Abstract
TRB3 has been implicated in the regulation of several biological processes in mammalian cells through its ability to influence Akt and other signaling pathways. In this study, we investigated the role of TRB3 in regulating adipogenesis and the activity of adipogenic transcription factors. We find that TRB3 is expressed in 3T3-L1 preadipocytes, and this expression is transiently suppressed during the initial days of differentiation concomitant with induction of C/EBPβ. This event appears to be a prerequisite for adipogenesis. Overexpression of TRB3 blocks differentiation of 3T3-L1 cells at a step downstream of C/EBPβ. Ectopic expression of TRB3 in mouse fibroblasts also inhibits the C/EBPβ-dependent induction of PPARγ2 and blocks their differentiation into adipocytes. This inhibition of preadipocyte differentiation by TRB3 appears to be the result of two complementary effects. First, TRB3 inhibits extracellular signal-regulated kinase activity, which prevents the phosphorylation of regulatory sites on C/EBPβ. Second, TRB3 directly interacts with the DR1 domain of C/EBPβ in the nucleus, further inhibiting both its ability to bind its response element and its ability to transactivate the C/EBPα and a-FABP promoters. Thus, TRB3 is an important negative regulator of adipogenesis that acts at an early step in the differentiation cascade to block the C/EBPβ proadipogenic function.
Obese individuals, especially those with abdominal adiposity, are prone to develop type 2 diabetes, hypertension, hyperlipidemia, and cardiovascular disease. This excess of adipose tissue can be the consequence of both hypertrophy (increased lipid accumulation) and hyperplasia (increased proliferation and/or differentiation) of adipose cells. The mechanisms controlling this process of preadipocyte proliferation and differentiation have been extensively studied in cell culture models, especially the 3T3-L1 and 3T3-F442A mouse preadipocyte cell lines (17, 18).
A number of signaling pathways are essential for the induction of the adipocyte differentiation, especially the insulin and insulin-like growth factor 1 (IGF1) signaling pathways. Disruption of the insulin receptor or its major substrates in adipocytes IRS-1 and IRS-3 results in the inhibition of the adipogenic capacity of preadipocytes (12, 13, 57). In vivo fat specific ablation of insulin receptor gene also leads to severe alteration of the adipose tissue development and function (3). This effect of insulin on adipocyte differentiation appears to be mediated through the phosphatidylinositol (PI) 3-kinase and Akt pathway and is blocked by wortmannin or dominant-negative Akt constructs (16, 33, 45, 54, 57). Another critical pathway for adipogenesis is the extracellular signal-regulated kinase (ERK) signaling pathway. This is apparent at the initiation of 3T3 adipocyte differentiation, which is marked by a growth event referred to as “clonal expansion” that is a prerequisite for terminal differentiation (52). This step is also activated by insulin, and inactivation of ERK1/2 prevents this proliferative step and inhibits adipocyte differentiation (4).
Coincident with these signaling events is a well-programmed series of transcriptional events that begins with the induction of two members of the CCAAT/enhancer binding protein family, C/EBPβ and C/EBPδ, by mechanisms that involve glucocorticoid receptors and stimulation of the cyclic AMP (cAMP) regulatory element binding protein CREB (1, 7). An important function of C/EBPβ and C/EBPδ is to directly activate expression of the two principal regulators of terminal adipogenesis, PPARγ and C/EBPα, as well as by stimulating expression of some of the growth-associated genes that are required for facilitating C/EBP activity (8, 11, 14, 20, 53, 59). Once PPARγ and C/EBPα are expressed, they cooperate to orchestrate expression of the full adipogenic program including induction of additional transcription factors, suppression of growth-associated genes and stimulation of insulin-dependent glucose transport (15, 44).
TRB3 (Tribbles homologue 3, NIPK, SKIP3) is a mammalian homologue of the Drosophila Tribbles gene (6, 32). TRB3 is a member of the recently defined family of pseudokinases (5), proteins that contain a serine/threonine kinase catalytic domain but lack an ATP binding site or at least one of the conserved catalytic residues essential for kinase activity. In the liver, it appears that TRB3 can as a dominant-negative regulator of several kinases, most notably Akt (10). Overexpression of TRB3 in liver completely inhibits insulin-stimulated S6 kinase 1 (S6K1) activation by mammalian target of rapamycin (mTOR), while knockdown of endogenous TRB3 increases both basal and insulin-stimulated activity (28). The combination of elevated TRB3 and constitutive S6K1 activity results in decreased insulin signaling via the IRS-1/PI 3-kinase/Akt pathway (31). Finally, TRB3 is also able to regulate the ERK signaling cascades by controlling both the extent and the specificity of MEK1/2 activation of ERK1/2 (27).
In the present study we have investigated a potential role for TRB3 in regulating the differentiation of adipocytes. We show that TRB3 is normally downregulated during the clonal expansion phase of adipogenesis in 3T3-L1 preadipocytes in direct response to dexamethasone (DEX) and 3-isobutyl-1-methyl-xanthine (MIX). Overexpression of TRB3 in 3T3-L1 preadipocytes or mouse mesenchymal stem cells blocks adipogenesis by preventing induction of C/EBPα and PPARγ. The inhibition of preadipocyte differentiation is accompanied by a TRB3-associated blockade of ERK signaling that prevents phosphorylation of regulatory phosphoacceptor sites within C/EBPβ. Moreover, TRB3 is present in both the nucleus and the cytoplasm of preadipocytes and physically interacts with C/EBPβ at repression domain 1, inhibiting its ability to bind DNA and transactivate downstream adipogenic promoters. Taken together, these data demonstrate that TRB3 is an important negative regulator of adipogenesis, acting to control the proadipogenic activity of C/EBPβ.
MATERIALS AND METHODS
TRB3 cloning.
mRNA of proliferating 3T3-L1 cells was extracted by using an RNeasy minikit (QIAGEN) and reverse transcripted into cDNA with a high-capacity cDNA reverse transcription kit (Applied Biosystems). TRB3 forward (5′-GAT-ATC-GGA-TCC-ACC-ATG-CGA-GCT-ACA-CCT-CTG-GCT-GCT-TCT-GCT-GAT-3′) and TRB3 reverse (5′-GAT-ATC-GTC-GAC-CTA-GCC-GTA-CAG-CCC-CAC-CTC-CCC-TTC-CTC-AGC-3′) primers were used to amplify TRB3 cDNA using Platinum Pfx DNA Polymerase (Invitrogen). The PCR fragment was cloned with a pGEM-T easy vector system I (Promega Corp.). TRB3 cDNA was subcloned into BamHI-XhoI-digested pCDNA3.1 vector (Invitrogen) and into pBABE-Neo vector using BamHI-SalI restriction sites.
Retroviral infection.
TRB3 was stably introduced into 3T3-L1 cells and Swiss-LAP cells by retroviral infection. Plates (10 cm) of human embryonic kidney 293T cells were transiently transfected with 10 μg of retroviral expression vectors and the viral packaging vectors SV-E-MLV-env and SV-E-MLV using TransIT-Express transfection reagent (Mirus Bio Corp.). At 48 h after transfection, virus-containing medium was collected and passed through a 0.45-μm-pore-size syringe filter. Filter-sterilized Polybrene (hexadimethrine bromide; 12 μg/ml) was added to the virus-loaded medium. This medium was then applied to proliferating (40% confluent) cells. At 24 h after infection, cells were treated with trypsin and replated in a medium supplemented with zeocin (Invitrogen) as aselection antibiotic.
Analysis of gene expression by quantitative PCR.
A total of 1 μg of total RNA was extracted by using an RNeasy minikit (QIAGEN) and was reverse transcribed in 20 μl by using an Advantage RT-for-PCR kit (BD Biosciences) according to the manufacturer's instructions. A portion (5 μl) of diluted (1/20) reverse transcription reaction was amplified with specific primers (300 nM each) in a 20-μl PCR with a SYBR green PCR master mix (Applied Biosystems). Analysis of gene expression was done in an ABI Prism 7000 sequence detector for an initial denaturation at 95°C for 10 min, followed by 40 PCR cycles, each cycle consisting of 95°C for 15 s, 60°C for 1 min, and 72°C for 1 min, and SYBR green fluorescence emissions were monitored after each cycle. For each gene, mRNA expression was calculated relative to TBP for murine samples. Amplification of specific transcripts was confirmed by melting-curve profiles (cooling the sample to 68°C and heating slowly to 95°C with measurement of fluorescence) at the end of each PCR. The specificity of the PCR was further verified by subjecting the amplification products to agarose gel electrophoresis. The primers used for quantitative PCR were TRB3 (5′-CTT-TTG-GAA-CGA-GAG-CAA-GG-3′ and 5′-GTG-TTG-TGG-GTA-TCT-GAA-GG-3′), C/EBPβ (5′-CCA-AGA-AGA-CGG-TGG-ACA-A-3′ and 5′-CAA-GTT-CCG-CAG-GGT-GCT-3′), C/EBPδ (5′-ATC-GAC-TTC-AGC-GCC-TAC-A-3′ and 5′-GCT-TTG-TGG-TTG-CTG-TTG-AA-3′), C/EBPα (5′-CAA-GAA-CAG-CAA-CGA-GTA-CCG-3′ and 5′-GTC-ACT-GGT-CAA-CTC-CAG-CAC-3′), PPARγ (5′-CCC-TGG-CAA-AGC-ATT-TGT-AT-3′ and 5′-GAA-ACT-GGC-ACC-CTT-GAA-AA-3′), FAS (5′-GAG-GAC-ACT-CAA-GTG-GCT-GA-3′ and 5′-GTG-AGG-TTG-CTG-TCG-TCT-GT-3′), Glut4 (5′-TGA-TTC-TGC-TGC-CCT-TCT-GT-3′ and 5′-GGA-CAT-TGG-ACG-CTC-TCT-CT-3′), aP2 (5′-CTG-GGC-GTG-GAA-TTC-GAT-3′ and 5′-GCT-CTT-CAC-CTT-CCT-GTC-GTC-T-3′), and TBP (5′-ACC-CTT-CAC-CAA-TGA-CTC-CTA-TG-3′ and 5′-TGA-CTG-CAG-CAA-ATC-GCT-TGG-3′).
