries a dominant-negative DRG1 allele which contains CAA in codon 617 instead of GAA. This allele was generated by site-specific mutagenesis and cloned into pAZ7 by fragment swapping.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| Nonfusion strains | ||
| BY4741 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 | 4 |
| W303 | MATaade2 ura3-Δ0 his3-Δ1 leu2-Δ0 trp1 | S. D. Kohlwein |
| FWY111 | MATaade2 his3 leu2 trp1 ura3 afg2-18 | 37 |
| DTY4 | MATaade2 his3 leu2 trp1 ura3 drg1-sup | 37 |
| LMA596 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 KanMX6::PGAL1-DRG1 | This study |
| MNY8 | MATaura3 his3 leu2 trp1 crm1T539C | 20 |
| TAP fusions | ||
| YSC1178-7501205 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 lys2-Δ0 met15-Δ0 ALB1-TAP::HIS3MX6 | 10 |
| YSC1178-7499424 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 lys2-Δ0 met15-Δ0 REI1-TAP::HIS3MX6 | 10 |
| LMA158 | MATatrp1-289 ura3-52 ade2 leu2-3,112 arg4 NOG1-TAP::TRP1MX6 | 28 |
| LMA636 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 KanMX6::PGAL1-DRG1 NOG1-TAP::TRP1MX6 | This study |
| LMA160 | MATatrp1-289 ura3-52 ade2 leu2-3,112 arg4 RLP24-TAP::TRP1MX6 | 28 |
| GZY24 | MATα ade2 ura3-Δ0 leu2-Δ0 trp1 RLP24-TAP::TRP1MX6 | This study |
| GZY2418 | MATα ade2 ura3-Δ0 leu2-Δ0 trp1 afg2-18 RLP24-TAP::TRP1MX6 | This study |
| LMA611 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 KanMX6::PGAL1-DRG1 RLP24-TAP::TRP1MX6 | This study |
| LMA397 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 ARX1-TAP::HIS3MX6 | 17 |
| LMA609 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 KanMX6::PGAL1-DRG1 ARX1-TAP::HIS3MX6 | This study |
| GZYX | MATα ura3-Δ0 his3-Δ1 leu2-Δ0 trp1 ARX1-TAP::TRP1MX6 | This study |
| GZYX18 | MATα ura3-Δ0 his3-Δ1 leu2-Δ0 trp1 afg2-18 ARX1-TAP::TRP1MX6 | This study |
| Nsa3-TAP | MATaura3-Δ0 his3-Δ1 leu2-Δ0 trp1 NSA3-TAP::TRP1MX6 | 21 |
| Lsg1-TAP | MATaura3-Δ0 his3-Δ1 leu2-Δ0 trp1 LSG1-TAP::TRP1MX6 | 21 |
| GFP fusions | ||
| Nog1-GFP | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 NOG1-GFP(S65T)::HIS3MX6 | 14 |
| BNY1 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 NOG1-GFP(S65T)::HIS3MX6 | This study |
| BNY118 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 afg2-18 NOG1-GFP(S65T)::HIS3MX6 | This study |
| LMA401 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 ARX1-GFP(S65T)::HIS3MX6 | 17 |
| LMA606 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 KanMX6::PGAL1-DRG1 ARX1-GFP(S65T)::HIS3MX6 | This study |
| Tif6-GFP | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 TIF6-GFP(S65T)::HIS3MX6 | 14 |
| ELY1 | MATaade2 ura3-Δ0 his3-Δ1 leu2-Δ0 trp1 TIF6-GFP(S65T)::HIS3MX6 | This study |
| ELY118 | MATaade2 ura3-Δ0 his3-Δ1 leu2-Δ0 trp1 afg2-18 TIF6-GFP(S65T)::HIS3MX6 | This study |
| LMA607 | MATaura3-Δ0 his3-Δ1 leu2-Δ0 met15-Δ0 KanMX6::PGAL1-DRG1 NOG2-GFP(S65T)::HIS3MX6 | This study |
Fluorescence and electron microscopy.
For temperature shift experiments, strains were grown at 25°C in synthetic complete medium to the early log phase (A600, 0.3 to 0.6). The culture was split, and one aliquot was shifted to 37°C. After different times of incubation at the restrictive temperature, cells were inspected by fluorescence microscopy using a narrow-band enhanced GFP filter (Zeiss) on a Zeiss Axioskop microscope. Leptomycin B was provided by Alexis Biochemicals. Other epifluorescence microscopy experiments were performed as described previously (17). For electron microscopy, cells were fixed and prepared for immunocytochemistry as described previously (34). Immunolabeling was performed on ultrathin sections of Unicryl-embedded cells, using an anti-Drg1 antibody and gold-conjugated goat anti-rabbit antibodies (34). Area measurement of the compartments was performed using the MetaMorph software (Universal Imaging Corp.).
rRNA labeling and pulse-chase analyses.
For the pulse-chase analyses, strains were grown at 25°C to an A600 of 1.2 in SD medium. When cells were analyzed at the restrictive temperature, they were preincubated at 37°C for 90 min. Cells corresponding to 3.5 A600 units were harvested by centrifugation and resuspended in 1 ml SD medium. Then, 5 μCi [14C]uridine/ml was added. After 2 min, a 50-fold excess of nonradioactive uridine was added to the culture and 0.5-ml samples were removed after different times of incubation. Cells were suspended in 5 M guanidinium-HCl buffer and broken by vigorous vortexing with glass beads. RNA was extracted and transferred onto nylon membranes as described previously (24). After UV cross-linking, the radioactivity on the membrane was detected by phosphorimaging using a tritium screen (Kodak). To trace the formation of low-molecular-weight RNAs, RNAs were separated on 8% acrylamide-8 M urea gels. The gels were dried, and the radioactivity was detected by exposure of X-ray films.
Polysome analyses.
Strains were grown at 25°C in 100 ml yeast extract-peptone-dextrose medium to early log phase (A600 of 0.6), and subsequently cycloheximide (final concentration, 100 μg/ml) was added. When incubated at the nonpermissive temperature, the cells were shifted to 37°C for 30 or 120 min before cycloheximide treatment. Extract preparation and sucrose gradient centrifugation were performed as described previously (27).
Cell fractionation.
Cell extracts were obtained by homogenization of spheroplasts suspended in Ficoll buffer (18% Ficoll, 20 mM potassium phosphate buffer, pH 7.4, 0.5 mM MgCl2) in a Potter-Elvehjem cell disintegrator. After Potter disintegration, an equal volume of sorbitol buffer (2.4 M sorbitol, 20 mM potassium phosphate buffer, pH 7.4, 0.5 mM MgCl2) was added and the cell debris was removed by centrifugation at 3,000 × g for 10 min. Nuclei were harvested by centrifugation at 12,000 × g and 4°C for 30 min.
TAP purification.
