Abstract
Cells can regulate their protein repertoire in response to extracellular stimuli via alternative splicing; however, the mechanisms controlling this process are poorly understood. The CD45 gene undergoes alternative splicing in response to T-cell activation to regulate T-cell function. The ESS1 splicing silencer in CD45 exon 4 confers basal exon skipping in resting T cells through the activity of hnRNP L and confers activation-induced exon skipping in T cells via previously unknown mechanisms. Here we have developed an in vitro splicing assay that recapitulates the signal-induced alternative splicing of CD45 and demonstrate that cellular stimulation leads to two changes to the ESS1-bound splicing regulatory complex. Activation-induced posttranslational modification of hnRNP L correlates with a modest increase in the protein's repressive activity. More importantly, the splicing factor PSF is recruited to the ESS1 complex in an activation-dependent manner and accounts for the majority of the signal-regulated ESS1 activity. The associations of hnRNP L and PSF with the ESS1 complex are largely independent of each other, but together these proteins account for the total signal-regulated change in CD45 splicing observed in vitro and in vivo. Such a combinatorial effect on splicing allows for precise regulation of signal-induced alternative splicing.
To maintain viability most, if not all, cells within an organism must be capable of responding to a changing environment. For example, neuronal and muscle cells must respond to activation to promote behaviors and movement, while lymphoid cells must respond to an immune challenge to prevent or control infection. Such flexibility requires that individual cells have the ability to change function rapidly and precisely in response to a given stimulus. In general, cellular responsiveness is accomplished through the activity of signal transduction cascades that ultimately alter protein expression or function via numerous mechanisms. Much of the previous work on signal-regulated protein expression has focused on the regulation of transcription; however, there is growing recognition of the abundance and importance of signal-induced changes in alternative splicing (2, 11, 13).
Alternative splicing is the process by which exons of a given gene are differentially included or excluded from the final message, such that a single gene can be processed into multiple discrete mRNAs (3, 16). Since each of these variant mRNAs typically encodes a unique protein of distinct function, changes in the splicing pattern of a gene can have a profound effect on protein expression and cellular activity. It has recently become apparent that numerous genes undergo changes in splicing patterns upon neuronal depolarization (2, 13) or T-cell activation (11), suggesting a broad role for this mode of gene regulation in determining the physiologic response of cells to stimulation. Thus, understanding the mechanisms by which extracellular stimuli can induce changes in the splicing regulatory machinery is of primary importance in deciphering the cellular response to environmental cues.
Alternative splicing is typically controlled by the binding of splicing regulatory proteins to sequences within or near the variable exon that, in turn, controls the recognition and use of the exon by the splicing machinery, or spliceosome (3, 16). Generally, splicing regulatory proteins fall into one of two categories, as follows: SR (serine-arginine-rich) proteins generally enhance exon inclusion, whereas hnRNP proteins typically promote exon skipping (3). Additional potential regulatory proteins also exist, such as the PTB-associated splicing factor (PSF), which has a general role as an essential spliceosomal component but has also been implicated in regulating numerous other nuclear processes such as mRNA localization, transcription, and DNA unwinding (reviewed in reference 29).
Despite the prevalence of signal-induced splicing regulation, little is known about how the substrate-specific binding or activity of splicing regulatory proteins is influenced by signal transduction pathways. A few studies have described changes in the phosphorylation of SR or hnRNP proteins in response to cellular stress or stimulation leading to altered activity and/or subcellular localization of these proteins (1, 4, 19, 20, 34). However, there has been little direct demonstration that altered phosphorylation of an SR/hnRNP protein confers a specific change in known signal-responsive alternative splicing events. Moreover, two major unanswered questions related to signal-induced alternative splicing are how specificity is achieved and why a larger number of genes are not affected by stimuli that alter the activity of ubiquitous SR or hnRNP proteins.
The CD45 gene, which encodes a hematopoiesis-specific transmembrane protein tyrosine phosphatase, provides an excellent model for signal-induced alternative splicing. Three variable exons of the CD45 gene are inducibly skipped upon T-cell activation, leading to decreased phosphatase activity and maintenance of T-cell homeostasis (8, 14). Importantly, while T-cell activation results in only a three- to fivefold change in the ratio of CD45 isoforms, the physiologic importance of this change is evidenced by the fact that naturally occurring polymorphisms within the CD45 gene that disrupt signal-induced alternative splicing correlate with susceptibility to a wide range of autoimmune diseases and viral infection in humans (5, 12, 32, 33).
In previous work, we have identified an exonic splicing silencer designated ESS1 within the CD45 gene's variable exon 4 that mediates both partial exon repression in resting cells and increased exon skipping upon cellular stimulation (26). We have furthermore shown that the partial exon repression in resting cells is conferred via the binding of hnRNP L to ESS1, which stalls the assembly of the U1 and U2 snRNP components of the spliceosome on the repressed exon in an exon-defined orientation unable to progress to a final catalytic complex (9, 27). Using an in vitro assay that recapitulates the signal-induced alternative splicing of the CD45 gene in T cells, we now show that T-cell activation induces two changes in the composition of the ESS1-bound regulatory complex: the posttranslational modification of hnRNP L and the addition of PSF. The change in modification of hnRNP L correlates with a modest increase in silencing activity, while the addition of PSF to the ESS1-bound complex has a larger effect on exon skipping. Interestingly, the activation-specific addition of PSF to the ESS1 regulatory complex is not a result of a global change in the nuclear concentration of PSF but, rather, is due to some change in PSF activity. Indeed, we find that recombinant PSF purified from resting and activated T cells exhibits differential activity in ESS1-dependent exon silencing. Finally, several lines of evidence suggest that the functional consequences of the modification of hnRNP L and the addition of PSF are largely separable but together combine to give the full increase in exon silencing that is observed both in vitro and in vivo upon T-cell stimulation. These data provide a new mechanism for signal-induced regulation of alternative splicing.
MATERIALS AND METHODS
Plasmids and RNA.
The WT, ΔESS1, and Mut1 minigenes have been described previously as WT, alt-ESS, and mESS1, (9) and the NS Control RNA sequence has been previously published as E14 (27). The Mut2 minigene was derived from the WT version by PCR-based mutagenesis as described previously (26) The EF-Flag-PSF vector for the expression of recombinant N-terminally tagged PSF in JSL1 cells was made by inserting the EF1alpha promoter and the sequence encoding a Flag tag into a pcDNA3 backbone to generate pEF-Flag. An XhoI-flanked cDNA encoding PSF, a gift from B. Blencowe (University of Toronto), was inserted in frame into the pEF-Flag vector by blunt ligation to the NdeI site downstream of the Flag sequence. For in vitro splicing experiments, minigene-derived RNAs were transcribed in vitro using T7 polymerase (Promega). 5′-biotinylated ESS1, Mut1, NS Control, and CA repeat region [(CA)20] RNA as well as nonbiotinylated ESS1, Mut1, Mut2, and NS Control RNA was chemically synthesized by Dharmacon.