Cell lysates and immunoblotting.
For total cell lysates, cells were washed once with cold phosphate-buffered saline (PBS) and scraped in radioimmunoprecipitation assay lysis buffer complemented with 1% sodium dodecyl sulfate, 10 mM β-glycerophosphate, 10 mM NaF, 0.1 mM sodium orthovanadate, and the complete protease inhibitor mixture (1:50 tablet per ml; Roche Applied Sciences). Lysis of cells was immediately followed by boiling for 3 min. Lysates were subsequently treated with benzonase nuclease (Emdbiosciences-Novagene). Whole-cell extracts were stored at −80°C. Nuclear and cytoplasmic extracts were prepared with an NE-PER nuclear and cytoplasmic extraction reagent kit (Pierce Biotechnologies). Protein concentrations were determined by the Bradford method (Bio-Rad). Equal amounts of protein were used for immunoblot analysis. Proteins were separated by electrophoresis in polyacrylamide gels. Protein was transferred to a polyvinylidene difluoride membrane (Amersham Biosciences) and immunoblotted with the appropriate antibodies. Secondary antibodies were horseradish peroxidase (HRP)-conjugated goat anti-rabbit, HRP-conjugated goat anti-mouse (Pierce Biotechnologies), and HRP-conjugated bovine anti-goat (Santa Cruz Biotechnologies) immunoglobulin G (IgG). Membranes were visualized by using Supersignal West Pico substrate or Supersignal West Dura extended duration substrate (Pierce Biotechnologies).
Coimmunoprecipitation.
NIH 3T3 cells were transiently transfected with TransIT-Express transfection reagent. Two days later, the cells were washed twice with cold PBS buffer, scraped in ice-cold lysis buffer (50 mM Tris·HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 50 mM NaF, 10 mM β-glycerophosphate, 0.5 mM Na3VO4, and protease inhibitor mixture), sonicated, and then centrifuged at 10,000 × g at 4°C for 20 min. Supernatants were recovered, and protein concentrations were determined. Antibodies (10 μg/ml) against the Flag tag were added to protein G-Sepharose beads equilibrated in the lysis buffer for 1 h at 4°C. Beads were washed three times with 1 ml of iced-cold lysis buffer and added to1 mg of protein lysate in a final volume of 0.5 ml, followed by incubation at 4°C for 4 h. Immunoprecipitates were recovered by centrifugation at 2,500 × g and washed three times with ice-cold lysis buffer. Immunoprecipitated proteins were eluted in Laemmli buffer, denatured at 100°C for 5 min, and subjected to Western blotting as described above.
Reporter assays.
Swiss LAP A cells were plated in 24-well plates and grown to ∼40% confluence, at which time they were changed to media in the presence or absence of tetracycline. Two days later, at ∼85% confluence, the cells were transfected with 2 μg of promC/EBPα-LUC, 2 μg of pcDNA3-control, or pcDNA3-TRB3 and 0.04 μg of pRL-CMV (Promega, Madison, WI) using TransIT-Express transfection reagent. The cells were harvested 48 h later and analyzed for luciferase activity. The luciferase activity was measured by using the DLRII kit (Promega). NIH 3T3 cells were also plated in 24-well plates, grown up to 80% confluence, and transfected with 200 ng of CEBPs constructs and/or 400 ng of pcDNA3.1-TRB3 with Lipofectamine 2000 transfection reagent (Invitrogen). Cells were then treated and analyzed as described above.
Antibodies.
Anti-TRB3 (ST-1032) antibody was purchased from Calbiochem. Antibodies to phospho-Thr235-C/EBPβ (3084), phospho-Ser473-Akt (catalog no. 9271), Akt (catalog no. 9272), phospho-Ser9-GSK3β (catalog no. 9336), phospho-Thr202/Tyr204-p44/42 mitogen-activated protein (MAP) kinase (catalog no. 9101), p44/42 MAP kinase (catalog no. 9102), and lamin A/C (catalog no. 2032) were purchased from Cell Signaling. Antibodies to fatty acid synthase (FAS; ab22759) and superoxide dismutase 4 (SOD4; ab16834) were purchased from Abcam. C/EBPβ (sc-150), C/EBPδ (sc-151), C/EBPα (sc-61), HRP-conjugated antiactin (sc-1616), and Glut4 (sc-7938) were purchased from Santa Cruz Biotechnologies. PPARγ antibody (catalog no. 07-466) was purchased from Upstate. Antiadiponectin (PA1-054) was purchased from Affinity Bioreagents. Anti-Flag M2 (F1804) was purchased from Sigma-Aldrich. Polyclonal antiperilipin was a gift from Andy Greenberg (New England Medical Center, Tufts University, Boston, MA).
Oil Red O staining.
Culture dishes were washed in PBS (pH 7.4), and cells were fixed in 3.7% formaldehyde for 1 h, followed by staining with Oil Red O for 1 h. Oil Red O was prepared by diluting a stock solution (0.5 g of Oil Red O [Sigma]) in 100 ml of isopropanol with water (60:40 [vol/vol]), followed by filtration. After staining, plates were washed twice in water and photographed.
Immunohistochemistry.
At 2 days postinduction the cells were fixed in 4% paraformaldehyde for 15 min at room temperature, permeabilized with 0.1% Triton X-100 for 10 min, and blocked in 0.1% bovine serum albumin for 30 min. Cells were then incubated with C/EBPβ antibody, followed by Alexa Fluor 594-conjugated secondary antibody. Preparations were mounted in DAPI (4′,6′-diamidino-2-phenylindole) mounting solution (Vector Laboratories).
Oligonucleotide pull-down assays.
Nuclear extracts were performed as described above after which 250 μg of the extract was mixed with oligonucleotides containing C/EBP consensus and mutant sequences that had been conjugated to agarose (Santa Cruz Biotechnologies). Purifications were performed as recommended by the manufacturer, and protein complexes were eluted in Laemmli buffer, boiled for 3 min, and analyzed by Western blot.
RESULTS
TRB3 expression is regulated during adipocyte differentiation.
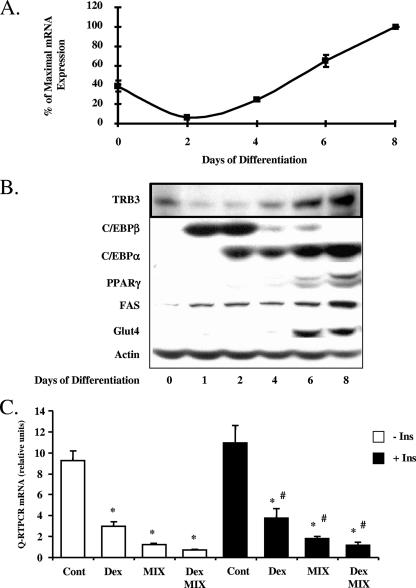
Insulin or IGF1, acting through PI 3-kinase and Akt, are principal extracellular regulators of the early phase of adipogenesis. Since TRB3 has been shown to be a potentially physiologically important regulator of insulin action and Akt signaling, we sought to determine whether TRB3 might participate in the differentiation of adipocyte cells. To this end, we first analyzed the expression of TRB3 mRNA and protein during the conversion of 3T3-L1 preadipocytes into adipocytes by using a protocol with 2 days of treatment by DEX, MIX, insulin, and fetal bovine serum, after which the cells were maintained in medium containing insulin and fetal bovine serum. By real-time PCR, TRB3 mRNA was easily detected in preadipocytes, and its expression was downregulated by 85% after exposure of the cells to DEX, MIX, and insulin during the initial 2 days of differentiation (Fig. 1A). This fall was reversed as soon as the induction cocktail was eliminated from the culture medium. TRB3 mRNA then increased to 2.5-fold above initial levels as the preadipocytes underwent terminal differentiation to adipocytes (days 2 to 8). Consistent with these data, Western blots showed a low but detectable level of TRB3 protein in preadipocytes, followed by a transient downregulation of TRB3 during the first 2 days of adipogenic induction and then a severalfold increase during terminal differentiation (Fig. 1B). These changes occurred in parallel with previously defined changes in adipogenic markers and the transcriptional cascade leading to adipocyte differentiation. Thus, during the first 2 days, there was a transient, but marked, increase in C/EBPβ and C/EBPδ, followed by gradual increases in C/EBPα, PPARγ, and ultimately FAS and GLUT4 (Fig. 1B).
FIG. 1.
TRB3 expression is regulated during adipocyte differentiation. (A) TRB3 mRNA expression during adipocyte differentiation of 3T3-L1 cells. TRB3 mRNA expression was measured by quantitative real-time PCR and normalized to TBP mRNA expression. The data are represented as the percentage of maximal expression. The results are representative of three independent experiments. (B) TRB3 protein expression during adipocyte differentiation of 3T3-L1 cells. A total of 40 μg of total protein extracts was loaded, and adipocyte differentiation markers expression were assessed by Western blotting. For TRB3, 100 μg of protein was loaded and analyzed in a 12.5% polyacrylamide gradient gel and assessed by Western blotting. The results are representative of three independent experiments. (C) TRB3 mRNA regulation by the adipogenic cocktail components. 3T3-L1 cells were treated during 24 h with DEX, MIX, and insulin separately or in combination as described in Materials and Methods. TRB3 mRNA expression was measured by quantitative real-time PCR and normalized to TBP mRNA expression. The data are represented in relative units. The results are representative of three independent experiments. *, P < 0.01 compared to the “control (Cont) − Ins” value; #, P < 0.01 compared to the “control + Ins” value.