Complexes were purified according to the standard TAP protocol (25), starting from 4 liters of yeast culture. The components of the fractions were analyzed by immunoblotting using polyclonal antibodies, after membrane staining with Ponceau red. For the preparation of tobacco etch virus (TEV) eluates only, 500 ml of yeast cultures grown to late log phase was used. Extracts were prepared in buffer A (20 mM HEPES-NaOH, pH 7.5, 10 mM KCl, 2.5 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and Complete protease inhibitor cocktail; Roche) and incubated with 40 μl of settled immunoglobulin G (IgG) beads (GE Healthcare) for 90 min at 4°C. Beads were washed three times with 1 ml of buffer A and once with 1 ml of buffer B (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, 0.075% NP-40). TEV elution was performed as described previously (25).
Western blotting.
Antibodies directed against Nog1, Rlp24 (28), Nog2 (27), Tif6 (29), and Arx1 and Rei1 (17) were described previously. Anti-Rpl1 antibody (a gift from F. Lacroute) was used at a 1:10,000 dilution, and anti-Drg1 rabbit polyclonal IgG was used at a 1:40,000 dilution. Polyclonal anti-Rpl16 antibody (a gift of S. Rospert) was used at a 1:16,000 dilution. GFP was detected with a polyclonal antibody (Sigma) at a 1:2,000 dilution. Secondary antibodies (goat anti-rabbit antibody-horseradish peroxidase conjugate from Bio-Rad) were used at a 1:10,000 dilution. The peroxidase activity was visualized with the Immobilon Western chemiluminescence kit (Millipore).
RESULTS
Functional Drg1 is required for 60S subunit formation.
Recently we demonstrated that the 60S ribosomal subunit formation defects caused by the drug diazaborine are compensated for by allelic forms of Drg1 (24). To test whether Drg1 plays a role in ribosome biogenesis, we obtained polysome profiles from the temperature-sensitive drg1-ts strain, the intragenic suppressor strain (drg1-sup), and the isogenic wild-type strain (Fig. 1A shows a schematic drawing of amino acid changes). As shown in Fig. 1B, the drg1-ts mutant already showed half-mers at the permissive temperature. Ribosome half-mers consist of mRNAs with a 43S translation initiation complex bound in addition to the already assembled 80S ribosome(s) and are often observed in mutants with defects in the maturation of the large ribosomal subunit. After 30 min of incubation at the restrictive temperature, the amount of half-mers increased and a reduction in free 60S subunits compared to 40S subunits was observed. After 2 h of incubation at 37°C, these effects were even more significant and the 80S ribosome peak was strongly reduced (data not shown). In contrast, the drg1-sup strain behaved similarly to the wild-type strain under all conditions (Fig. 1B and data not shown).
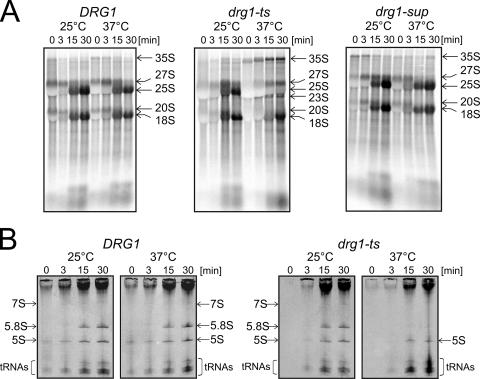
Pulse-chase analyses showed that the drg1-ts mutant exhibited a slightly slower formation of the 25S pre-rRNA even at 25°C compared to the wild-type strain (Fig. 2A; 15 min of chase). Incubation of the mutant at 37°C for 90 min resulted in a dramatic inhibition of the formation of 25S rRNA from its 27S pre-rRNA precursor (Fig. 2A). In addition, the formation of 7S pre-rRNA and 5.8S rRNA was blocked (Fig. 2B). These results show that the drg1-ts mutant is defective for processing of the 27S pre-rRNA to its products. Furthermore, the 35S precursor accumulated (Fig. 2A), indicating that in response to the inhibition of 27S pre-rRNA processing, a slowdown in early processing steps occurred. This assumption is supported by the appearance of a signal for the 23S RNA (Fig. 2A), an aberrant product of premature processing of the 35S pre-rRNA which is indicative of a slowdown in early processing steps. Formation of 23S RNA is frequently observed when processing of the 27S pre-rRNA is blocked (33). As a consequence of the buildup of 35S pre-rRNA, we detected a slightly slower formation of 20S pre-rRNA and 18S rRNA. In contrast to the drg1-ts strain, processing of the drg1-sup strain showed kinetics similar to those of the wild-type strain (Fig. 2A and data not shown).
FIG. 2.
Functional Drg1 is required for correct 27S pre-rRNA processing. Cells from the wild-type strain W303, the drg1-ts mutant FWY111, and the drg1-sup suppressor mutant DTY4 were incubated at 25°C and 37°C for 90 min, pulse-labeled with [14C]uridine for 2 min, and chased with a 50-fold excess of nonradioactive uridine for 0, 3, 15, and 30 min. RNA was extracted by the hot-phenol method. (A) The RNA was separated on 1.2% formaldehyde agarose gels to follow the processing of high-molecular-weight rRNA precursors. The gel was blotted, and the radioactivity was detected by phosphorimaging. (B) The RNAs of W303 (DRG1) and FWY111 (drg1-ts) were separated on 8% acrylamide-8 M urea gels to detect the formation of low-molecular-weight RNAs. The gel was dried, and radioactivity was detected by exposure of an X-ray film.
To define the exact step in 27S pre-rRNA processing that is blocked in the drg1-ts mutant, we performed Northern blotting and primer extension experiments (see Fig. S1 in the supplemental material). A slight accumulation of the 27SB pre-rRNA in the drg1-ts mutant at the permissive temperature and a significant decrease of the signal of a metastable processing intermediate of 25S synthesis at the restrictive temperature indicate that processing of the 27SB pre-rRNA at site C2 is affected in the drg1-ts mutant (see Fig. S1 in the supplemental material).
Drg1 is localized in the cytoplasm.