Cell culture, transfections, and RNA isolation.
JSL1 cells have been described previously (15). Stable cell lines expressing minigenes were prepared and analyzed as described by Rothrock et al. (26). Antisense knockdown of proteins was performed by electroporation of 20 million JSL1 cells with the indicated amounts of morpholino oligonucleotides, as follows: hnRNP L, CGCCCGCCGCCGCCATCTTCACCAT; PTB, CTATATCTGGGACAATGCCGTCCAT; PSF, CTCCGGAACCGATCCCGAGACATGT; and p54nrb, TATTACTCTGCATTTTTGCACCCTC (Gene-Tools, Inc.). Cells were allowed to recover overnight in RPMI medium plus 5% fetal calf serum before stimulation with phorbol myristate acetate (PMA) and then harvested for RNA and protein 48 h after treatment with PMA (20 ng/ml). Combination knockdown experiments were performed as described above, using 5 nmol of the hnRNP L morpholino in combination with either 10 nmol of the PSF morpholino or 5 nmol of the p54nrb morpholino.
Nuclear extract and purified proteins.
JSL1 cells were grown in roller bottles, either untreated (resting) or treated with 50 ng/ml PMA (stimulated) for approximately 60 h, and nuclear extract (NE) was purified as described previously (27). Recombinant glutathione S-transferase (GST)-hnRNP L was purified from SF9 cells, using glutathione-Sepharose 4B resin (GE Biosciences) according to the manufacturer's protocol and as described by Rothrock et al. (26). Recombinant His-tagged PSF was purified using the previously described pET-PSF construct (a kind gift from J. Patton, Vanderbilt). Bacterial cultures containing the pET-PSF were grown at 37°C to an optical density at 600 nm of 0.5 and induced with isopropyl-β-d-thiogalactopyranoside (1 mM) and incubated for an additional 7 h at 37°C. Bacteria were treated with 5 mg/ml lysozyme and sonicated to lyse. His-tagged PSF was then purified in batch using Ni-nitrilotriacetic acid resin (QIAGEN). After extensive washing, protein was eluted using increasing concentrations of imidazole (0 to 0.5 M), and fractions were dialyzed overnight against BC100 (20 mM Tris-HCl [pH 7.5], 100 mM KCl, 0.2 mM EDTA, 20% glycerol). The protocol was adapted from the study by Patton et al. (21). Purification of hnRNP L from either the resting or the stimulated JSL1 nuclear extract was performed as described previously (10). Five hundred picomoles of 5′-biotinylated (CA)20 RNA (Dharmacon) was coupled to 20 μl of streptavidin-conjugated acrylamide resin (Pierce). Resting or stimulated JSL1 NE (approximately 1.5 mg of NE) was incubated with 40 μl of RNA-conjugated resin. After extensive washing, hnRNP L was eluted using buffer containing 6 M urea, and samples were dialyzed overnight against 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 20% glycerol, 1 mM EDTA, and 1 mM dithiothreitol (DTT). Flag-tagged PSF was purified from nuclear extracts derived from JSL1 cells stably expressing Flag-PSF. Nuclear extracts were prepared as described above. Flag-PSF was immunopurified by incubation of 40 to 50 μg of nuclear extract with 40 μl of EZ-View Red FLAG-conjugated resin (Sigma) in buffer containing 20 mM Tris-HCl (pH 7.5), 100 mM KCl, and 1 mM EDTA. After extensive washing in binding buffer, protein was eluted by incubation with 500 ng/μl 3× Flag peptide (Sigma).
RT-PCR.
Reverse transcription (RT)-PCR and analysis of cellular minigenes were performed as described previously (15). Total RNA from JSL1 cells was harvested using RNA Bee (Tel-Test) according to the manufacturer's protocol. One microgram of total RNA was heated to 90°C in the presence of 1 ng of vector-specific reverse primer (5′-GCGAGCTTAGTGATACTTGTGGGCC-3′), 300 mM NaCl, 10 mM Tris-HCl (pH 7.5) and 2 mM EDTA and allowed to cool to 43°C. The reaction mixture was diluted into an RT mixture to a final concentration of 10 mM Tris-HCl (pH 7.5), 6 mM MgCl2, 10 mM DTT, 50 mM NaCl, 1 mM of each dNTP, and Moloney murine leukemia virus. The RT reaction mixture was incubated for an additional 30 min at 43°C, boiled for 5 min, and placed on ice. PCRs were performed using one-fourth of the RT reaction mixture in 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 0.2 mM of each dNTP, 5 ng of reverse primer, 2.5 ng of unlabeled forward primer (5′-GGTTCGGCTTCTGGCGTGTGACCG-3′), 2.5 ng of 32P-labeled forward primer, and Taq polymerase. PCR was performed by heating samples to 94°C for 2 min, followed by 16 cycles of 1 min at 94°C, 1 min at 70°C, and 1.5 min at 72°C. Samples were suspended in formamide buffer and analyzed on 5% denaturing polyacrylamide gels. Quantitation was performed by densitometry using a Typhoon PhosphorImager (Amersham Biosciences).
In vitro splicing.
In vitro splicing reactions were carried out as described by Rothrock et al. (26). Approximately 1 fmol of unlabeled, capped RNA substrate was incubated with 30% JSL1 nuclear extract in a total volume of 12.5 μl containing 3.2 mM MgCl2, 20 mM phosphocreatine, 1 mM ATP, 3% polyvinyl alcohol, 1 mM DTT, 0.25 U RNasin (Promega), 90 mM KCl, 10 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, and 7% glycerol. Antibodies or proteins were added to the reaction mixture prior to the addition of substrate RNA where indicated. Reaction mixtures were incubated for 2 h at 30°C. RNA was recovered following treatment of reaction mixtures with proteinase K, phenol-chloroform extraction, and ethanol precipitation. RNA was analyzed by RT-PCR as described above with an increase in the number of cycles to 20.
RNA affinity purification.
Direct RNA affinity purifications were performed as described by Rothrock et al. (26). 5′-biotinylated RNA was incubated with JSL1 nuclear extract in buffer containing 3.2 mM MgCl2, 20 mM phosphocreatine, 1 mM ATP, 1.3% polyvinyl alcohol, 25 ng yeast tRNA, 1 mM DTT, 0.25 U RNasin (Promega), 75 mM KCl, 10 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, and 10% glycerol for 30 min at 30°C. RNA-protein complexes were then isolated by incubation with streptavidin-agarose beads (Pierce). After incubation, samples were washed in 20 mM Tris-HCl (pH 7.5), 100 mM KCl, 4 mM MgCl2, and 1 mM EDTA. Competition RNA affinity purifications were performed as described above with the addition of competitor RNA to final concentrations of 0.1 μM, 0.3 μM, 1.0 μM, and 3.0 μM.