To identify which components of the adipogenic cocktail were responsible for the transient suppression of TRB3 expression during the initial 2 days of induction and/or differentiation, we treated 3T3-L1 preadipocytes with DEX, MIX, or insulin, either separately or in combination, for 24 h. TRB3 mRNA expression was inhibited 67% by DEX, 80% by MIX, and 91% by both. These effects occurred both in the presence and in the absence of insulin. Thus, DEX and MIX appeared to act in a synergistic manner to suppress TRB3 expression, and this effect was independent of the effect of insulin on the differentiation process (Fig. 1C).
TRB3 overexpression blocks the differentiation of 3T3-L1 cells into adipocytes.
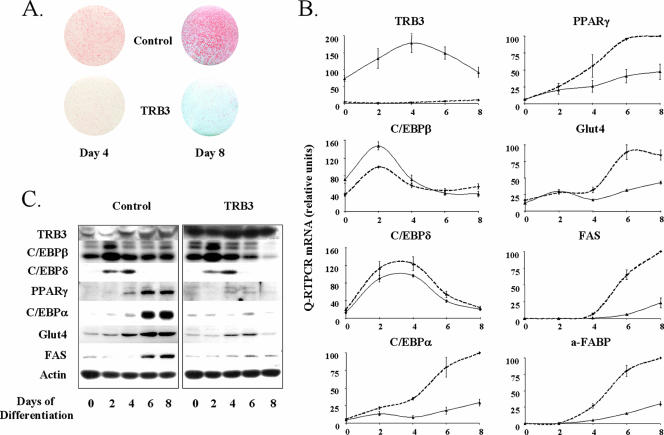
The data in Fig. 1 suggested that one of the early events in adipogenesis is a transient inhibition of TRB3 expression by glucocorticoids and cAMP-mediated activators in the adipogenic cocktail. To determine whether this downregulation of TRB3 was a prerequisite or important in the adipogenic differentiation process, we stably expressed TRB3 in 3T3-L1 cells by using retroviral gene transduction and then tested the ability of the resulting cell line to differentiate into adipocytes. During the entire time course of adipocyte differentiation, the 3T3-L1-TRB3 (Fig. 2B, solid line) displayed at least a 20-fold overexpression of TRB3 compared to the 3T3-L1 infected with the control retrovirus (dashed line) (Fig. 2B). For the cells overexpressing TRB3, we observed a dramatic reduction in their capacity to achieve adipogenesis. This deficiency was characterized by their inability to accumulate triglyceride containing lipid droplets, as visualized by the Oil Red O staining of cells 8 days after induction (Fig. 2A) and could be observed as early as by 4 days of induction.
FIG. 2.
TRB3 overexpression blocks the differentiation of 3T3-L1 cells into adipocytes. (A) Oil Red O staining of lipid droplets in 3T3-L1 cells overexpressing or not TRB3 at day 4 or 8 of differentiation. Cells were fixed overnight in 10% formalin, and lipid droplets were stained with Oil Red O for 1 h. Pictures represent scans of the stained plates. (B) Characterization of mRNA expression of different adipocyte differentiation markers during the conversion of 3T3-L1 cells overexpressing TRB3 or not. Cells were induced to differentiate, and mRNA extracts were prepared at the time indicated and analyzed by quantitative PCR. The results are normalized to TBP mRNA expression and are expressed in relative units. Dotted lines represent control cell values, whereas solid lines represent TRB3-overexpressing cells. The data are represented as the percentage of maximal expression obtained in control cells. The results are representative of three independent experiments. (C) Characterization of protein expression of different adipocyte differentiation markers during the conversion of 3T3-L1 cells overexpressing TRB3 or not. A total of 40 μg of total protein extracts was loaded, and adipocyte differentiation marker expression was assessed by Western blotting. The results are representative of three independent experiments.
This failure of the 3T3-L1-TRB3 preadipocytes to differentiate into mature adipocytes was confirmed by real-time PCR analysis of differentiation markers. At day 8 postinduction, there was a 79% reduction in Glut4 mRNA expression, a 54% reduction for FAS mRNA expression and a 75% reduction in the adipocyte fatty acid binding protein (A-FABP or aP2) mRNA expression (Fig. 2B). This inhibition was easily detected as early as day 4 with reductions of Glut4, FAS. and A-FABP mRNA expression, by 92, 62, and 87%, respectively.
In an attempt to identify potential targets of the inhibitory action of TRB3, we analyzed the transcription factors regulating expression of the mature adipocyte markers. In cells overexpressing TRB3, the expression of both C/EBPα and PPARγ mRNAs were already decreased at day 4 by 80 and 63%, respectively. This decrease persisted throughout the entire time course of differentiation, with 70 and 50% reductions in C/EBPα and PPARγ expression, respectively, on day 8 (Fig. 2B). These results suggested that TRB3 must have been acting at some site early in the adipogenic cascade to antagonize induction of the C/EBPα and PPARγ genes. In 3T3-L1 cell differentiation, the genes immediately upstream of C/EBPα and PPARγ are C/EBPβ and C/EBPδ. In contrast to C/EBPα and PPARγ, C/EBPδ expression was unchanged in the 3T3-L1-TRB3 cells, and C/EBPβ mRNA expression was actually increased by 50% over that in control cells at days 0 and 2 and showed no significant difference at other time points. In addition, both of these transcription factors were induced during the first days of differentiation with kinetics identical to those in the control cells. Western blots experiments confirmed the gene expression patterns (Fig. 2C). Thus, both Glut4 and FAS protein expression were almost abolished in the TRB3-overexpressing cells, as was the expression of C/EBPα and PPARγ protein, whereas both C/EBPβ and C/EBPδ proteins displayed a normal transient expression pattern with a slight increase in the amount of C/EBPβ at day 2 in cells overexpressing TRB3. Taken together with previous studies indicating that C/EBPβ and C/EBPδ are key intermediates during the mitotic clonal expansion of preadipocytes during induction by DEX and MIX and function together to induce expression of both C/EBPα and PPARγ (15), these data suggest that TRB3 can block adipogenesis by acting at a step between these C/EBPβ and C/EBPδ in the induction of PPARγ and C/EBPα.
TRB3 specifically inhibits C/EBPβ transcriptional activity.
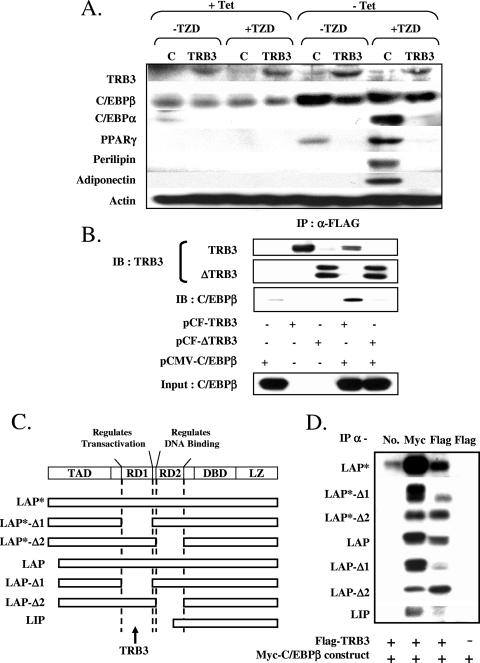
To investigate how TRB3 might regulates C/EBPβ activity to produce these effects, we used a Swiss 3T3 cell line in which ectopic production of C/EBPβ is facilitated by a Tet-off conditional expression system (Swiss-LAP cells) (60). Culture of these cells in tetracycline suppresses C/EBPβ and maintains their growth as fibroblasts, while removal of tetracycline from the culture media leads to a robust induction of exogenous C/EBPβ, which in turn promotes PPARγ expression, allowing full induction of adipogenesis following in the presence of a potent PPARγ ligand such as rosiglitazone (Fig. 3A). To test the effect of TRB3 on C/EBPβ, we infected the Swiss-LAP cells with a retroviral vector containing TRB3 cDNA, leading to a cell line that constitutively expresses TRB3 but produces C/EBPβ conditionally in response to tetracycline (Swiss-LAP-TRB3). Western blot experiments were performed on Swiss-LAP 8 days after differentiation induction in the presence or the absence of the thiazolidinedione rosiglitizone (TZD), tetracycline, and TRB3 overexpression. As expected, in the presence of tetracycline, the endogenous levels of C/EBPβ in Swiss-LAP cells were very low or undetectable and were not able to induce the expression of PPARγ or C/EBPα or to promote differentiation of these cells into adipocytes even in the presence of the TZD (Fig. 3A). As soon as the tetracycline was removed, the expression of C/EBPβ was upregulated resulting in an increase in PPARγ expression, and when TZDs were added to the differentiation medium, this led to the differentiation of Swiss-LAP cells into mature adipocytes, characterized by lipid accumulation and the expression of adiponectin and perilipin (Fig. 3A). TRB3 overexpression in these cells did not prevent the overexpression of exogenous C/EBPβ in the absence of tetracycline but blocked its ability to induce PPARγ expression and therefore prevented the TZD from triggering differentiation. Indeed, even in the presence of TZD and the absence of tetracycline, Swiss-LAP cells overexpressing TRB3 were not able to differentiate into mature adipocytes or increase their expression of adiponectin or perilipin (Fig. 3A).
FIG. 3.