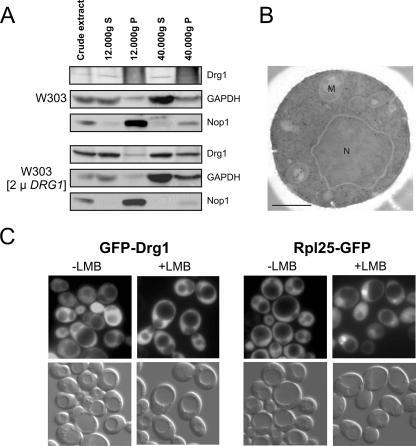
As the pre-rRNA processing steps blocked in the drg1-ts mutant are performed in the nucleus, we investigated the cellular distribution of Drg1 by cell fractionation, fluorescence microscopy, and immune gold labeling electron microscopy. Since chromosomally encoded Drg1 was barely detectable in crude extracts, we also performed the fractionations with cells overexpressing the wild-type gene. As shown in Fig. 3A, Drg1 was mainly present in the crude extract, in the 12,000 × g supernatant, and in the 40,000 × g supernatant representing the cytosol fraction. No signal for Drg1 could be seen in the nuclear preparation of the wild-type strain (12,000 × g pellet), and only a faint signal was observed in the overexpressing strain. Since a weak signal for glyceraldehyde-3-phosphate dehydrogenase was also found in the nuclear preparations, the Drg1 signal in the 12,000 × g pellet likely arises from contamination with cytosol. To investigate the localization of Drg1 in detail, we used electron microscopy and immune gold labeling (Fig. 3B). Because of the low abundance of Drg1, we did not obtain sufficient labeling with the wild-type strain. Therefore, we performed this experiment with the Drg1-overexpressing strain. The vast majority of gold grains (79.1%) were detected in the cytosol, 9.6% were in the nuclei, 7.3% were in the vacuole, and 4% were in the mitochondria as determined for 103 cells. Calculation of the mean label density by quantification and area measurement for the individual compartments in 53 cells that displayed nuclei, vacuole, and mitochondria gave values of 0.40, 0.46, and 0.47 gold particles per square μm, respectively. For the cytosol, we calculated a label density of 0.94 gold particles per square μm. Due to the similar label densities of nuclei, vacuole, and mitochondria, we cannot exclude nonspecific labeling by the antibody or overexpression-dependent mislocalization of Drg1. We therefore set out to test the significance of nuclearly located Drg1 by an independent approach.
FIG. 3.
Drg1 localization. (A) Cell fractionation. Crude extracts from the wild-type strain W303 and from W303 carrying the DRG1 gene on a 2μm plasmid (pGZ252) were fractionated by centrifugation. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted. Drg1, Nop1 (control for nuclear localization), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; control for cytosol) were detected in the resulting fractions by Western blot analysis using specific antibodies. Pellet fractions are marked by P, and supernatant fractions are marked by S. (B) Drg1 in an overexpressing strain visualized by immune gold labeling and electron microscopy. Nuclei and mitochondria are denoted with N and M, respectively. Bar, 0.5 μm. (C) The leptomycin-sensitive crm1 strain expressing GFP-Drg1 or Rpl25-GFP fusions was grown to mid-log phase and treated with 100 ng/ml leptomycin B (+LMB) for 1 h. Then cells were inspected by fluorescence microscopy for the localization of the fusion proteins (upper panels). In the lower panels, the corresponding Nomarski images are shown. An untreated culture of each strain served as control.
For this purpose, we constructed N- and C-terminal GFP fusions of Drg1. Both fusions complemented the drg1-null allele, although the C-terminal fusion showed slightly slower growth than did the wild-type strain (data not shown). The experiments shown below were therefore performed with the GFP-Drg1 fusion, although the same results were obtained with the C-terminal fusion (data not shown). Fluorescence microscopy showed that GFP-Drg1 localized in the cytoplasm (Fig. 3C, leftmost panel). The vacuole and the nucleus were devoid of the fusion protein.
All these experiments assessed the steady-state distribution of Drg1. To test if Drg1 might be a shuttling protein present at low levels within the nucleus, the GFP-Drg1 fusion was expressed in strain MNY8. This strain contains a T539C exchange in the exportin Crm1/Xpo1 which renders it sensitive to leptomycin B (20). Treatment of the sensitive strain with the inhibitor prevents export of preribosomal particles and causes their accumulation in the nucleus (13). As shown in Fig. 3C, the localization of GFP-Drg1 was not affected after 1 h of leptomycin B treatment, while a strong accumulation of the Rpl25-GFP control in the nucleus was observed. In addition, the GFP-Drg1 fusion did not accumulate in the nucleus upon 60S export inhibition by expression of the dominant-negative nmd3-Δ100 allele (13) (data not shown). These results strongly suggest that Drg1 is exclusively cytoplasmic. We conclude that the effects of the drg1-ts mutant on nuclear rRNA maturation are most probably due to a defect in the cytoplasmic function of the protein.
Drg1 binds to late pre-60S particles.
To gain further knowledge about the cytoplasmic role of Drg1 in ribosome biogenesis, we attempted TAP purifications of Drg1 and tested the eluate for the presence of preribosome maturation factors by Western blotting. Unfortunately, C-terminally TAP-tagged Drg1 was not functional. An N-terminal TAP fusion was able to complement the absence of Drg1. The purified complexes were enriched for all the pre-60S factors that we tested, with the exception of Mak11 (data not shown). Since the bait protein was overexpressed, some of these interactions could be nonspecific. That is why we did not make further use of this result.
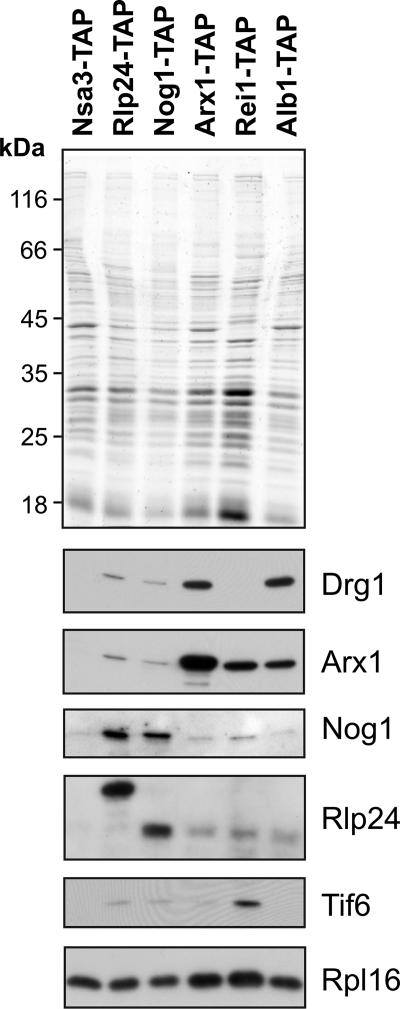
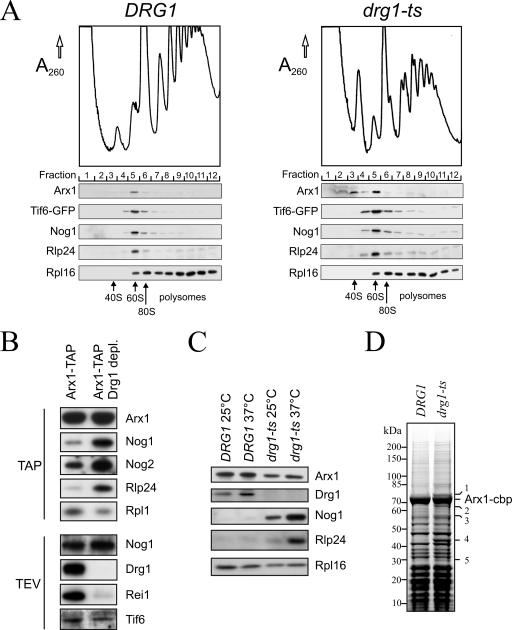
To test the binding of Drg1 to preribosomal particles with a different method that did not rely on tagged versions of Drg1, we screened for the presence of the protein in affinity purifications of factors involved in 60S maturation. For this purpose, we used TAP tag fusions of Nsa3, Rlp24, Nog1, Arx1, Alb1, and Rei1 as baits that are known to purify early, intermediate, and late pre-60S particles (17, 21). The isolated preribosomal particles were tested for the presence of Drg1 by Western blotting. As shown in Fig. 4, minor amounts of Drg1 were found in the Rlp24 and Nog1 purifications. This finding indicates that Drg1 binds only to a small subset of Rlp24- and Nog1-containing preribosomal particles. In contrast, a stronger signal for Drg1 was detected in the more mature Arx1- and Alb1-containing preribosomal particles. Arx1 joins the preribosomal particle within the nucleus and accompanies it into the cytoplasm (21). Alb1 was shown recently to bind to the preribosomal particle at a maturation step similar to the one at which Arx1 does (17). Surprisingly, no Drg1 was detected in the purification of the cytoplasmic pre-60S factor Rei1, although Rei1 is present in Arx1-containing pre-60S particles (17). This result suggests that Drg1 leaves the Arx1 particle before the arrival of Rei1. Consistent with this hypothesis, affinity purifications with TAP-tagged Lsg1, a protein that joins the preribosomal particle late in the cytoplasm, did not contain Drg1 (data not shown).