Mass spectrometry.
Mass spectrometry (MS) was performed by the Protein Technology Core at UT Southwestern Medical Center, by digesting proteins in solution with porcine trypsin. Tryptic peptides were dissolved and injected into reverse-phase high-performance liquid chromatography/ion trap with a nanospray source, using a ThermoFinnigan LCQ Deca XP MS instrument and Xcalibur 1.3 software. Tandem MS-MS files were used to search against NCBI nonredundant protein sequence databases, using the Sonar database software (GenomicSolutions, Inc.).
Two-dimensional electrophoresis.
A 7-cm Immobiline DryStrip pH 3-10 NL (GE Biosciences) was rehydrated for 15 h in 8 M urea, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 0.5% IPG buffer (IPG buffer 3-10 NL; GE Biosciences), and 20 mM DTT. Approximately 30 μg of JSL1 nuclear extract was loaded onto the strip during rehydration. Isoelectric focusing was performed using an Ettan IPGphor II isoelectric focusing unit (Amersham Biosciences) according to the manufacturer's recommendations, with the first step extended to 60 min at 300 V. DryStrips were then incubated for 15 min at room temperature in 0.5 M Tris-HCl (pH 6.8), 8 M urea, 40% glycerol, 2.5% sodium dodecyl sulfate (SDS), and 65 mM DTT and sealed to the top of a 10% SDS-polyacrylamide gel using 0.5% agarose. An SDS-polyacrylamide gel electrophoresis was run at 200 V, and products were transferred to nitrocellulose. Membranes were then subjected to Western blotting analysis as described by Lynch and Weiss (15).
Antibodies.
Antibodies used for the various applications were as follows: anti-hnRNP L (4D11; Abcam), anti-hnRNP E2 (rabbit polyclonal; a gift from R. Andino), anti-PTB N-term (rabbit polyclonal; a gift from D. Black), anti-hnRNP A1 (4B10; ImmunoQuest), anti-hnRNP A2/B1 (DP3B3; ImmunoQuest), anti-hnRNP K/J (3C2; ImmunoQuest), anti-hnRNP D (rabbit polyclonal; BioLegend), anti-U1A (rabbit polyclonal; a gift from I. Mattaj), and anti-nmt55/p54nrb (78-1C; Affinity BioReagents). Two anti-PSF antibodies were used for PSF: B92 (Sigma) and 6D7 (Abnova).
RESULTS
In vitro splicing recapitulates the signal-responsive splicing pattern of CD45.
Previously, we have shown that the regulated splicing of the CD45 variable exons can be mimicked using minigenes expressed in the T-cell-derived JSL1 cell line (15). Consistent with the pattern of splicing of the endogenous CD45 gene, RNA derived from a minigene in which the CD45 variable exon 4 is flanked by constitutive exons 3 and 7 (Fig. 1A, WT) shows only partial inclusion of exon 4 when expressed in resting JSL1 cells (basal exon repression) and a further three- to fivefold increase in exon 4 skipping when expressed in activated JSL1 cells (activation-induced exon repression) (26). The ESS1 splicing silencer sequence is necessary and sufficient for both the basal and activation-induced repression of exon 4 in vivo, as evidenced in part by the fact that mutation or deletion of the ESS1 results in significantly higher levels of exon 4 inclusion in both resting and stimulated cells (26). Moreover, heterologous exons show no change in splicing upon activation, unless the ESS1 is introduced into the exonic sequence (26).
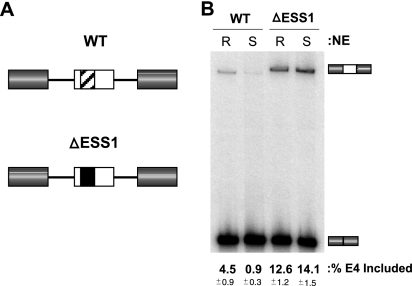
FIG. 1.
Endogenous CD45 splicing regulation is reproduced in vitro. (A) Schematic representation of the wild-type (WT) and ΔESS1 exon 4 minigenes. Variable exon 4 of CD45 is flanked by constitutive exons 3 and 7 of CD45. Exons and introns are represented by boxes and lines, respectively. The ESS1 silencer element is represented by the crosshatched region within exon 4 of the WT minigene. This element has been removed and replaced with heterologous sequence from the CD45 exon 14 in the ΔESS1 minigene. (B) In vitro splicing of WT and ΔESS1 minigenes. Using an assay established previously (9, 27), capped, unlabeled RNA derived from the WT and ΔESS1 constructs was incubated with nuclear extract prepared from either the resting (R) or the stimulated (S) JSL1 cells. Splice products were then analyzed by RT-PCR (see Materials and Methods) to calculate the percentage of exon 4 (E4) inclusion. This quantification and all others in this study are derived from at least three independent experiments, with averages (bold) and standard deviations (small type) given.
As a first step toward characterizing the changes in splicing regulation that occur upon cellular activation, we sought to determine if nuclear extracts from resting and stimulated JSL1 cells contain the minimal factors necessary to confer the differences in isoform expression observed in vivo. Previously, we have developed an in vitro splicing assay for CD45 and have demonstrated that silencing of the CD45 exon 4 in extracts from resting cells is dependent on the presence of the ESS1 motif (9, 27) (Fig. 1; and see Materials and Methods for more detail). We have further shown that skipping of the CD45 exon 4 under resting conditions is due to a stall in spliceosome assembly at a step in which the U1 and U2 snRNPs are bound to the splice sites flanking the repressed exon, but are inhibited from progressing in the assembly pathway to tri-snRNP addition and “B” complex (9). This stall in spliceosome assembly, at a complex we have termed the A-like exon complex (“AEC”), is dependent on both the ESS1 sequence and on hnRNP L (9).
Importantly, by comparing the splicing of the CD45 exon 4 in extracts from resting cells with that in extracts from stimulated JSL1 cells, we find that the signal-induced alternative splicing of the CD45 exon 4 can also be fully recapitulated in vitro. As shown in Fig. 1B, exon 4 is included in ∼4% of products derived from in vitro splicing of the WT minigene in nuclear extract from resting cells (R-NE), while incubation of the same construct in nuclear extract from PMA-stimulated cells (S-NE) results in only ∼1% inclusion of exon 4. This difference in exon 4 inclusion between the resting and the stimulated extracts correlates to the same three- to fivefold decrease in exon inclusion that is observed in vivo for both the minigenes and the endogenous CD45 pre-mRNA. Interestingly, analysis of spliceosome assembly in resting and activated extracts reveals the same point of inhibition under both conditions (i.e., blockage from the AEC to the B complex; data not shown), suggesting that the increased repression of exon 4 upon stimulation is due to strengthening of the AEC stall, not the acquisition of a new mechanistic block.