TRB3 inhibits C/EBPβ transcriptional activity. (A) Characterization of the ability of the exogenous C/EBPβ to induce PPARγ in fibroblastic cells Swiss 3T3 in the presence or absence of TRB3. A total of 40 μg of total protein extracts was loaded, and adipocyte differentiation marker expression was assessed by Western blotting. The results are representative of two independent experiments. (B) Coimmunoprecipitation experiments of C/EBPβ with TRB3 or a N-terminal truncated form of TRB3. A total of 500 μg of protein was used to immunoprecipitate FLAG-tagged TRB3 or FLAG-tagged ΔTRB3. The elution was loaded equally on two gels to assess TRB3 and C/EBPβ protein amounts. The results are representative of two independent experiments. (C) Schematic of the structures of the C/EBPβ mutant isoforms used in the deletion analysis. TAD, transactivation domain; RD1, repression domain 1; RD2, repression domain 2; DBD, DNA-binding domain; LZ, leucine zipper. (D) Coimmunoprecipitation experiments of deletion mutants of C/EBPβ isoforms with TRB3. A total of 250 μg of protein were used to perform the different immunoprecipitations. The elutions were analyzed by Western blotting to assess TRB3 and C/EBPβ isoform protein amounts.
TRB3 is known to interact with leucine-zipper transcription factors such as ATF-4 (CREB-2) (36). Since C/EBPβ is a member of the same family of leucine-zipper proteins, we questioned whether there might be a direct physical association between TRB3 and C/EBPβ. Accordingly, we transfected HEK-293 cells with cDNAs expressing a Myc-tagged C/EBPβ alone or in combination with either a Flag-tagged TRB3 or a Flag-tagged N-terminal truncated form of TRB3 and performed a series of immunoprecipitations with a polyclonal anti-Flag antibody. Western blot analysis of the immunoprecipitates with polyclonal anti-TRB3 and C/EBPβ antibodies demonstrated that the anti-Flag antibody precipitated each of the corresponding Flag-tagged forms of TRB3, but not the Myc-tagged C/EBPβ, when transfected separately (Fig. 3B). However, when TRB3 and C/EBPβ were coexpressed in the HEK-293 cells, Western blot analysis of the immunoprecipitates with anti-C/EBPβ revealed that TRB3 was coimmunoprecipitated with the full-length C/EBPβ (Fig. 3B), This appeared to be mediated via a C-terminal domain of TRB3, since no interaction was observed between C/EBPβ and a C-terminal truncated form of TRB3 (ΔTRB3), despite similar levels of expression of C/EBPβ in the cell lysates (Fig. 3B). To address the specificity of this interaction, we performed similar experiments with cDNAs for full-length C/EBPα and C/EBPδ, but in neither case were there any interactions with either C/EBPα or C/EBPδ (data not shown).
This last result raised the question about how does TRB3 achieve specificity toward C/EBPβ. As reviewed by Ramji and Foka (40), all C/EBP members share high sequence identity in the C-Terminal regions containing the bZIP structure (basic DNA binding domain and leucine zipper). In contrast, members of the C/EBP family are quite divergent concerning in the N-terminal sequence, especially the repression domains that are not conserved between C/EBP members. C/EBPβ possesses two major repression domains, RD1 and RD2, which have been involved, respectively, in the regulation of its transactivation function and its DNA-binding activity. To study the importance of these two domains in the interaction between TRB3 and C/EBPβ, we created several mutated Myc-tagged isoforms of C/EBPβ LAP*, LAP and LIP, truncated from either RD1 (Δ1) or RD2 (Δ2) (Fig. 3C). Each of the C/EBPβ constructs was then transfected into HEK-293 cells alone or with a Flag-tagged TRB3 construct, and their ability to interact with TRB3 was determined by coimmunoprecipitation. Each sample was then incubated with either a normal mouse IgG as a negative control (No), an anti-Myc IgG as a positive control (Myc), or an anti-Flag IgG (Flag), and immunoprecipitates were analyzed by Western blotting. As shown in Fig. 3D, the interaction between C/EBPβ and TRB3 was confirmed with both full-length cDNA encoding LAP* and LAP isoforms. More interestingly, the deletion of the first repression domain RD1 of those two isoforms (LAP*-Δ1 and LAP-Δ1) markedly impaired their ability to interact with TRB3, whereas the deletion of the second repression domain, RD2, did not (LAP*-Δ2 and LAP-Δ2). As a confirmation, the LIP isoform of C/EBPβ, which lacks entirely RD1 and lacks partially RD2, was unable to interact with TRB3 (LIP). These results indicate that TRB3 specifically interacts with the repression domain RD1 of C/EBPβ isoforms.
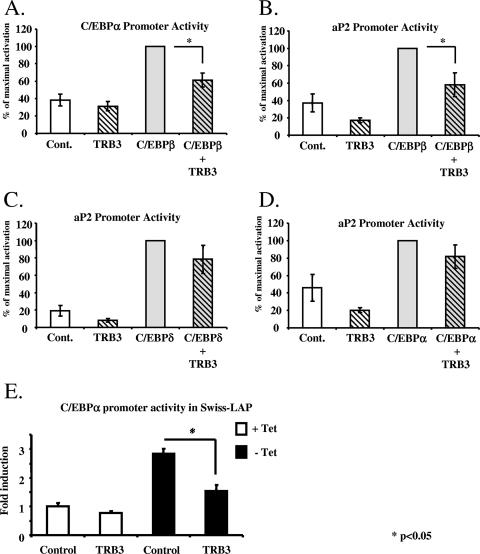
To address whether the interaction between TRB3 and C/EBPβ affects the transcriptional activity of C/EBPβ, we performed a series of reporter gene assays using constructs in which the a-FABP promoter and C/EBPα promoter can drive the expression of luciferase in response to C/EBPβ activity due to the presence a C/EBP binding element in these promoters (8). The assays were performed by transfecting the a-FABP or C/EBPα promoter/reporter construct in HEK-293 cells with or without a TRB3 and/or LAP-expressing constructs (Fig. 4A and B). Both promoters were transactivated in response to LAP overexpression, whereas the overexpression of TRB3 tended to diminish their basal activity. More interestingly, the coexpression of TRB3 inhibited C/EBPβ ability to transactivate the a-FABP and C/EBPα promoters by 42% (P < 0.04) and 39% (P < 0.05). It should be noted that the specificity of interaction of TRB3 for C/EBPβ was also confirmed in these experiments, since TRB3 was not able to inhibit either C/EBPδ (Fig. 4C) or C/EBPα (Fig. 4D) effects on the C/EBPα promoter. Similar assays were also performed by transfecting the C/EBPα promoter/reporter construct into Swiss-LAP cells in the presence or absence of an ectopic TRB3 with or without exposure to tetracycline. Simple expression of ectopic TRB3 did not significantly modify the basal activity of C/EBPα promoter. Furthermore, culture of the transfected Swiss-LAP cells in the absence of tetracycline led to a threefold activation of the C/EBPα promoter reflecting the expression of the exogenous C/EBPβ. However, in cells overexpressing TRB3, the level of C/EBPα promoter activation by C/EBPβ was reduced by 50% (P < 0.05), indicating that TRB3 is capable of blocking C/EBPβ transcriptional activity (Fig. 4E). Taken together, these results are consistent with the observations above that the overexpression of TRB3 prevents the expression of C/EBPα and a-FABP.
FIG. 4.
TRB3 inhibits C/EBPβ transcriptional activity on a-FABP and C/EBPα promoters. (A) Characterization of the ability of the exogenous C/EBPβ to activate the C/EBPα promoter in HEK-293 cells in the presence or the absence of TRB3 overexpression. The cells were harvested 48 h after transfection and analyzed for luciferase activity. The results are representative of four independent experiments. *, P < 0.05. (B) Characterization of the ability of the exogenous C/EBPβ to induce a-FABP promoter in HEK-293 cells in the presence or the absence of TRB3 overexpression. The cells were harvested 48 h after transfection and analyzed for luciferase activity. The results are representative of four independent experiments. *, P < 0.05. (C) Characterization of the ability of the exogenous C/EBPδ to induce a-FABP promoter in HEK-293 cells in the presence or absence of TRB3 overexpression. The cells were harvested 48 h after transfection and analyzed for luciferase activity. The results are representative of four independent experiments. (D) Characterization of the ability of the exogenous C/EBPα to induce a-FABP promoter in HEK-293 cells in the presence or absence of TRB3 overexpression. The cells were harvested 48 h after transfection and analyzed for luciferase activity. The results are representative of four independent experiments. (E) Characterization of the ability of the exogenous C/EBPβ to induce C/EBPα promoter in fibroblastic Swiss 3T3 cells in the presence or absence of TRB3 overexpression. The cells were harvested 48 h after transfection and analyzed for luciferase activity. The results are representative of three independent experiments. *, P < 0.05.
TRB3 does not prevent the translocation of C/EBPβ into centromeric regions of the nucleus but impairs its ability to bind DNA.