FIG. 4.
Drg1 interacts with late preribosomal particles. Cultures of strains expressing TAP-tagged versions of Nsa3, Rlp24, Nog1, Arx1, Rei1, and Alb1 were grown to late log phase, and the proteins were affinity purified on IgG beads as described in Materials and Methods. Aliquots of the TEV eluates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted. Western blot analyses with polyclonal antibodies directed against Drg1, Arx1, Nog1, Rlp24, and Tif6 are shown below the Coomassie blue-stained gel. A Western blot with a polyclonal antibody directed against the ribosomal protein Rpl16 served as loading control.
All these results suggest that Drg1 binds to a subpopulation of preribosomal particles that contain Arx1, Alb1, Rlp24, and Nog1 but lack Nsa3, Rei1, and Lsg1. Interestingly, Drg1 has a cytoplasmic localization but is present in complexes purified in association with Nog1, a nuclear factor previously not known to shuttle between the nucleus and the cytoplasm. These results suggest that Nog1 accompanies the preribosomal particle into the cytoplasm where it quickly has to be released and reimported.
Inactivation of Drg1 results in cytoplasmic accumulation of Nog1, Rlp24, Arx1, and Tif6.
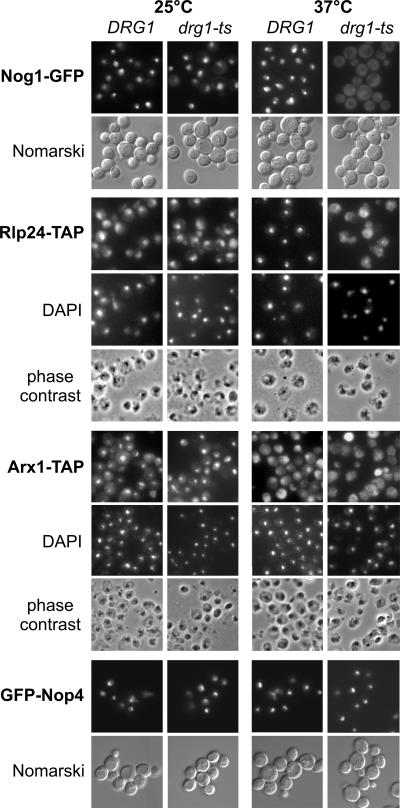
As Drg1 is cytoplasmic, but 27SB pre-rRNA processing, which is inhibited in the drg1-ts mutant, is a nuclear event, the observed defects have to be the consequence of a later, cytoplasmic disturbance in pre-60S maturation. One plausible model for the function of Drg1 would be that the protein promotes the release or recycling of pre-60S maturation factors from the cytoplasm back into the nucleus. To test this hypothesis, we wanted to investigate whether the localization of proteins involved in pre-60S maturation is different in the drg1-ts mutant. In contrast to the localizations of Nop1, Nop4, Ssf1, Drs1, Rlp7, and Nop7, which were similar to those of the wild type, Nog1-GFP, Rlp24-TAP, and Arx1-TAP were relocalized to the cytoplasm in the drg1-ts mutant after 2 hours of incubation at 37°C (Fig. 5 and data not shown). In addition, some Tif6-GFP also mislocalized to the cytoplasm in the drg1-ts strain after incubation at 37°C, but this effect was less pronounced than that for the other proteins (data not shown).
FIG. 5.
Nog1, Rlp24, and Arx1 accumulate in the cytoplasm of the drg1-ts mutant after incubation at the restrictive temperature. Nog1-GFP, Rlp24-TAP, Arx1-TAP, and GFP-Nop4 fusion proteins were expressed in the wild-type strain and the drg1-ts strain. The strains were grown to early log phase at 25°C and incubated for 2 hours at 37°C before they were analyzed by fluorescence microscopy (GFP constructs) or fixed and processed for indirect immune fluorescence and DAPI (4′,6′-diamidino-2-phenylindole) staining (TAP constructs). The indirect immune fluorescence assay was performed according to the method in reference 17 using a polyclonal antibody directed to protein A and a rhodamine-conjugated secondary antibody.