The specificity of the differential processing of exon 4 in extracts from resting and activated cells is demonstrated by the fact that the replacement of the ESS1 element with an unrelated sequence of similar length (Fig. 1, ΔESS1) leads to increased inclusion of exon 4 in extract from resting cells and abolishes the decrease in exon inclusion between resting and stimulated extracts. These results indicate that the signal-induced repression of exon 4 in vitro is dependent on the ESS1 regulatory motif and is not simply a result of weak splice sites or inefficient exon inclusion.
Therefore, both the specificity of regulation and the magnitude of the change in isoform expression of CD45, observed upon T-cell activation in vivo, can be entirely accounted for by a direct regulation of splicing. Importantly, this finding allows us to rule out potential alternative mechanisms of the CD45 regulation by coordinated events such as transcription or mRNA export. In addition, these extracts provide a valuable tool for dissecting the changes in the ESS1-bound splicing regulatory complex that are induced upon activation to cause the signal-dependent silencing of exon 4.
HnRNP L shows differential mobility and an increase in activity following stimulation.
HnRNP L is the main functional component of the ESS1-binding complex present in resting cells, although this complex also contains PTB and hnRNP E2 (27). Given the activity of this hnRNP L-PTB-E2 complex in resting cells, we first sought to determine if the increase in exon repression in activated cells correlates with an increase in expression of some or all of these proteins upon activation. However, Western blotting analysis of the expression of hnRNP L, PTB, and hnRNP E2 as well as several other splicing regulatory proteins in serial dilutions of resting and stimulated nuclear extract showed no significant change in nuclear protein concentration following stimulation (see Fig. S1 in the supplemental material), nor is there any detectable increase in the affinity of these three proteins for the ESS1 element (see discussion below).
Since the levels of the ESS1-binding proteins do not change significantly, we next evaluated the modification state of these proteins by two-dimensional (2D) gel analysis of the nuclear extracts to determine if differential posttranslational modification might affect the silencing activity of these proteins. While hnRNP E2 and PTB show no significant changes in migration patterns in resting versus stimulated nuclear extracts (see Fig. S2B in the supplemental material and data not shown), Western blotting of 2D gels for hnRNP L reveals a differential pattern of migration (Fig. 2A). In nuclear extract prepared from resting cells, the bulk of hnRNP L migrates close to its predicted pI of 6.7, with only a minor population migrating at a more acidic pI. In contrast, in extract from stimulated cells, a larger proportion of hnRNP L is observed for the acidic population (Fig. 2A). The differences in the pI profiles of hnRNP L in resting and stimulated extracts suggest differential posttranslational modification of at least a portion of the nuclear hnRNP L pool in activated cells. While an increase in the acidic population of hnRNP L is consistent with induced phosphorylation upon T-cell stimulation, treatment of the extracts with the phosphatase CIP (see Fig. S2 in the supplemental material) or BAP (data not shown) results in only a partial loss of the acidic population of hnRNP L and a general dispersion of the protein across the pH gradient, complicating interpretation of the data. Therefore, we are unable to conclusively determine whether phosphorylation is the sole cause of the shift in the charge of hnRNP L upon activation.
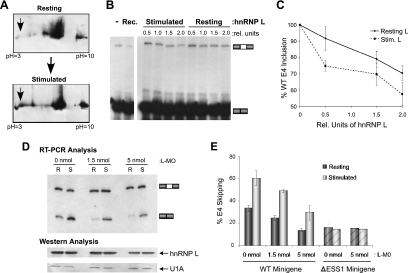
FIG. 2.
Increased silencing activity of hnRNP L following stimulation accounts for a portion of the activation-induced skipping of the CD45 exon 4. (A) Analysis of hnRNP L by 2D electrophoresis. Resting or stimulated JSL1 NE was subjected to separation by 2D electrophoresis and blotted with hnRNP L antibody (see Materials and Methods). pH labels correspond to the pI predicted for proteins migrating at that position. (B) Functional evaluation of hnRNP L purified from either resting or stimulated NE. Equal amounts of purified hnRNP L from either resting or stimulated NE (see Fig. S3A in the supplemental material) were titrated into in vitro splicing reaction mixtures containing the WT CD45 minigene and NE prepared from resting JSL1 cells (as shown in Fig. 1). Purification was done by affinity to poly(CA) RNA as described by Hui et al. (10). Addition of 100 ng of recombinant GST-hnRNP L (Rec. lane) is shown as a positive control. (C) Quantitation of in vitro splicing assays as shown in panel B. All values were normalized to exon 4 (E4) inclusion in the absence of hnRNP L addition (set to 100% inclusion). Statistical analysis reveals that the difference between the activity of resting hnRNP L and that of stimulated hnRNP L is significant (P < 0.05). (D) Antisense knockdown of hnRNP L using morpholino oligonucleotides in JSL1 cells. Shown are RT-PCR analysis of the WT minigene RNA isolated from JSL1 cells treated with the indicated amount of the hnRNP L morpholino oligonucleotide (L-MO) under either resting (R) or stimulated (S) conditions and Western blotting analysis of hnRNP L and U1A (loading control) from protein samples isolated from the cells used for RT-PCR analysis. (E) Bar graph represents the quantitation of the RNA splicing analysis for the WT minigenes as well as results from parallel experiments with a minigene lacking the ESS1 sequence (ΔESS1).
To determine if the change in the apparent pI of hnRNP L correlates with altered silencing activity of this protein, we purified endogenous hnRNP L from resting and stimulated cells and tested the relative activities of these proteins in in vitro splicing assays. As we have reported previously, the addition of baculovirus-produced recombinant hnRNP L to resting nuclear extract increases repression of the WT CD45 exon 4, resulting in a decrease in three-exon product (27) (Fig. 2B). Similar results were obtained upon addition of endogenous hnRNP L purified from JSL1 cells by the method described by Hui et al. (10) (Fig. 2B and C; and see Fig. S3A in the supplemental material). Notably, hnRNP L purified from stimulated JSL1 cells exhibits an increase in repressive activity compared to that of protein purified from resting JSL1 cells (Fig. 2B and C, resting versus stimulated cells). Quantification of the effect of hnRNP L titration over multiple independent experiments confirms that the differences in the repressive activities of hnRNP L from resting and activated cells, while relatively modest, are statistically significant (P < 0.05) (Fig. 2C). Thus, the change in migration of hnRNP L by 2D gel analysis correlates with an increase in the silencing activity of the hnRNP L protein in in vitro splicing assays, suggesting that differential modifications of hnRNP L influence its repressive activity.