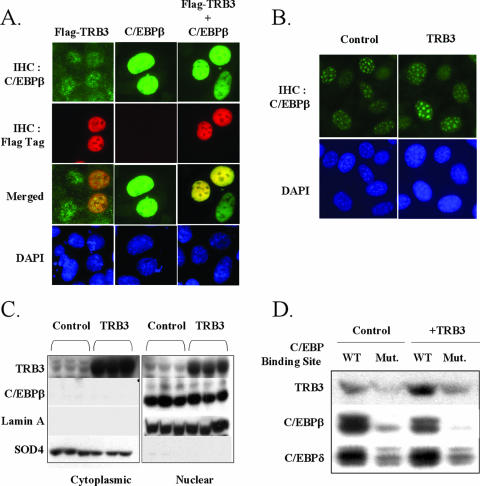
During mitotic clonal expansion, C/EBPβ is translocated into the nucleus, where it can transactivate expression of target genes, including PPARγ and C/EBPα. One possible mechanism by which TRB3 might inhibit C/EBPβ activity is to prevent its translocation into the nucleus. To assess this possibility, we examined the cellular localization of C/EBPβ by immunohistochemistry in HEK-293 cells transfected with a construct expressing a FLAG-tagged TRB3. As expected, endogenous C/EBPβ localization was mainly nuclear in HEK-293 cells (Fig. 5A). When TRB3 was overexpressed in these cells, it displayed a clear nuclear localization and, in fact, appeared to colocalize with the endogenous C/EBPβ in the nucleus, as shown by the merged pictures (Fig. 5A, left panels). To confirm this result, we overexpressed C/EBPβ in HEK-293 cells in the presence or the absence of ectopic TRB3. In the absence of ectopic TRB3 expression, the same nuclear localization was observed for the exogenous C/EBPβ compared to the signal observed for the endogenous C/EBPβ (Fig. 5A, middle panels). When both TRB3 and C/EBPβ were ectopically expressed, they both colocalized within the nucleus, confirming that TRB3 interaction with C/EBPβ did not modify C/EBPβ nuclear localization (Fig. 5A, right panels).
FIG. 5.
TRB3 colocalize with C/EBPβ within the nucleus and does not prevent its localization within centromeric regions in preadipocytes. (A) Immunohistochemistry of C/EBPβ (green) and exogenous TRB3 (red) in HEK-293 cells. Cells were transfected with 4 μg of DNA, and exogenous protein localization was assessed 48 h posttransfection. Nuclear visualization is assessed with DAPI staining (blue). (B) Immunohistochemistry of endogenous C/EBPβ (green) in 3T3-L1 preadipocytes overexpressing TRB3 or not. Cells were fixed 2 days after induction. Nuclear visualization is assessed with DAPI staining (blue). The results are representative of three independent experiments. (C) C/EBPβ protein nuclear localization in 3T3-L1 preadipocytes overexpressing TRB3 or not. Nuclear and cytoplasmic extracts were prepared 24 h postinduction, and 30 μg of protein was loaded into each lane and analyzed by Western blotting. Nuclear and cytoplasmic extracts quality was determined with lamin A and SOD4 protein. The results are representative of three independent experiments. (D) C/EBP consensus binding site oligonucleotide pull-down assays of 3T3-L1 preadipocytes nuclear extracts, with or without the overexpression of TRB3. Nuclear extracts were prepared 48 h postinduction, and 250 μg was used for the oligonucleotide pull-down assays. Elutions were analyzed by Western blotting.
To determine whether TRB3 might alter C/EBPβ localization during the adipocyte differentiation process, we examined the cellular localization of endogenous C/EBPβ by immunohistochemistry in 3T3-L1 preadipocytes overexpressing TRB3. Two days after exposure to induction cocktail, the immunohistology demonstrated that C/EBPβ was nuclear and strongly localized to centromeric regions of the nuclei of the control 3T3-L1 cells, and this distribution was unchanged in preadipocytes overexpressing TRB3 (Fig. 5B). Analysis of the distribution of C/EBPβ between cytoplasmic and nuclear compartments by Western blot analysis after subcellular fractionation revealed that C/EBPβ sedimented exclusively with nuclei during the fractionation of the 3T3-L1 preadipocytes and that TRB3 had no effect on its distribution (Fig. 5C). The purity of these cellular fractions was verified by immunoblotting with anti-lamin A (nuclear) and anti-SOD4 (cytoplasmic) antibodies. Taken together, our data suggest that TRB3 can interact directly with C/EBPβ and inhibit its transcriptional activity. Furthermore, TRB3 is located in both the cytoplasmic and the nuclear compartments but does not inhibit C/EBPβ translocation into the nucleus of 3T3-L1 preadipocytes during clonal expansion and adipocyte differentiation.
Another possible mechanism by which TRB3 might inhibit C/EBPβ activity is to prevent its ability to bind its response element. To address this possibility, we performed an oligonucleotide-pulldown assay with consensus and mutant C/EBP oligonucleotides. Nuclear extracts from 3T3-L1 preadipocytes overexpressing or not TRB3 were prepared 2 days after induction of adipocyte differentiation and incubated with the consensus and mutant C/EBP oligonucleotides conjugated to agarose beads. The protein complexes bound to those elements were analyzed by Western blotting. As expected, in the control conditions, both C/EBPβ and C/EBPδ were able to bind the C/EBP response element in a specific manner, as demonstrated by the markedly reduced binding of both C/EBPβ and C/EBPδ with the C/EBP mutant consensus oligonucleotides. Under these conditions, we also observed a detectable signal of endogenous TRB3 in the proteins complexed to the C/EBP response element, and this was strongly reduced with the mutant oligonucleotide, probably reflecting the interaction between the endogenous TRB3 and the endogenous C/EBPβ (Fig. 5D). In these experiments, the overexpression of TRB3 reduced the amount of C/EBPβ bound the C/EBP response element by 28% (P = 0.05), whereas no changes in the amount of C/EBPδ were observed. Note that the overexpression of TRB3 was also reflected by an increase in the amount of TRB3 complexed to the C/EBP response element. These results demonstrate that some of the inhibitory effect of TRB3 on C/EBPβ is due to an inhibition of its ability to bind DNA, but another additional mechanism was probably involved.
TRB3 inhibits phosphorylation of C/EBPβ during the early phase of adipogenesis.
TRB3 has been shown to inhibit both the Akt and the MAP kinase pathways in response to insulin signaling in several cell types (10, 27). As noted above, these signaling pathways are also known to be essential for preadipocyte differentiation. To determine whether TRB3 might be affecting C/EBPβ activity through perturbation of one of these pathways, we assessed the activity of the Akt and MAP kinase pathways in cultured 3T3-L1 cells ectopically expressing TRB3. In 3T3-L1 overexpressing TRB3, we observed a significant inhibition of basal and insulin stimulated phosphorylation of ERK1/2 phosphorylation state, as well as an inhibition of basal and insulin stimulated phosphorylation of GSK3 and activation of Akt (Fig. 6A).
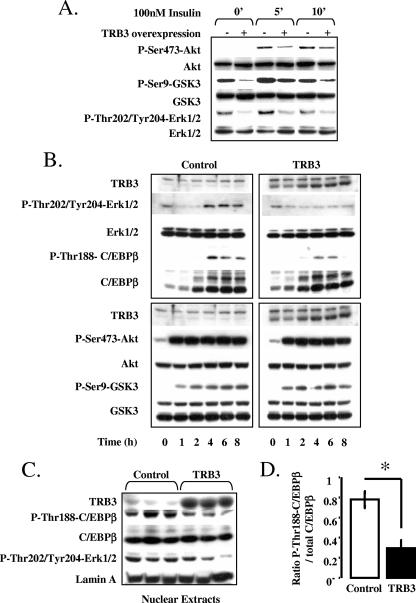
FIG. 6.
TRB3 inhibits ERK-dependent phosphorylation of C/EBPβ during the early phase of adipogenesis. (A) Effect of TRB3 overexpression in 3T3-L1 cells on insulin-dependent activation of Akt/GSK3 and ERK pathways. Cells were grown to confluence and induced with 100 nM insulin for 5 or 10 min. Protein extracts were prepared at the times indicated. A total of 30 μg of total protein extracts was loaded per lane and analyzed by Western blotting. (B) Effect of TRB3 overexpression in 3T3-L1 cells on early activation of Akt/GSK3 and ERK pathways by the adipogenic cocktail. Cells were induced to differentiate, and protein extracts were evaluated at the times indicated. A total of 30 μg of total protein extracts was loaded per lane and analyzed by Western blotting. The results are representative of three independent experiments. (C) TRB3 regulation of C/EBPβ phosphorylation in nuclear extracts of 3T3-L1 preadipocytes. Nuclear and cytoplasmic extracts were prepared at 24 h postinduction, and 30 μg of protein were loaded in each lane and analyzed by Western blotting. Nuclear and cytoplasmic extract quality was determined with lamin A and SOD4 protein. The results are representative of three independent experiments. Quantification of the phosphorylation of C/EBPβ in 3T3-L1 preadipocytes nuclear extracts. Western blot results were analyzed with numeric quantification program (ImageQuant). The results are representative of the three independent experiments presented in Fig. 5B and are expressed as the ratio of phosphorylated C/EBPβ on total C/EBPβ.
Since both GSK3 and ERK1/2 have been shown to phosphorylate C/EBPβ and thereby regulate its transcriptional activity, it was important to analyze the activation of these specific signaling molecules during the first few hours of differentiation in 3T3-L1 cells to determine which might be the earliest target of TRB3. During the first hour of differentiation after exposure to DEX, IBMX, and insulin, there was a robust induction of phosphorylation of Akt (P-Ser473 Akt) and GSK3 (P-Ser9-GSK), and this was maintained during the first 8 h of induction (Fig. 6B, low left panel). Somewhat surprisingly, at these early time points in differentiation, we observed that overexpression of TRB3 had no effect on the phosphorylation of Akt or GSK3 in response to the adipogenic cocktail in these cells (Fig. 6B, low right panel). In control cells, stimulation of the ERK pathway was first detected at 4 h postinduction as indicated by an extensive phosphorylation of ERK1/2 (P-Thr202/Tyr204-ERK1/2). In addition, in control cells during this induction, C/EBPβ expression was already induced by 2 h and concomitant with the activation of the ERK1/2, C/EBPβ was phosphorylated on Thr188 (Fig. 6B, upper left panel). Overexpression of TRB3, on the other hand, inhibited the phosphorylation and activation of the ERK1/2 and, in doing so, strongly reduced the phosphorylation of C/EBPβ (Fig. 6B, upper right panel).