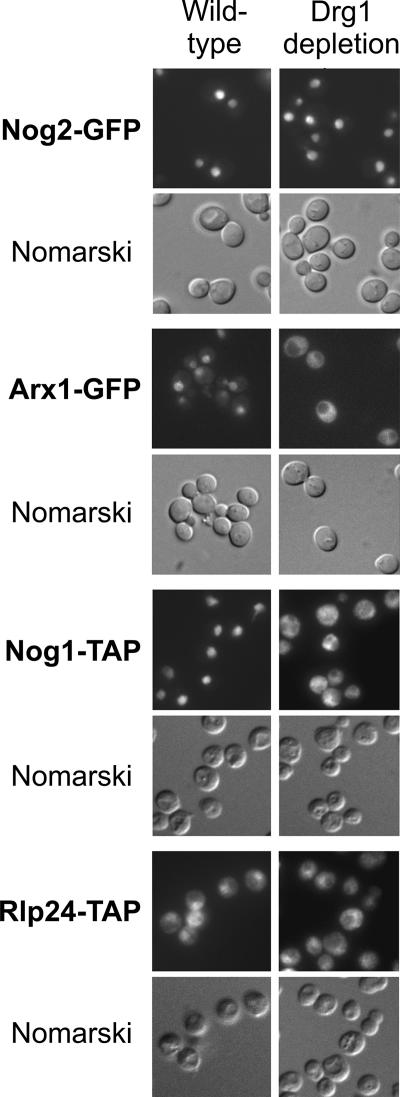
Since elevated temperature caused retention of Rlp24 in the nucleolus of the wild-type strain (Fig. 5), we tested the influence of Drg1 depletion on shuttling protein localization in a system independent of a temperature shift. For this purpose, we constructed a strain that conditionally expressed the DRG1 gene under the control of the GAL1 promoter. Upon shift to glucose-containing growth medium, the expression of DRG1 is repressed. As a consequence, this strain stopped growing after 16 h of incubation in glucose-containing medium and showed half-mers when tested by sucrose gradient ultracentrifugation (data not shown). As in the temperature-sensitive mutant, a shutdown of DRG1 expression resulted in the accumulation of Nog1-TAP and Arx1-GFP in the cytoplasm within 20 h, while no mislocalization of Nog2-GFP was observed (Fig. 6). The effects on Rlp24-TAP localization were less obvious than in the case of the drg1-ts mutant. A complete relocalization of Tif6-GFP to the cytoplasm could also be detected after 30 h of repression (data not shown). These results strongly suggest a role of Drg1 in the recycling of pre-60S factors, probably by direct interaction with the nascent particles in the cytoplasm. Arx1 and Tif6 remain bound to pre-60S particles until a late maturation step in the cytoplasm and are still found in Rei1-TAP purifications (Fig. 4), when Drg1 has already left the preribosomal particles. For this reason, Arx1 and Tif6 are probably not the direct targets of Drg1.
FIG. 6.
Nog1 and Arx1 accumulate in the cytoplasm when Drg1 is depleted. Nog1-TAP (LMA636), Rlp24-TAP (LMA611), Arx1-GFP (LMA606), and Nog2-GFP (LMA607) fusion proteins were produced in strains expressing DRG1 under the control of the GAL1 promoter. Strains were grown in galactose-containing medium and shifted to glucose-containing synthetic medium for 20 h at 30°C to shut down expression of DRG1. The corresponding wild-type strains were grown in glucose-containing synthetic complete medium at 30°C. Thereafter, Nog2-GFP- and Arx1-GFP-expressing strains were examined by epifluorescence microscopy. Nog1-TAP- and Rlp24-TAP-expressing cells were fixed and processed for immune fluorescence using an anti-protein A antibody as described previously (17). Fluorescence and Nomarski images for each strain are shown.
Drg1 is essential to release preribosome maturation factors from late pre-60S particles.
To further test whether Drg1 is involved in pre-60S maturation factor recycling, we investigated whether Nog1, Rlp24, Arx1, and Tif6 accumulate in soluble or ribosome-bound form in the cytoplasm of the drg1-ts strain. If inactivation of Drg1 blocked import of these factors, they should accumulate in the cytoplasm in soluble form and sediment in the upper fractions of sucrose gradients. In contrast, a defect in their release would result in sedimentation in the fractions around the 60S peak. We used a Tif6-GFP-expressing strain to facilitate detection of Tif6. As shown in Fig. 7A, Nog1, Rlp24, and Tif6-GFP sedimented in the range of the 60S peak in both wild-type and mutant strains incubated at 37°C. Similar results were obtained after depletion of Drg1 (data not shown). These results indicate that Nog1, Rlp24, and Tif6 remain bound to the preribosomal particle and fail to be released upon Drg1 inactivation. Thus, the cytoplasmic accumulation of Nog1, Rlp24, and, to a smaller extent, Tif6 in the drg1-ts mutant that we detected by fluorescence microscopy does not result from a defect in import of these proteins into the nucleus but from a defect in releasing them from the preribosomal particles in the cytoplasm. In the case of Arx1, most of the protein was detected in the 60S fractions in the drg1-ts mutant, but a small amount of the protein was found to be soluble. Previously we showed that Rei1 depletion leads to the accumulation of a small Arx1-containing complex (17). We assume that the altered sedimentation behavior of Arx1 in the drg1-ts strain is due to the fact that Rei1 pre-60S association is lost upon Drg1 inactivation (Fig. 7B).
FIG. 7.
Nog1, Rlp24, Arx1, and Tif6 remain associated with pre-60S particles in the drg1-ts mutant and upon depletion of Drg1. (A) Polysome profiles from the temperature-sensitive drg1-ts mutant (ELY118) and the isogenic wild-type strain (ELY1) expressing the Tif6-GFP fusion are shown. The strains were grown at 25°C and shifted to 37°C for 2 h. Extracts were prepared, and 6.5 A260 units each was loaded on 15 to 50% sucrose gradients. After ultracentrifugation for 2.5 h at 200,000 × g, polysome profiles were recorded, and fractions of 800 μl were collected. The proteins present in the individual fractions were precipitated with trichloroacetic acid, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and blotted. Western blot analyses with polyclonal antibodies directed against Arx1, GFP, Nog1, Rlp24, and the ribosomal protein Rpl16 are shown. (B) TAP purification of Arx1 containing preribosomal particles after depletion of Drg1. The wild-type strain (LMA397) and the PGAL1::DRG1 (LMA609) strain expressing Arx1-TAP fusions were grown in galactose-containing synthetic medium. Mid-log cells were transferred to glucose-containing synthetic medium for 19 h to deplete cells of Drg1. Thereafter, TAP purification was performed as described previously (25). Proteins in the eluates were separated by SDS-PAGE and blotted. Western blotting results with antibodies directed against Arx1, Drg1, Rei1, Nog2, Nog1, Rlp24, Tif6, and the ribosomal protein Rpl1 are shown either on TEV eluates or on TAP purifications. (C) The wild-type and drg1-ts strains expressing chromosomally TAP-tagged Arx1 were grown to early log phase and incubated for 60 min at 37°C. After cell lysis, Arx1-containing preribosomal particles were affinity purified on IgG beads and eluted by TEV cleavage. Samples were analyzed by SDS-PAGE and Western blotting. Twice as much eluate was loaded from the wild-type strain to facilitate detection and quantification of Nog1 and Rlp24. (D) Arx1-containing preribosomal particles were TAP purified from the drg1-ts mutant and the isogenic wild-type strain after incubation at 37°C for 1 h. Proteins in the eluates were separated on a 4 to 12% gradient sodium dodecyl sulfate-polyacrylamide gel and stained with Coomassie blue. Prominent proteins present in increased amounts in the eluate from the mutant are indicated by numbers and were identified by mass spectrometry.