Although the change in hnRNP L activity in activated cells is consistent with increased repression of the CD45 exon 4, this difference is sufficiently small that it is unlikely to account for more than a minor portion of the three- to fivefold change in exon inclusion observed upon stimulation. HnRNP L knockdown experiments with JSL1 cells further suggest that additional mechanisms contribute to the increase in exon silencing following stimulation. Previously, we have shown that a 50% reduction in hnRNP L by RNA interference (RNAi) in 293T cells results in a significant reduction in the skipping of the CD45 variable exon 4 (27). The JSL1 cells are refractory to RNAi; however, we can achieve a similar level of hnRNP L knockdown in JSL1 cells by using morpholino antisense oligonucleotides that are complementary to the mRNA sequence surrounding the translation start site. Such oligonucleotides are efficiently taken up by lymphocytes and block translation (17, 18). Upon knockdown of hnRNP L with the morpholino oligonucleotide, we observe a marked reduction in the silencing of exon 4 in both the wild-type minigene as well as the endogenous CD45 gene in resting cells (Fig. 2D and E, R lanes, and data not shown). This effect of hnRNP L depletion on splicing is ESS1 dependent, as no change is observed for the splicing of a construct lacking the ESS1 sequence (Fig. 2D, ΔESS1, graph).
In activated cells, we also observe a decrease in WT exon skipping when comparing morpholino-treated cells and mock-treated controls (Fig. 2D, WT versus the hnRNP L morpholino oligonucleotide), consistent with the hnRNP L contribution to the overall exon repression under both resting and activated conditions. In contrast, knockdown of PTB in JSL1 cells with a morpholino oligonucleotide, in which we can achieve >75% reduction in protein, has no effect on the basal or activation-induced splicing pattern (see Fig. S3B in the supplemental material), in agreement with our previous in vitro studies (27). However, despite the fact that the absolute level of exon 4 skipping is decreased upon hnRNP L knockdown in both resting and stimulated cells, the ESS1 sequence in the morpholino-treated cells retains the ability to respond to cellular activation, as evidenced by the difference in splicing in the absence and presence of PMA (Fig. 2D, compare the hnRNP L morpholino oligonucleotide and WT lanes R versus S). The ability of cells to support signal-induced changes in CD45 splicing under conditions of limited hnRNP L is consistent with the modest contribution of hnRNP L to hyper-exon repression in activated cells. Together, the results presented in Fig. 2 indicate that while hnRNP L plays a major role in basal exon skipping in resting cells and does contribute to exon 4 skipping in activated cells, additional proteins or mechanisms must exist which confer the majority of the increase in exon repression observed after cellular activation.
PSF mediates ESS1-dependent exon silencing specifically under stimulated conditions.
We have previously been unable to detect differences by silver stain between the protein composition of the ESS1-bound complex purified from resting cell extracts and that from stimulated cell extracts (27) (see Fig. S4A in the supplemental material). However, since differences in protein composition could be below the detection limits of a silver stain assay, we repeated the ESS1 affinity purification and subjected the entire sample to in-solution digestion with trypsin and analysis by mass spectrometry. The ESS1-associated proteins identified by multiple high-confidence tryptic peptides in the mass spectrometry analysis are listed in Fig. 3A [log(e) confidence scores are listed in Fig. S4B in the supplemental material]. As anticipated from previous data, peptides derived from hnRNP L, PTB, and hnRNP E2 were among those most frequently identified in both resting and stimulated purifications, and several other members of the hnRNP family (hnRNP K, hnRNP D, and hnRNP A1) were also identified in both resting and stimulated purifications (Fig. 3A). In addition to these hnRNPs, 10 to 20 other proteins were identified by a single peptide in the mass spectrometry analysis and thus deemed low-confidence peptides and/or assumed to be present in the purification at substoichiometric concentrations. Remarkably, however, PSF and the highly related protein p54nrb were both identified by multiple high-confidence peptides in the ESS1 affinity purifications from stimulated nuclear extract, without a single peptide discovered for these proteins in the purification from resting cells (Fig. 3A; also see Fig. S4B and C in the supplemental material).
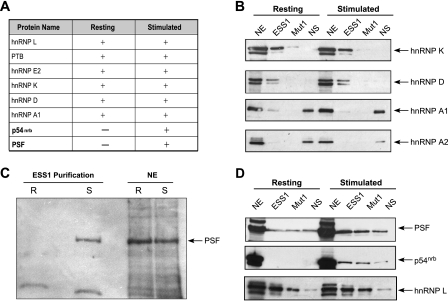
FIG. 3.
PSF and p54nrb are added to the ESS1-binding complex following stimulation. (A) The table shows the proteins identified by mass spectrometry analysis of proteins isolated from resting or stimulated NE following RNA affinity purification with ESS1. + indicates the positive identification of multiple peptides by mass spectrometry and − indicates the absence of peptides as found by mass spectrometry. (B) Western blotting analysis of total nuclear protein (NE) or direct RNA affinity purifications from either resting or stimulated NE using either wild-type ESS1 RNA (ESS1), mutant RNA (Mut1; see Fig. 5A), or nonspecific control RNA (NS), using antibodies against hnRNP K, hnRNP D, hnRNP A1, and hnRNP A2 as indicated. (C) Western blotting analysis of PSF binding to wild-type ESS1 RNA from resting (R) or stimulated (S) NE. Equal concentrations of total NE were analyzed in parallel. (D) Western blotting analysis of direct RNA affinity purifications from resting or stimulated NE, as described in panel B, using antibodies to PSF, p54nrb, and hnRNP L (loading control) as indicated.
To confirm and evaluate the specificity of the mass spectrometry results, RNA affinity purification experiments were repeated using a mutant version of ESS1 (Mut1) which abolishes both basal and activation-induced silencing activities of the sequence (9, 26, 27 [Mut1 was referred to as “mESS1” in these earlier studies; also see discussion below]) and an unrelated RNA sequence that we have previously used as a nonspecific control (NS) (26, 27). As reported previously, hnRNP L, PTB, and hnRNP E2 each bind with higher affinity to ESS1 than to Mut1 or NS RNAs (27) (see Fig. S4D in the supplemental material). hnRNPs K and D exhibit a similar specificities for ESS1, while hnRNP A1 shows a greater affinity for the control RNA sequence, as does hnRNP A2, used as a negative control (Fig. 3B), suggesting that the presence of hnRNP A1 in the ESS1 purification is due to weak background association. Notably, however, the nuclear expression level, the ESS1 association, and the ESS1 specificity for each of these seven hnRNP proteins remain unchanged between resting and stimulated conditions (Fig. 3B; also see Fig. S4D in the supplemental material).