To confirm this observation, we prepared nuclear extracts of three independent preadipocyte cell lines 1 day postinduction and studied the phosphorylation state of nuclear C/EBPβ in the presence or the absence of TRB3 ectopic expression. In the control cells, we obtained a robust activation of C/EBPβ on its ERK consensus site, whereas in the 3T3-L1-TRB3 cells, the activation of both ERK1/2 and C/EBPβ were strongly reduced (Fig. 6C). TRB3 overexpression resulted in at least a 60% reduction in the phosphorylation of C/EBPβ (Fig. 6D). Taken together, these data indicate that TRB3 can block C/EBPβ activity directly by interacting with the protein and reducing its ability to bind DNA, thereby suppressing its transcriptional activity, but also by suppressing the MAP kinase signaling pathway that normally leads to the phosphorylation and activation of C/EBPβ.
DISCUSSION
There are three isoforms of TRB in mammalian cells, which represent orthologues of Drosophila Tribbles, a protein that inhibits mitosis early in development (19, 30, 46). All three mammalian TRB isoforms, as well as Tribbles, have homology to serine/threonine kinases but lack catalytic residues, making them members of the pseudokinase family (5). Trb3 is highly expressed in some human tumors (6) and has been shown to be increased under fasting conditions in the adipose tissue, where it functions to inhibit lipid synthesis by stimulating the degradation of the rate-limiting enzyme of fatty acid synthesis, acetyl coenzyme A (39). Trb3 has also been reported to be increased by fasting in the liver, where it suppresses insulin signal transduction by inhibiting the activity of the protein kinase Akt, as well as potentially other signaling pathways (10, 27, 48). In the present study, we explored the role of TRB3 in adipogenesis and defined a new activity of this protein as a new negative modulator of adipocyte differentiation as it acts an inhibitor of C/EBPβ transcriptional activity.
The commitment of precursor cells to differentiate along a particular lineage is determined by the interplay of a series of negative and positive effectors controlled by a number of hormonal and growth factor signals (41). In cultured models, the hormonal signals include activation of the insulin or IGF1, glucocorticoid and cAMP signaling pathways, which act to regulate a cascade of transcription factors controlling differentiation, especially members of the C/EBP family and PPARγ. In the present study, we find that TRB3 is normally expressed in preadipocytes and is transiently downregulated during the clonal expansion phase of adipogenesis by induced by both glucocorticoids and agents that increase cAMP. This downregulation appears to be an important step toward adipogenic differentiation by allowing a MAP kinase-dependent phosphorylation and activation of C/EBPβ, which in turn induces expression of C/EBPα, PPARγ, and essential proteins for the adipocyte function, such as a-FABP, resulting in terminal differentiation of the adipocyte. Overexpression of TRB3 in both 3T3-L1 preadipocytes and in embryonic fibroblasts, such as Swiss fibroblasts, completely inhibits the induction of proadipogenic genes by C/EBPβ and, consequently, blocks terminal adipogenesis.
Recent studies have shown that C/EBPβ, along with C/EBPδ, is a key player in the early stages of adipogenesis. Not only does C/EBPβ induce expression of C/EBPα and PPARγ, it also regulates the expression and activity of additional factors that cooperate with C/EBPα and PPARγ to ensure terminal differentiation. Most notably, C/EBPβ stimulates clonal expansion, induces the proadipogenic transcription factor KLF5, and is required for the synthesis of PPARγ ligands (20, 35, 51, 61). The transcriptional activity of C/EBPβ is regulated by several mechanisms, including association with other proteins, such as DIPA, DeltaFosB, ETO, CHOP-10, or GATA (2, 9, 29, 42, 50, 55), as well as posttranslational modifications, including phosphorylation, acetylation, and sumoylation (23, 37, 38, 47, 49).
Several reports have demonstrated a role for phosphorylation in regulating C/EBPβ activity in response to a variety of effectors in different cell types. In fact, the data suggest strongly that C/EBPβ normally exists in a repressed state and is activated by phosphorylation of the regulatory domain (26, 37, 38, 49). This domain contains several serines and threonines, all of which are phosphorylated to some extent by a constitutive process. The site encompassing threonine 188 (SPPGT188PSP) is a consensus site for both GSK3 and ERK1/2. Modification of this site by mutation of Thr188 to alanine inhibits the C/EBPβ-associated transcription of a c-fos reporter in response to growth hormone signaling and prevents the induction of C/EBPα and adiponectin expression by C/EBPβ (37, 38). In addition, a recent study has identified a proline-directed phosphoacceptor site at serine 64, which appears to be a target of Cdk2 and Cdc2. The authors of that study found that Thr188 can be phosphorylated by CDKs in addition to ERK1/2 and GSK3 and demonstrated that cell cycle-dependent phosphorylation of C/EBPβ on Ser64 and Thr188 is required to promote Ras-induced transformation of NIH 3T3 cells (47).
In vitro experiments with C/EBPβ show that phosphorylation of Thr188 by ERK1/2 primes C/EBPβ for subsequent phosphorylation on Ser184 and Thr179 by GSK3β, acquisition of DNA-binding function, and transactivation of the C/EBPα and PPARγ genes (49). Our data indicate that TRB3 is inhibiting C/EBPβ activity by directly interacting with C/EBPβ on its RD1 domain and also preventing its phosphorylation by ERK1/2 and possibly GSK3 on critical regulatory phosphoacceptor sites. In fact, the data show that TRB3 significantly attenuates the proadipogenic, insulin-dependent activation of ERK1/2. Interestingly, in these cells, under early differentiation conditions, we see no effect of TRB3 overexpression to block Akt or GSK3 phosphorylation or activation. On the other hand, we find that TRB3 is capable of specifically interacting with C/EBPβ through its repression domain 1, preventing its ability to bind DNA and transactivate the a-FABP gene and C/EBPα gene promoters. Our results indicate that TRB3 is able to regulate C/EBPβ activity through two distinct mechanisms: an upstream regulation of its phosphorylation or activation by MAP kinase and a direct physical interaction between TRB3 and C/EBPβ (Fig. 7), although the relative importance of these two mechanisms in the final inhibition of C/EBPβ still needs to be determined. Interestingly, these results demonstrate that, in addition to the role of TRB3 in regulating insulin signaling and lipid mobilization in mature adipocytes (39), TRB3 also can control the transcriptional cascade of early adipogenesis and, thus, TRB3 is an important determinant of the adipocyte differentiation process.
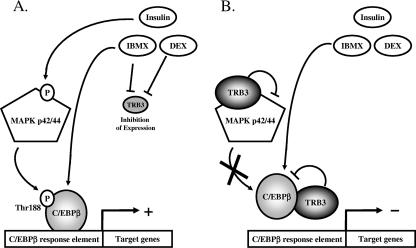
FIG. 7.
Schematic diagram of TRB3-mediated inhibition of adipocyte differentiation. (A) Induction of adipocyte differentiation requires low or absent TRB3. (B) Normal or increased TRB3 blocks C/EBPβ-mediated adipocyte differentiation.
A similar dual function, i.e., effects on differentiation and mature cell function, has been assigned to some other regulators of adipogenesis, including ERK (4, 21) and ADD1/SREBP1 (24, 25, 56). Indeed, ERK proteins are able to regulate the activity of key transcription factors that are essential to adipocyte differentiation such as C/EBPβ or PPARγ and also regulate the insulin-stimulated glucose transport in mature adipocyte. ADD1/SREBP1 is also able to promote adipocyte differentiation and is involved in the insulin-mediated regulation of fatty acid synthetase and leptin genes expression in mature adipocytes.
In this regard, it is worth noting that an additional function of the proadipogenic hormones, DEX and cAMP, is to suppress TRB3 expression coincident with their induction of C/EBPβ. Such a coordinated mechanism must exist since the subsequent induction of PPARγ and C/EBPα depends on optimum C/EBPβ activity. Our data are consistent with the notion that TRB3 is a negative regulator of C/EBPβ, and, consequently, its expression and/or activity needs to be attenuated to permit C/EBPβ target gene expression. Once C/EBPβ has fulfilled its commitments during early adipogenesis its expression is terminated, and TRB3 expression or activity can be restored to fulfill its functions in the mature adipocyte.
As noted above, Tribbles proteins belong to a new family of proteins termed pseudokinases (5). At present some 48 different proteins have been identified as having kinase homology sequences but lacking catalytic activity due to amino acid sequence alterations in the kinase catalytic site. For example, the “kinase-like” domain of TRB3 lacks the consensus ATP binding site essential for any kinase activity, and the kinase consensus motif (H/Y)RDL(K/R)XXN in the catalytic core domain is not conserved. Thus, TRB3 is not a functional kinase. However, pseudokinases such as TRB3 still possess the protein-protein interaction domains required for substrate regulation, allowing them to act as dominant negative in several signaling pathways. Indeed, the Tribbles family has been extensively studied for its implication in the regulation of several key signaling pathways such as the Akt/GSK3, ERK, and S6K1-mTOR pathways, together with their potential involvement in diabetes and cancer (10, 27, 31).