A defect in Nog1 and Rlp24 release from pre-60S particles should result in increased levels of these proteins in purifications of late preribosomal particles. As shown in Fig. 7B, depletion of Drg1 resulted in increased levels of Nog1, Rlp24, and Nog2 in Arx1-TAP purifications compared to the signal for the ribosomal protein Rpl1. The increase in Nog2 amounts seems rather surprising, given the observed nuclear signal obtained with Nog2-GFP under similar conditions (Fig. 6). However, we cannot totally exclude the possibility that a minor fraction of Nog2 could also be present in the cytoplasm. Alternatively, a small fraction of Arx1 might be blocked in the nucleus in Nog2-containing complexes. Interestingly, the copurifying amount of Rei1 was strongly decreased, indicating that Rei1 is not associated stably with the pre-60S complexes in Drg1-depleted strains. These results show that functional Drg1 is required to promote the release of the shuttling proteins Nog1 and Rlp24 and allows the transition of the preribosomal particles to later maturation forms that bind Rei1.
To distinguish if the defect in release of shuttling proteins in the drg1-ts strain results from a failure of the altered protein to bind to the preribosomal particles or if the protein can still bind but not carry out its function, we affinity purified Arx1-TAP from the mutant. As in the case of Drg1 depletion, incubation of the thermosensitive drg1-ts mutant at 37°C resulted in a strong enrichment of Nog1 and Rlp24 in the Arx1-TAP purification (Fig. 7C). Slightly increased amounts of Nog1 and Rlp24 were detected in the Arx1-TAP preparation from the mutant already at the permissive temperature. This finding correlates with our results showing that at the permissive temperature the drg1-ts mutant had half-mers and slightly slower pre-rRNA processing (Fig. 1B and 2). Reduced amounts of Drg1-ts protein copurified with Arx1 from the mutant even when grown at 25°C. As expression levels of Drg1 are equal in the wild-type and thermosensitive strains (data not shown), the reduced amounts are not a consequence of reduced cellular levels of Drg1 in the mutant but reflect the inability of the altered protein to bind efficiently to the preribosomal particles. To investigate whether additional proteins accumulate on late preribosomal particles upon Drg1 inactivation, we purified TAP-tagged Arx1 from the drg1-ts mutant incubated at 37°C and stained the gel with Coomassie blue. As shown in Fig. 7D, several protein bands were present in increased amounts in the drg1-ts mutant. These bands were identified by mass spectrometry as proteins Nog1 and Lsg1 (band 1), Nug1 (band 2), Nog2 (band 3), Act1 (band 4), and the actin binding protein Cap1 (band 5). The identification of Nog1 confirms our results from the Western blot assay (Fig. 7C), although we cannot exclude the possibility that Lsg1 is also enriched in the drg1-ts mutant. Similarly, the increased purification of Nog2 confirms the observation that Nog2 is enriched in the Arx1-TAP purification of a Drg1-depleted strain (Fig. 7B). The increased association of Nug1 with Arx1 particles in the drg1-ts mutant was not further investigated in this work.
The significance of the presence of actin and Cap1 is not clear at the moment but is currently subject to closer investigation. Due to the prominent presence of ribosomal proteins in the low-molecular-weight range, we were not able to identify Rlp24 on the stained gel. However, its accumulation was confirmed by Western blotting from the same preparation (data not shown).
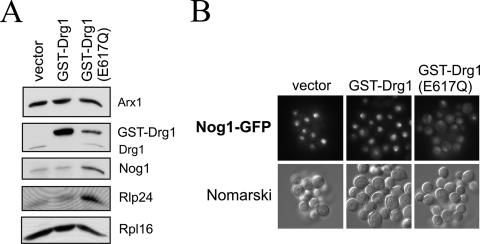
To test whether the ATPase activity of Drg1 is required for the release reaction, we generated a variant of Drg1 containing an E617Q exchange in the Walker B motif of the second AAA domain. An E-to-Q exchange in the Walker B motif was shown previously in different AAA proteins to abolish ATP hydrolysis and to exert a dominant-negative effect (e.g., references 1, 7, and 36). As expected, expression of the Drg1(E617Q) variant was not able to complement the drg1 null allele, indicating that ATP hydrolysis by D2 is required for Drg1 function (data not shown). To test whether the protein is still able to bind to the preribosomal particle, we generated a GST fusion of the E617Q variant and ectopically expressed it under the control of the CUP1 promoter in the Arx1-TAP strain. As the fusion with GST results in an increased molecular weight, this strategy allows the simultaneous detection of the endogenous wild-type protein and the GST-Drg1(E617Q) variant and the estimation of whether it binds to the preribosomal particle. Although the GST-Drg1(E617Q) protein was expressed at much lower levels than was the GST fusion with the wild-type Drg1 protein (data not shown), it clearly bound to late preribosomal particles (Fig. 8A), demonstrating that ATP hydrolysis is not required for binding to the 60S precursor. However, upon induction, the E617Q variant showed a dominant-negative effect on growth and on the release of Rlp24 and Nog1 from the Arx1 particle (Fig. 8A and data not shown). Concomitantly, the formation of half-mers in polysome profiles was observed (data not shown). In addition overexpression of the E617Q variant resulted in the accumulation of Nog1-GFP in the cytoplasm as shown by fluorescence microscopy (Fig. 8B). These results show that ATP hydrolysis by the D2 AAA domain of Drg1 is required for the release of shuttling proteins in the cytoplasm.
FIG. 8.
ATP hydrolysis by D2 of Drg1 is required for the release of shuttling proteins. (A) Overexpression of a Drg1 variant containing an E617Q exchange in the Walker B motif of D2 causes accumulation of Nog1 and Rlp24 on late pre-60S particles. Late preribosomal particles were affinity purified from the Arx1-TAP strain after overexpression of a GST fusion of Drg1(E617Q) under the control of the CUP1 promoter. The wild-type GST-Drg1 fusion and the cloning vector served as controls. The strains were grown to early log phase, and expression was induced by addition of 0.5 mM copper sulfate for 3 h. Western blots of the TEV eluates are shown. (B) The Drg1(E617Q) protein exerts a dominant-negative effect on Nog1-GFP localization. The Nog1-GFP strain carrying a plasmid with a GST fusion of Drg1(E617Q) under the control of the CUP1 promoter was grown to early log phase, and the expression of the fusion protein was induced by 0.5 mM copper sulfate for 6 h. Thereafter, cells were inspected by fluorescence microscopy. The expression vector (pYEX4-T1) and the vector expressing the GST fusion with wild-type Drg1 (pAZ7) served as controls.
Taken together, our results show that Drg1 binds to preribosomal particles and that its ATPase activity is required for the cytoplasmic release of the shuttling proteins Nog1 and Rlp24. This release in turn is a prerequisite for transition to later maturation forms that bind Rei1 and allow the recycling of Arx1 and Tif6.