In striking contrast to results with the hnRNP proteins, Western blotting analysis of PSF binding to the wild-type ESS1 probe confirms robust PSF association with ESS1 only in purifications from stimulated extract (Fig. 3C, ESS1 purification). We note that this change in ESS1-PSF association is independent of the level of nuclear PSF protein, since Western blots of total nuclear extract revealed no reproducible changes in the expression of PSF protein between resting and stimulated conditions (Fig. 3C, lanes NE; also see Fig. S1 in the supplemental material). Further evaluation demonstrated that a weak nonspecific interaction of PSF with ESS1 can be observed in resting nuclear extract upon overexposure; however, this level of bound PSF is similar to that associated with the NS RNA (Fig. 3D). In contrast, in activated extracts, PSF exhibits enhanced binding to the ESS1 RNA relative to that of both the Mut1 and the NS RNAs as well as relative to the binding observed for any of the RNAs in resting cells (Fig. 3D, also see below). A similar specificity of ESS1 binding was obtained for p54nrb, a protein highly homologous to PSF and often found together with PSF in cellular complexes. However, since PSF has been more clearly implicated in splicing than p54nrb and preliminary data suggest that p54nrb does not function to repress exon 4 (see below), our subsequent experiments focused solely on PSF.
To determine if the association of PSF with the ESS1 sequence contributes to the increased repression of the CD45 exon 4 in extract from activated cells, we evaluated the effect of inhibiting PSF in in vitro splicing assays. Complete depletion of PSF from extracts results in a general block in splicing (21); however, we can titrate limiting concentrations of anti-PSF antibody into extracts, which should specifically block those processes that are most sensitive to levels of PSF activity. Strikingly, titration of a monoclonal antibody directed against PSF into stimulated extract results in a dose-dependent increase in exon 4 inclusion in in vitro splicing assays, corresponding to a loss of exon 4 silencing (Fig. 4A, stimulated). In contrast, no change in the level of exon 4 inclusion is observed when the PSF antibody is titrated into resting extract (Fig. 4A, resting). This effect is consistent with the fact that PSF is bound to ESS1 only under stimulated conditions and, by extension, can only affect ESS1-dependent splicing under these conditions. Quantitation of the PSF antibody effect on splicing is represented in Fig. 4B, demonstrating a dose-dependent and statistically significant (P < 0.05) increase in exon inclusion under stimulated conditions (Fig. 4B). It should be noted, however, that the maximal effect of PSF antibody addition does not return the level of variable exon inclusion to the levels of inclusion seen under resting conditions (Fig. 4A, compare first and last lanes).
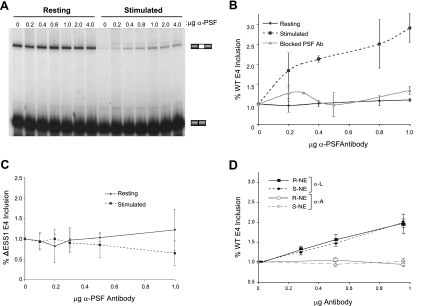
FIG. 4.
PSF increases ESS1-dependent silencing under stimulated conditions. (A) In vitro splicing of the wild-type ESS1 minigene in either resting or stimulated NE in the presence of the indicated amounts of monoclonal PSF antibody. (B) Quantitation of the CD45 exon 4 (WT E4) inclusion in in vitro splicing assays following the addition of PSF antibody. Levels of WT E4 inclusion were normalized to the level of E4 inclusion in the absence of antibody (set at 1.0) in the corresponding resting (solid line) or stimulated (dashed line) NE. Data are also shown for preincubation of PSF antibody with bacterially purified, inactive His-tagged PSF prior to addition into splicing reaction mixtures with stimulated (S) NE (gray line). (C) Quantitation of the effect of PSF antibody on the splicing of a minigene lacking the ESS1 sequence (ΔESS1) in resting or stimulated NE. Analysis was done as described in the legend to panel B. (D) Quantitation of the effect of hnRNP L and hnRNP A2/B1 antibody on the splicing of the WT minigene in resting or stimulated NE. Analysis was done as described in the legend to panel B.
The specificity of the PSF antibody effect was confirmed by preblocking the antibody with inactive bacterium-produced recombinant PSF protein, which abrogates any effect of the antibody on splicing in stimulated extract (Fig. 4B; also see Fig. S5A in the supplemental material). Furthermore, the PSF antibody has no effect on the splicing of the ΔESS1 construct in either the resting or the stimulated extracts, demonstrating that the antibody only relieves ESS1-dependent exon silencing in stimulated extracts (Fig. 4C; also see Fig. S5B in the supplemental material). In contrast to the addition of PSF antibody, the addition of hnRNP L antibody into in vitro splicing assays causes a decrease in the level of exon 4 skipping under both resting and stimulated conditions, whereas the addition of hnRNP A2 antibody causes no change in the level of variable exon inclusion in either extract (Fig. 4D).
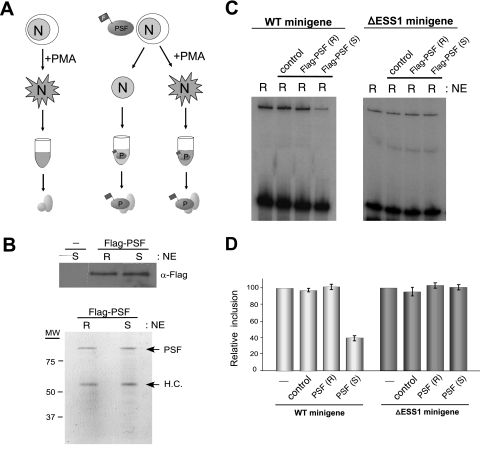
As mentioned above, recombinant PSF purified from bacterial cells has no exon 4 silencing activity (see Fig. S5A in the supplemental material). We reasoned that this lack of activity, together with the absence of PSF function in exon 4 splicing in resting extracts, may reflect the absence of some modification or interaction of PSF unique to stimulated T cells. To investigate this possibility we stably expressed a Flag-tagged recombinant version of PSF at nearly endogenous levels in JSL1 cells and then grew these cells under resting and activated conditions and generated nuclear extract (Fig. 5A). The nuclear extracts containing the Flag-PSF have the same differential splicing activity as those shown in Fig. 1, indicating that the expression of the recombinant protein does not disrupt splicing regulation (data not shown). To specifically characterize the influence of cell stimulation on PSF activity, equal amounts of Flag-PSF protein were isolated from the resting or stimulated extracts by the use of anti-Flag-coated beads (Fig. 5A and B; also see Materials and Methods). As a control, these same beads were used in a mock purification from extracts of stimulated cells not expressing any Flag protein (Fig. 5A and B).