More recently, TRB3 has also been implicated in the regulation of other proteins, including transcriptional factors, especially the leucine-zipper transcription factors such as members of the ATF family (34, 36). Indeed, TRB3 has been shown to interact and inhibits both ATF-4 and ATF-5 by regulating their association with the transcriptional apparatus. It has been proposed that the association of TRB3 with ATF-4 or -5 triggers their ubiquitination and therefore their degradation by the proteasome (36). C/EBPs are also members of the leucine zipper family of transcription factors, and TRB3 appears to be involved in their regulation as well. For example, in Drosophila Tribbles interacts with Slbo, a b-ZIP transcription factor homologous to C/EBP protein, and this interaction facilitates the ubiquitination and degradation of Slbo (43). In mammalian cells TRB3 can also interact with CHOP (C/EBP-homologous protein). In this case, TRB3 inhibits CHOP activity but does not promote its degradation (34). Furthermore, C/EBPα has been shown to be a target of the Tribbles homolog TRB2, which inactivates C/EBPα in the pathogenesis of acute myelogenous leukemia (22). In our data, no alteration of either the total amount of C/EBPβ or C/EBPβ protein half-life (data not shown) was observed in response to TRB3 overexpression, indicating that TRB3 is not triggering C/EBPβ ubiquitination and therefore its degradation by the proteasome. Furthermore, under the conditions of these experiments, TRB3 is a specific inhibitor of C/EBPβ and not of C/EBPα or C/EBPδ. An explanation for this specificity was revealed by the deletion analysis that defined the repression domain RD1 of C/EBPβ as the major binding site for TRB3. Despite the fact that all C/EBP family members share a high identity in their b-ZIP (involved in the specificity of the DNA binding and the dimerization) and in their transactivation domains (involved in the recruitment of the basal transcription apparatus), the other domains, including RD1, are quite divergent.
In the present study, we demonstrate that TRB3 is a direct regulator and inhibitor of C/EBPβ. In this case, TRB3 inhibits C/EBPβ in part by preventing its phosphorylation by MAP kinases, as well as by associating with C/EBPβ in the nucleus through its repression domain 1, resulting in a blockade of its transcriptional activity. The same kind of regulation has been observed for p65/RelA. TRB3 interacts with p65/RelA in 293 cells and prevents its protein kinase A-dependent phosphorylation but not the nuclear translocation of p65/RelA (58). Together, these data strongly support the emerging perspective that TRB3 and other pseudokinases participate as a scaffolding protein involved in the assembly of multiprotein complexes involved in signaling processes and transcriptional regulation (5).
In conclusion, the mammalian homologue of Tribbles, TRB3, is an important negative regulator of adipogenesis, acting to block the activity of a critical adipogenic transcription factor, C/EBPβ. This effect occurs through multiple mechanisms, including a direct interaction between the two proteins. The fact that C/EBPβ functions during very early steps in the commitment of mesenchymal stem cells to adipocytic and other lineages suggests that TRB3 may play a more general role in the determination of the fate of mesenchymal progenitors. Further delineation of the role of TRB3 in regulating processes controlling the expansion (proliferation) and differentiation of adipogenic progenitors should lead to the identification of additional targets for therapeutic approaches to combat obesity and its associated disorders.
Acknowledgments
We thank Marc Montminy for providing the Flag-TRB3 constructs and Shuai Wangand for the C/EBPβ deletion constructs.
This work was supported by National Institutes of Health grants DK33201 and DK55545 and Joslin Diabetes and Endocrinology Research Center grant DK34834 (to C.R.K.).
Footnotes
Published ahead of print on 23 July 2007.
REFERENCES
- 1.Belmonte, N., B. W. Phillips, F. Massiera, P. Villageois, B. Wdziekonski, P. Saint-Marc, J. Nichols, J. Aubert, K. Saeki, A. Yuo, S. Narumiya, G. Ailhaud, and C. Dani. 2001. Activation of extracellular signal-regulated kinases and CREB/ATF-1 mediate the expression of CCAAT/enhancer binding proteins beta and delta in preadipocytes. Mol. Endocrinol. 15:2037-2049. [DOI] [PubMed] [Google Scholar]
- 2.Bezy, O., C. Elabd, O. Cochet, R. K. Petersen, K. Kristiansen, C. Dani, G. Ailhaud, and E. Z. Amri. 2005. Delta-interacting protein A, a new inhibitory partner of CCAAT/enhancer-binding protein beta, implicated in adipocyte differentiation. J. Biol. Chem. 280:11432-11438. [DOI] [PubMed] [Google Scholar]
- 3.Bluher, M., M. D. Michael, O. D. Peroni, K. Ueki, N. Carter, B. B. Kahn, and C. R. Kahn. 2002. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev. Cell 3:25-38. [DOI] [PubMed] [Google Scholar]
- 4.Bost, F., M. Aouadi, L. Caron, and B. Binetruy. 2005. The role of MAPKs in adipocyte differentiation and obesity. Biochimie 87:51-56. [DOI] [PubMed] [Google Scholar]
- 5.Boudeau, J., D. Miranda-Saavedra, G. J. Barton, and D. R. Alessi. 2006. Emerging roles of pseudokinases. Trends Cell Biol. 16:443-452. [DOI] [PubMed] [Google Scholar]
- 6.Bowers, A. J., S. Scully, and J. F. Boylan. 2003. SKIP3, a novel Drosophila tribbles ortholog, is overexpressed in human tumors and is regulated by hypoxia. Oncogene 22:2823-2835. [DOI] [PubMed] [Google Scholar]
- 7.Cao, Z., R. M. Umek, and S. L. McKnight. 1991. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 5:1538-1552. [DOI] [PubMed] [Google Scholar]
- 8.Christy, R. J., K. H. Kaestner, D. E. Geiman, and M. D. Lane. 1991. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc. Natl. Acad. Sci. USA 88:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, K. A., A. W. Harmon, J. B. Harp, and Y. M. Patel. 2004. Rb regulates C/EBPβ-DNA-binding activity during 3T3-L1 adipogenesis. Am. J. Physiol. Cell Physiol. 286:C349-C354. [DOI] [PubMed] [Google Scholar]
- 10.Du, K., S. Herzig, R. N. Kulkarni, and M. Montminy. 2003. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300:1574-1577. [DOI] [PubMed] [Google Scholar]
- 11.Elberg, G., J. M. Gimble, and S. Y. Tsai. 2000. Modulation of the murine peroxisome proliferator-activated receptor gamma 2 promoter activity by CCAAT/enhancer-binding proteins. J. Biol. Chem. 275:27815-27822. [DOI] [PubMed] [Google Scholar]
- 12.Entingh, A. J., C. M. Taniguchi, and C. R. Kahn. 2003. Bi-directional regulation of brown fat adipogenesis by the insulin receptor. J. Biol. Chem. 278:33377-33383. [DOI] [PubMed] [Google Scholar]
- 13.Entingh-Pearsall, A., and C. R. Kahn. 2004. Differential roles of the insulin and insulin-like growth factor-I (IGF-I) receptors in response to insulin and IGF-I. J. Biol. Chem. 279:38016-38024. [DOI] [PubMed] [Google Scholar]
- 14.Farmer, S. R. 2005. Regulation of PPARγ activity during adipogenesis. Int. J. Obesity 29(Suppl. 1):S13-S16. [DOI] [PubMed] [Google Scholar]
- 15.Farmer, S. R. 2006. Transcriptional control of adipocyte formation. Cell Metab. 4:263-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gagnon, A., C. S. Chen, and A. Sorisky. 1999. Activation of protein kinase B and induction of adipogenesis by insulin in 3T3-L1 preadipocytes: contribution of phosphoinositide-3,4,5-trisphosphate versus phosphoinositide-3,4-bisphosphate. Diabetes 48:691-698. [DOI] [PubMed] [Google Scholar]
- 17.Green, H., and O. Kehinde. 1976. Spontaneous heritable changes leading to increased adipose conversion in 3T3 cells. Cell 7:105-113. [DOI] [PubMed] [Google Scholar]
- 18.Green, H., and M. Meuth. 1974. An established pre-adipose cell line and its differentiation in culture. Cell 3:127-133. [DOI] [PubMed] [Google Scholar]
- 19.Grosshans, J., and E. Wieschaus. 2000. A genetic link between morphogenesis and cell division during formation of the ventral furrow in Drosophila. Cell 101:523-531. [DOI] [PubMed] [Google Scholar]
- 20.Hamm, J. K., B. H. Park, and S. R. Farmer. 2001. A role for C/EBPβ in regulating peroxisome proliferator-activated receptor gamma activity during adipogenesis in 3T3-L1 preadipocytes. J. Biol. Chem. 276:18464-18471. [DOI] [PubMed] [Google Scholar]
- 21.Harmon, A. W., D. S. Paul, and Y. M. Patel. 2004. MEK inhibitors impair insulin-stimulated glucose uptake in 3T3-L1 adipocytes. Am. J. Physiol. Endocrinol. Metab. 287:E758-E766. [DOI] [PubMed] [Google Scholar]
- 22.Keeshan, K., Y. He, B. J. Wouters, O. Shestova, L. Xu, H. Sai, C. G. Rodriguez, I. Maillard, J. W. Tobias, P. Valk, M. Carroll, J. C. Aster, R. Delwel, and W. S. Pear. 2006. Tribbles homolog 2 inactivates C/EBPα and causes acute myelogenous leukemia. Cancer Cell 10:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, J., C. A. Cantwell, P. F. Johnson, C. M. Pfarr, and S. C. Williams. 2002. Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J. Biol. Chem. 277:38037-38044. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J. B., P. Sarraf, M. Wright, K. M. Yao, E. Mueller, G. Solanes, B. B. Lowell, and B. M. Spiegelman. 1998. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J. Clin. Investig. 101:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, J. B., and B. M. Spiegelman. 1996. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 10:1096-1107. [DOI] [PubMed] [Google Scholar]