DISCUSSION
Our data show that the AAA protein Drg1 plays an essential role in ribosome biogenesis. Drg1 binds to late preribosomal particles, which are characterized by the presence of the pre-60S maturation factors Arx1 and Alb1. These proteins join the preribosomal particle in the nucleus and escort it into the cytoplasm (17, 21). Our experiments show that Drg1 joins the pre-60S particle only after its export into the cytoplasm. Moreover, we found that Drg1 is absent from the late-cytoplasmic Rei1- and Lsg1-associated pre-60S particles. These results indicate that Drg1 binds to the pre-60S subunits soon after export from the nucleus and is liberated before Lsg1 or Rei1 loading.
In affinity purifications of TAP-tagged preribosomal factors, we could detect Drg1 in complexes isolated using Rlp24, Nog1, and Arx1 as baits. Rlp24 and Arx1 are known shuttling proteins (21, 28) and are localized in the nucleus and the cytoplasm. In contrast, Nog1 is localized in the nucleolus and the nucleoplasm (14, 28) and is not detected in the cytoplasm at steady state. As Drg1 joins the preribosomal particle after export from the nucleus, the presence of Drg1 in the Nog1-TAP preparation suggests that Nog1 accompanies the preribosomal particle into the cytoplasm. This ability of Nog1 to shuttle between the nuclear and cytoplasmic compartments is further supported by the mislocalization of Nog1-GFP to the cytoplasm in a drg1-ts mutant strain grown at restrictive temperature and upon overexpression of the dominant-negative Drg1(E617Q) protein. In contrast, Nog1-GFP does not mislocalize to the cytoplasm in a rei1Δ strain, which provides further evidence that Rei1 acts downstream of Drg1 (A. Lebreton, C. Saveanu, and M. Fromont-Racine, unpublished results).
Inactivation of Drg1 by incubation of the temperature-sensitive drg1-ts mutant at 37°C or depletion of the protein leads to a decrease in the amounts of several shuttling pre-60S factors (i.e., Rlp24, Nog1, Arx1, and Tif6) in the nucleus and their accumulation in the cytoplasm. The effect of Drg1 on the localization of these proteins is unlikely to be due to a simple “leaking out” of the preribosomal particles from the nucleus, since the localization of other proteins involved in similar stages of pre-60S maturation, like Drs1, Nop7, or Rlp7, remained unaffected in the mutant. We and others previously showed that Nog1, Rlp24, and Tif6 are needed for 27SB pre-rRNA processing (2, 28). Therefore, similar or identical processing steps are affected in Nog1-, Rlp24-, and Tif6-depleted strains and in the drg1-ts mutant. This observation suggests that the pre-rRNA processing defects of the drg1-ts mutant arise from the depletion of these proteins from the nucleus.
Our fluorescence microscopy, polysome profile, and affinity purification experiments show that Nog1, Rlp24, Arx1, and, to a smaller extent, Tif6 remain bound to the pre-60S particle in the cytoplasm of the drg1-ts mutant. We conclude that the activity of Drg1 is needed for the release of these proteins from the preribosomal particles. Two additional preribosome maturation factors, which had not been included in our original studies, Nug1 and Nog2, were also enriched in Arx1-TAP purifications from the drg1-ts mutant incubated at 37°C. Since AAA-ATPases usually act on specific proteins, it is unlikely that Drg1 is directly responsible for the release of all of these proteins. We assume that Drg1 triggers the release of one of these proteins, a process which might be crucial to set free the other preribosome maturation factors. This hypothesis is supported by the observed effect on Arx1 and Tif6. Since Arx1 and Tif6 copurify with Rei1 and Rei1 binds pre-60S particles only after dissociation of Drg1, the failure to release these proteins in the drg1-ts mutant has to be a later consequence of Drg1 inactivation. We therefore propose the order of early cytoplasmic pre-60S maturation events shown in Fig. 9. Soon after export of pre-60S subunits Drg1 associates with the particles and induces the release of Nog1, Rlp24, or one of the other preribosome maturation factors identified as being enriched in our TAP purification from the drg1-ts mutant. Drg1 dissociates from the particles either upon the release of Nog1 and/or Rlp24 or shortly after, a process which might be required for loading of Rei1. In consecutive steps, Arx1 is released and recycled in a Rei1-dependent manner (15, 17) and Tif6 is released by the GTPase Efl1 prior to ribosomal subunit joining (29). The fact that Arx1 and Tif6 are blocked on pre-60S particles in the drg1-ts mutant although they are not direct substrates of Drg1 indicates that the protein performs a conversion of the pre-60S particle which is crucial for the later release of Arx1 and Tif6. This crucial step could, for example, be the release of Nog1 or Rlp24 or one of the other proteins identified in our TAP purification from the drg1-ts mutant. Nevertheless, our results indicate a strict order of disassembly of the shuttling preribosome maturation factors in the cytoplasm. We assume that Drg1 catalyzes the first release reaction, which in turn is the prerequisite for further maturation steps.
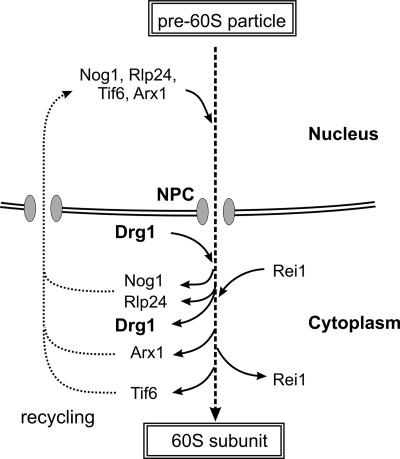
FIG. 9.
Model for Drg1 function in late stages of pre-60S biogenesis. Only factors relevant for this work are shown. Tif6, Rlp24, Nog1, and Arx1 bind to the early pre-60S particles in the nucleus and accompany them into the cytoplasm, where Drg1 associates. Drg1 is required for the release of Nog1 and Rlp24 from pre-60S particles and for the subsequent binding of Rei1. Consequently, Arx1 and Tif6 can be released and recycled back to the nucleus.
AAA proteins are regarded as specific chaperones which change their conformations during their ATPase cycle. The generated force may apply tension to bound proteins and so allows the ATP-dependent disruption of molecular or macromolecular structures, resulting in unfolding of proteins and/or disassembly of protein complexes (11, 19, 26). Our observation that the Drg1(E617Q) protein exhibits a dominant-negative effect on the release of at least Nog1 and Rlp24 shows that ATP hydrolysis in the D2 domain of Drg1 is required for the release of shuttling proteins and therefore is necessary for maturation of the 60S subunit in the cytoplasm.