FIG. 5.
PSF from resting and activated JSL1 cells exhibits differential silencing activity. (A) Schematic of the experimental outline for testing the effect of cellular stimulation on PSF activity (see Materials and Methods for details). Unlabeled ovals in the final step indicate the potential presence of coassociated proteins in our purification scheme that are below the level of detection as shown in panel B. Mock purification from stimulated extracts lacking Flag-PSF is used as a control for nonspecific proteins that could associate with the anti-Flag beads. (B) Anti-Flag Western blotting and the Coomassie-stained gel of protein isolated in the purifications are shown in panel A. The far left lane in the anti-Flag blot is the mock purification. PSF is not visible by silver stain despite its clear abundance as shown by Western blotting and Coomassie stain. In the Coomassie-stained gel, the second prominent band corresponds to anti-Flag antibody heavy chain (H.C.), which routinely gets stripped off the beads to some extent during the purification procedure. (C) In vitro splicing, with WT and ΔESS1 minigenes, as shown in Fig. 1. All reaction mixtures were done with extracts from resting cells not expressing Flag-PSF, either alone or supplemented with purification reaction mixtures shown in panel B. (D) Quantification of three independent experiments, as described in the legend to panel C. Exon inclusion in the extract-only lane is set to 100% in each case.
Strikingly, while the absolute concentrations of Flag-PSF isolated from resting or stimulated extracts were equivalent, only the protein isolated from stimulated cells was able to induce skipping of WT exon 4 when added to extracts from resting cells [Fig. 5C and D, Flag-PSF (S)]. Flag-PSF purified from resting cells had no effect on exon 4 inclusion, nor did the mock purification sample from stimulated cells (Fig. 5C and D). In addition, the silencing activity of the Flag-PSF isolated from stimulated cells is specific for the ESS1-dependent regulation, as this protein had no effect on the ΔESS1 minigene (Fig. 5C and D). Therefore, we conclude that cellular stimulation alters the functionality of PSF, thus allowing it to bind to the ESS1 element and induce activation-specific exon silencing. Whether this altered functionality is a result of a direct change in PSF (i.e., modification) or a change conferred by a variation in PSF-associated proteins is a question open for further investigation (see Discussion).
In vivo studies of PSF have been complicated by the essential role of PSF in numerous nuclear processes. A morpholino that causes partial reduction of PSF protein does reduce the change in CD45 splicing observed upon activation, but interpretation of these results is complicated by a number of pleiotropic effects including marked decrease in cell viability and a general alteration in splicing (data not shown). A requirement for normal expression of PSF to maintain viability is consistent with studies implicating PSF as an essential factor in numerous nuclear processes, including splicing, transcription, DNA unwinding, and nuclear retention of mRNA (29), and emphasizes the importance of our in vitro system which uncouples PSF-dependent regulation of splicing from other functional roles of this protein. We have also utilized a second morpholino that does not reduce PSF protein significantly and has no global effects on cellular splicing or viability. Not surprisingly, this second morpholino does not influence activation-induced splicing of CD45 on its own; however, when CD45 splicing is sensitized by the reduction of hnRNP L, treatment of cells with the second morpholino does specifically reduce activation-induced silencing of the CD45 exon 4 (see below). Together, the studies shown in Fig. 3 to 5 confirm a significant role for PSF in associating with ESS1 under activated conditions and regulating the signal-induced increase in ESS1-dependent silencing.
hnRNP L and PSF demonstrate combinatorial regulation of stimulated exon repression.
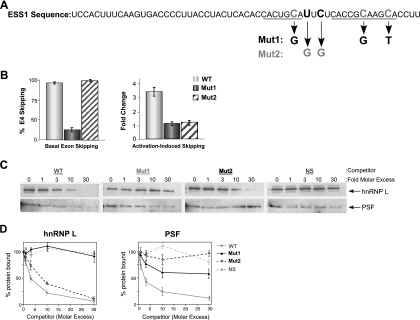
Given that we observe signal-induced exon 4 repression mediated by both an increase in hnRNP L activity and the addition of PSF to the ESS1-binding complex, we wanted to evaluate the interplay between these two factors by using various ESS1 mutants. The full 60-nucleotide ESS1 sequence contains short and long copies of the activation-responsive sequence (ARS) motif (Fig. 6A, the ARS motif is underlined), so defined because mutation of the conserved Cs in each ARS repeat abolishes both basal and activation-induced skipping of exon 4 in vivo and in vitro (9, 26) (Fig. 6B, Mut1). However, the ARS motif alone is not sufficient to confer the complete activity of the ESS1 element, even when present in multiple copies (A. Tong and K. W. Lynch, unpublished). Further mutational analysis of the ESS1 element reveals an important role for the sequence that acts as a spacer between the two copies of the ARS motif. Remarkably, substitution of two nucleotides between the short and the long copies of the ARS motif has little effect on basal repression but significantly reduces the change in exon repression between resting and stimulated extract (Fig. 6A and B, Mut2, change in repression, 1.6-fold versus 3.5-fold) as well as in activated cells (data not shown). Thus, basal silencing activity of the ESS1 sequence can be at least largely separated from the signal-induced silencing activity of this regulatory element both in vivo and in vitro.
FIG. 6.
hnRNP L and PSF contribute independently to the signal-induced increase in ESS1 silencing. (A) The complete sequence of the 60-nucleotide ESS1 element is shown with the short and long copies of the ARS motif underlined. The base changes within the two mutants of ESS1 (Mut1 and Mut2) are shown. (B) Quantitation of in vitro splicing of WT (white), Mut1 (gray), and Mut2 (crosshatched) minigene RNA in resting extract (basal skipping) or a comparison of resting versus stimulated extracts (activation-induced skipping) is shown. Change (n-fold) is a measure of the difference in splicing between resting and activated conditions and is calculated as the (percentage of exon 4 [E4] inclusion/percentage of E4 exclusion)resting/(percentage of E4 inclusion/percentage of E4 exclusion)stimulated. (C) Western blotting analysis of hnRNP L and PSF in ESS1 affinity purifications from stimulated NE done in the absence or presence of a nonbiotinylated RNA competitor as indicated (ESS1, Mut1, Mut2, and NS). (D) Quantitation of the Western blots in panel C. Values were normalized to the amount of protein bound in the absence of competitor RNA (set to 100%).