- 26.Kim, J. W., Q. Q. Tang, X. Li, and M. D. Lane. 2007. Effect of phosphorylation and S-S bond-induced dimerization on DNA binding and transcriptional activation by C/EBPβ. Proc. Natl. Acad. Sci. USA 104:1800-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss-Toth, E., S. M. Bagstaff, H. Y. Sung, V. Jozsa, C. Dempsey, J. C. Caunt, K. M. Oxley, D. H. Wyllie, T. Polgar, M. Harte, L. A. O'Neill, E. E. Qwarnstrom, and S. K. Dower. 2004. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J. Biol. Chem. 279:42703-42708. [DOI] [PubMed] [Google Scholar]
- 28.Koo, S. H., H. Satoh, S. Herzig, C. H. Lee, S. Hedrick, R. Kulkarni, R. M. Evans, J. Olefsky, and M. Montminy. 2004. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat. Med. 10:530-534. [DOI] [PubMed] [Google Scholar]
- 29.Kveiborg, M., G. Sabatakos, R. Chiusaroli, M. Wu, W. M. Philbrick, W. C. Horne, and R. Baron. 2004. DeltaFosB induces osteosclerosis and decreases adipogenesis by two independent cell-autonomous mechanisms. Mol. Cell. Biol. 24:2820-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mata, J., S. Curado, A. Ephrussi, and P. Rorth. 2000. Tribbles coordinates mitosis and morphogenesis in Drosophila by regulating string/CDC25 proteolysis. Cell 101:511-522. [DOI] [PubMed] [Google Scholar]
- 31.Matsushima, R., N. Harada, N. J. Webster, Y. M. Tsutsumi, and Y. Nakaya. 2006. Effect of TRB3 on insulin and nutrient-stimulated hepatic p70 S6 kinase activity. J. Biol. Chem. 281:29719-29729. [DOI] [PubMed] [Google Scholar]
- 32.Mayumi-Matsuda, K., S. Kojima, H. Suzuki, and T. Sakata. 1999. Identification of a novel kinase-like gene induced during neuronal cell death. Biochem. Biophys. Res. Commun. 258:260-264. [DOI] [PubMed] [Google Scholar]
- 33.Nakae, J., T. Kitamura, Y. Kitamura, W. H. Biggs III, K. C. Arden, and D. Accili. 2003. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell 4:119-129. [DOI] [PubMed] [Google Scholar]
- 34.Ohoka, N., S. Yoshii, T. Hattori, K. Onozaki, and H. Hayashi. 2005. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J. 24:1243-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oishi, Y., I. Manabe, K. Tobe, K. Tsushima, T. Shindo, K. Fujiu, G. Nishimura, K. Maemura, T. Yamauchi, N. Kubota, R. Suzuki, T. Kitamura, S. Akira, T. Kadowaki, and R. Nagai. 2005. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 1:27-39. [DOI] [PubMed] [Google Scholar]
- 36.Ord, D., and T. Ord. 2003. Mouse NIPK interacts with ATF4 and affects its transcriptional activity. Exp. Cell Res. 286:308-320. [DOI] [PubMed] [Google Scholar]
- 37.Park, B. H., L. Qiang, and S. R. Farmer. 2004. Phosphorylation of C/EBPβ at a consensus extracellular signal-regulated kinase/glycogen synthase kinase 3 site is required for the induction of adiponectin gene expression during the differentiation of mouse fibroblasts into adipocytes. Mol. Cell. Biol. 24:8671-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piwien-Pilipuk, G., O. MacDougald, and J. Schwartz. 2002. Dual regulation of phosphorylation and dephosphorylation of C/EBPβ modulate its transcriptional activation and DNA binding in response to growth hormone. J. Biol. Chem. 277:44557-44565. [DOI] [PubMed] [Google Scholar]
- 39.Qi, L., J. E. Heredia, J. Y. Altarejos, R. Screaton, N. Goebel, S. Niessen, I. X. Macleod, C. W. Liew, R. N. Kulkarni, J. Bain, C. Newgard, M. Nelson, R. M. Evans, J. Yates, and M. Montminy. 2006. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science 312:1763-1766. [DOI] [PubMed] [Google Scholar]
- 40.Ramji, D. P., and P. Foka. 2002. CCAAT/enhancer-binding proteins: structure, function and regulation 4. Biochem. J. 365:561-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ringold, G. M., A. B. Chapman, D. M. Knight, and F. M. Torti. 1986. Hormonal control of adipogenesis. Ann. N. Y. Acad. Sci. 478:109-119. [DOI] [PubMed] [Google Scholar]
- 42.Rochford, J. J., R. K. Semple, M. Laudes, K. B. Boyle, C. Christodoulides, C. Mulligan, C. J. Lelliott, S. Schinner, D. Hadaschik, M. Mahadevan, J. K. Sethi, A. Vidal-Puig, and S. O'Rahilly. 2004. ETO/MTG8 is an inhibitor of C/EBPβ activity and a regulator of early adipogenesis. Mol. Cell. Biol. 24:9863-9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rorth, P., K. Szabo, and G. Texido. 2000. The level of C/EBP protein is critical for cell migration during Drosophila oogenesis and is tightly controlled by regulated degradation. Mol. Cell 6:23-30. [DOI] [PubMed] [Google Scholar]
- 44.Rosen, E. D., and B. M. Spiegelman. 2000. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 16:145-171. [DOI] [PubMed] [Google Scholar]
- 45.Sakaue, H., W. Ogawa, M. Matsumoto, S. Kuroda, M. Takata, T. Sugimoto, B. M. Spiegelman, and M. Kasuga. 1998. Posttranscriptional control of adipocyte differentiation through activation of phosphoinositide 3-kinase. J. Biol. Chem. 273:28945-28952. [DOI] [PubMed] [Google Scholar]
- 46.Seher, T. C., and M. Leptin. 2000. Tribbles, a cell-cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation. Curr. Biol. 10:623-629. [DOI] [PubMed] [Google Scholar]
- 47.Shuman, J. D., T. Sebastian, P. Kaldis, T. D. Copeland, S. Zhu, R. C. Smart, and P. F. Johnson. 2004. Cell cycle-dependent phosphorylation of C/EBPβ mediates oncogenic cooperativity between C/EBPβ and H-RasV12. Mol. Cell. Biol. 24:7380-7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung, H. Y., S. E. Francis, D. C. Crossman, and E. Kiss-Toth. 2006. Regulation of expression and signalling modulator function of mammalian tribbles is cell type specific. Immunol. Lett. 104:171-177. [DOI] [PubMed] [Google Scholar]
- 49.Tang, Q. Q., M. Gronborg, H. Huang, J. W. Kim, T. C. Otto, A. Pandey, and M. D. Lane. 2005. Sequential phosphorylation of CCAAT enhancer-binding protein beta by MAPK and glycogen synthase kinase 3beta is required for adipogenesis. Proc. Natl. Acad. Sci. USA 102:9766-9771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang, Q. Q., and M. D. Lane. 2000. Role of C/EBP homologous protein (CHOP-10) in the programmed activation of CCAAT/enhancer-binding protein-beta during adipogenesis. Proc. Natl. Acad. Sci. USA 97:12446-12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang, Q. Q., T. C. Otto, and M. D. Lane. 2003. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc. Natl. Acad. Sci. USA 100:850-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang, Q. Q., T. C. Otto, and M. D. Lane. 2003. Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc. Natl. Acad. Sci. USA 100:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang, Q. Q., J. W. Zhang, and L. M. Daniel. 2004. Sequential gene promoter interactions by C/EBPβ, C/EBPα, and PPARγ during adipogenesis. Biochem. Biophys. Res. Commun. 318:213-218. [DOI] [PubMed] [Google Scholar]
- 54.Tomiyama, K., H. Nakata, H. Sasa, S. Arimura, E. Nishio, and Y. Watanabe. 1995. Wortmannin, a specific phosphatidylinositol 3-kinase inhibitor, inhibits adipocytic differentiation of 3T3-L1 cells. Biochem. Biophys. Res. Commun. 212:263-269. [DOI] [PubMed] [Google Scholar]
- 55.Tong, Q., J. Tsai, G. Tan, G. Dalgin, and G. S. Hotamisligil. 2005. Interaction between GATA and the C/EBP family of transcription factors is critical in GATA-mediated suppression of adipocyte differentiation. Mol. Cell. Biol. 25:706-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tontonoz, P., J. B. Kim, R. A. Graves, and B. M. Spiegelman. 1993. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol. Cell. Biol. 13:4753-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tseng, Y. H., K. M. Kriauciunas, E. Kokkotou, and C. R. Kahn. 2004. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol. Cell. Biol. 24:1918-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, M., L. G. Xu, Z. Zhai, and H. B. Shu. 2003. SINK is a p65-interacting negative regulator of NF-κB-dependent transcription. J. Biol. Chem. 278:27072-27079. [DOI] [PubMed] [Google Scholar]
- 59.Wu, Z., N. L. R. Bucher, and S. R. Farmer. 1996. Induction of peroxisome proliferator activated receptor y during conversion of 3T3-L1 fibroblasts into adipocytes is mediated by C/EBPΒ, C/EBP/δ, and glucocorticoids. Mol. Cell. Biol. 16:4128-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, Z., Y. Xie, N. L. Bucher, and S. R. Farmer. 1995. Conditional ectopic expression of C/EBPβ in NIH 3T3 cells induces PPARγ and stimulates adipogenesis. Genes Dev. 9:2350-2363. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, J. W., Q. Q. Tang, C. Vinson, and M. D. Lane. 2004. Dominant-negative C/EBP disrupts mitotic clonal expansion and differentiation of 3T3-L1 preadipocytes. Proc. Natl. Acad. Sci. USA 101:43-47. [DOI] [PMC free article] [PubMed] [Google Scholar]