Two other AAA proteins were shown to be involved in 60S subunit formation in yeast. Rea1 and Rix7 are both required for late steps in maturation of the pre-60S subunit within the nucleus (8, 22). The exact role of these proteins in 60S maturation is unclear, but they are proposed to function in the remodeling of the pre-60S particle, preparation for export, and/or dissociation of nonribosomal proteins. However, factors released by Rea1 or Rix7 have yet to be identified. We show here that the ATPase activity of the essential AAA-ATPase Drg1 is required to dissociate the shuttling preribosome maturation factors Rlp24 and Nog1 from pre-60S particles, which triggers the final maturation steps of the large ribosomal subunits before they enter into translation. Generally, our results support a model in which AAA proteins are required for the release of preribosomal factors from the pre-60S particle. In contrast to Rea1 and Rix7, Drg1 performs this activity in the cytoplasm. Thus, the activity of AAA-ATPases is required at different levels in the maturation pathway of the 60S ribosomal subunit, both in the nucleus and after its export into the cytoplasm.
Supplementary Material
Acknowledgments
We thank Günther Daum, Sabine Rospert, Franco Fasiolo, François Lacroute, Johannes H. Hegemann, Sepp D. Kohlwein, and Ed Hurt for providing antibodies and strains. We are indebted to Samantha Clay for critically reading the manuscript. The expert help of Ron Booij, Christina Morgenstern, Michael Durchschlag, and Eva Wehrschütz-Sigl during early phases of this work is kindly acknowledged.
This work was supported by grants P15458 (G.H.) and P19791 (H.B.) from the Austrian Science Foundation (FWF) and grant GIP ANR-06-BLAN-0037 (M.F.-R.) from the Ministère de la Recherche, France.
Footnotes
Published ahead of print on 23 July 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Babst, M., T. K. Sato, L. M. Banta, and S. D. Emr. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basu, U., K. Si, J. R. Warner, and U. Maitra. 2001. The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 21:1453-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berges, T., E. Petfalski, D. Tollervey, and E. C. Hurt. 1994. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 13:3136-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 5.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 6.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 7.Fujita, H., M. Yamanaka, K. Imamura, Y. Tanaka, A. Nara, T. Yoshimori, S. Yokota, and M. Himeno. 2003. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E vps phenotype in mammalian cells. J. Cell Sci. 116:401-414. [DOI] [PubMed] [Google Scholar]
- 8.Gadal, O., D. Strauss, J. Braspenning, D. Hoepfner, E. Petfalski, P. Philippsen, D. Tollervey, and E. Hurt. 2001. A nuclear AAA-type ATPase (Rix7p) is required for biogenesis and nuclear export of 60S ribosomal subunits. EMBO J. 20:3695-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadal, O., D. Strauss, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 11.Hanson, P. I., and S. W. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519-529. [DOI] [PubMed] [Google Scholar]
- 12.Hedges, J., M. West, and A. W. Johnson. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho, J. H., G. Kallstrom, and A. W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 15.Hung, N. J., and A. W. Johnson. 2006. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:3718-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kallstrom, G., J. Hedges, and A. Johnson. 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 23:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lebreton, A., C. Saveanu, L. Decourty, J. C. Rain, A. Jacquier, and M. Fromont-Racine. 2006. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 173:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, Y., J. Black, N. Kisiel, and M. F. Kulesz-Martin. 2000. SPAF, a new AAA-protein specific to early spermatogenesis and malignant conversion. Oncogene 19:1579-1588. [DOI] [PubMed] [Google Scholar]
- 19.Lupas, A. N., and J. Martin. 2002. AAA proteins. Curr. Opin. Struct. Biol. 12:746-753. [DOI] [PubMed] [Google Scholar]
- 20.Neville, M., and M. Rosbash. 1999. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 18:3746-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nissan, T. A., K. Galani, B. Maco, D. Tollervey, U. Aebi, and E. Hurt. 2004. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol. Cell 15:295-301. [DOI] [PubMed] [Google Scholar]
- 23.Nunnari, J., W. F. Marshall, A. Straight, A. Murray, J. W. Sedat, and P. Walter. 1997. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 8:1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pertschy, B., G. Zisser, H. Schein, R. Köffel, G. Rauch, K. Grillitsch, C. Morgenstern, M. Durchschlag, G. Högenauer, and H. Bergler. 2004. Diazaborine treatment of yeast cells inhibits maturation of the 60S ribosomal subunit. Mol. Cell. Biol. 24:6476-6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 26.Rouiller, I., B. DeLaBarre, A. P. May, W. I. Weis, A. T. Brunger, R. A. Milligan, and E. M. Wilson-Kubalek. 2002. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat. Struct. Biol. 9:950-957. [DOI] [PubMed] [Google Scholar]
- 27.Saveanu, C., D. Bienvenu, A. Namane, P. E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saveanu, C., A. Namane, P. E. Gleizes, A. Lebreton, J. C. Rousselle, J. Noaillac-Depeyre, N. Gas, A. Jacquier, and M. Fromont-Racine. 2003. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 23:4449-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senger, B., D. L. Lafontaine, J. S. Graindorge, O. Gadal, A. Camasses, A. Sanni, J. M. Garnier, M. Breitenbach, E. Hurt, and F. Fasiolo. 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell 8:1363-1373. [DOI] [PubMed] [Google Scholar]
- 30.Sun, C., and J. L. Woolford, Jr. 1994. The yeast NOP4 gene product is an essential nucleolar protein required for pre-rRNA processing and accumulation of 60S ribosomal subunits. EMBO J. 13:3127-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorsness, P. E., K. H. White, and W. C. Ong. 1993. AFG2, an essential gene in yeast, encodes a new member of the Sec18p, Pas1p, Cdc48p, TBP-1 family of putative ATPases. Yeast 9:1267-1271. [DOI] [PubMed] [Google Scholar]
- 32.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 33.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 34.Waterham, H. R., V. I. Titorenko, P. Haima, J. M. Cregg, W. Harder, and M. Veenhuis. 1994. The Hansenula polymorpha PER1 gene is essential for peroxisome biogenesis and encodes a peroxisomal matrix protein with both carboxy- and amino-terminal targeting signals. J. Cell Biol. 127:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendler, F., H. Bergler, K. Prutej, H. Jungwirth, G. Zisser, K. Kuchler, and G. Högenauer. 1997. Diazaborine resistance in the yeast Saccharomyces cerevisiae reveals a link between YAP1 and the pleiotropic drug resistance genes PDR1 and PDR3. J. Biol. Chem. 272:27091-27098. [DOI] [PubMed] [Google Scholar]
- 36.Whiteheart, S. W., K. Rossnagel, S. A. Buhrow, M. Brunner, R. Jaenicke, and J. E. Rothman. 1994. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J. Cell Biol. 126:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zakalskiy, A., G. Högenauer, T. Ishikawa, E. Wehrschütz-Sigl, F. Wendler, D. Teis, G. Zisser, A. C. Steven, and H. Bergler. 2002. Structural and enzymatic properties of the AAA protein Drg1p from Saccharomyces cerevisiae. Decoupling of intracellular function from ATPase activity and hexamerization. J. Biol. Chem. 277:26788-26795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.