We have previously shown that the Mut1 mutation of the ESS1 element reduces the affinity of hnRNP L for this sequence by ∼10-fold in resting cells, consistent with the loss of basal silencing of this mutation (27). The affinity purifications shown in Fig. 3D and Fig. S4D in the supplemental material strongly suggest that hnRNP L as well as PSF also has reduced binding to the Mut1 RNA under stimulated conditions. However, competition is a more reliable and quantitative measure of relative affinity. Therefore, to assess the affinity of hnRNP L and PSF for the mutant versus the wild-type ESS1 RNAs in stimulated extracts, we tested the ability of various RNAs to compete for binding to a wild-type biotinylated ESS1 probe. As anticipated, the addition of a wild-type competitor RNA to a binding reaction mixture in which stimulated nuclear extract is incubated with the ESS1 probe results in a dose-dependent decrease in the levels of hnRNP L and PSF associated with the biotinylated ESS1 RNA (Fig. 6C and D, WT). By contrast, titration of the nonspecific control RNA into parallel reaction mixtures has little or no effect on the binding of these proteins, even at a 30-fold molar excess of competitor (Fig. 6C and D, NS). In agreement with the direct affinity experiments in Fig. 3D, the Mut1 RNA also has little ability to compete for the binding of hnRNP L to the biotinylated ESS1 and has reduced affinity for PSF (Fig. 6C and D, Mut1), consistent with the inability of this mutant sequence to support either basal or activation-induced exon repression. In striking contrast to Mut1, Mut2 efficiently recruits hnRNP L, but not PSF, away from the biotinylated ESS1 RNA (Fig. 6C and D). Therefore, we conclude that Mut2 maintains the ability to bind to hnRNP L, consistent with the basal activity of this sequence; however, PSF is unable to associate with the Mut2 sequence and confer ESS1-mediated activation-induced exon skipping.
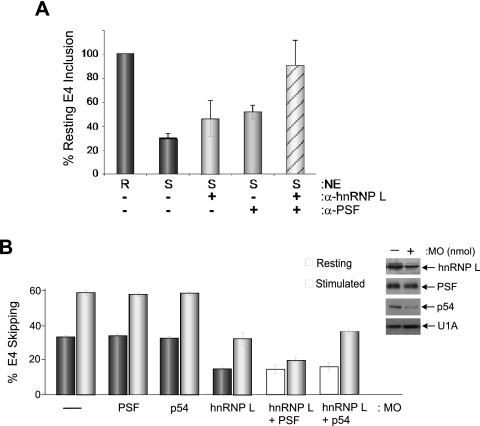
The experiments with Mut1 and Mut2 suggest that the binding of hnRNP L and binding of PSF to ESS1 are largely independent of each other. The autonomy of PSF and hnRNP L functions is further implied by the residual activation-induced repression observed under conditions in which PSF activity is reduced by antibody or the Mut2 mutation (Fig. 4A and B and 6B), consistent with the remaining activation-induced increase mediated by an increase in hnRNP L function (Fig. 2B and C). Moreover, knockdown of hnRNP L does not significantly weaken the activation-induced repression of ESS1-dependent splicing (Fig. 2D and E), presumably due to the continued activity of PSF. If hnRNP L and PSF do indeed function separately to repress the ESS1-containing exon 4, then we reasoned that blocking both hnRNP L and PSF in extract from activated cells should function in a combinatorial manner to return the level of exon inclusion to that of the resting extract. As predicted, while the addition of either hnRNP L or PSF antibody alone to stimulated extract causes a partial increase in the level of variable exon 4 inclusion, the addition of both antibodies has a markedly greater effect on exon inclusion, returning the level of exon inclusion in stimulated extract to nearly that observed under resting conditions (Fig. 7A). In contrast, the addition of an equal mass of hnRNP A2 antibody to the reaction mixture does not alter splicing, nor is there a similar combinatorial increase in exon inclusion upon addition of hnRNP L and PSF antibodies to resting extract (data not shown). Further evidence for the combinatorial regulation of the CD45 exon 4 by hnRNP L and PSF comes from the treatment of JSL1 cells with morpholino oligonucleotides targeting both proteins. As discussed above, a morpholino targeting PSF that does not cause global changes in splicing and cell viability also has no effect on CD45 splicing on its own (Fig. 7B, PSF). However, the treatment of cells with this PSF morpholino, in combination with the morpholino targeting hnRNP L, results in a specific decrease in the silencing of exon 4 in activated cells, with little alteration of splicing in resting cells (Fig. 7B, L versus PSF + L). This combinatorial effect is not observed for the closely related protein p54nrb (Fig. 7B) or for PTB (data not shown), despite the fact that we can deplete both of these proteins considerably more efficiently than PSF (Fig. 7B; see Fig. S3B in the supplemental material). The specific effect of the combined inhibition of PSF and hnRNP L under activated conditions both in vivo and in vitro strongly argues that the changes we have identified in the activity of hnRNP L and PSF upon cellular stimulation can fully account for the ESS1-dependent alternative splicing of the CD45 exon 4 that is induced upon T-cell activation.
FIG. 7.
Combined inhibition of hnRNP L and PSF abolishes activation-induced exon repression. (A) Quantitation of in vitro splicing of WT exon 4 minigene RNA in either resting (R) or stimulated (S) NE with antibodies to hnRNP L and PSF added alone or in combination. The inclusion of exon 4 (E4) was normalized relative to that observed in resting extracts in the absence of antibody (set at 100%). + represents the addition of 2 μg of hnRNP L antibody or 2 μg of PSF antibody. (B) Antisense knockdown of hnRNP L, PSF, or p54nrb, alone or in combination in JSL1 cells. Bar graph represents the level of exon skipping of the WT minigene in either resting or stimulated cells after treatment with the indicated morpholino oligonucleotide. Western blots of representative samples are shown with U1A as a loading control.
DISCUSSION
While signal-induced alternative splicing of the CD45 exon 4 via the ESS1 sequence is well documented, there has been no report of the mechanisms by which the activity of this silencer is induced upon cellular activation. In this report we demonstrate that the signal-induced change in isoform expression observed in vivo is due to a direct effect on splicing and can be recapitulated with extracts from resting and activated cells. We have further identified the two major contributors to the activation-induced increase in exon repression mediated by the ESS1 element (Fig. 8). A change in the modification state of hnRNP L upon stimulation correlates with an increase in the repressive activity of this protein, producing a modest increase in the skipping of the CD45 exon 4. Thus, hnRNP L plays a major role in the basal level of exon repression under resting conditions and contributes to the signal-induced exon skipping following stimulation. A second, and more sizable, effect on activation-induced repression is produced by the addition of PSF to the ESS1-binding complex following activation. Importantly, the combined contributions of hnRNP L and PSF to ESS1-dependent silencing fully account for the total activation-induced splicing change observed both in vitro and in vivo, suggesting that they confer most, if not all, of the mechanisms of signal-induced regulation in this system. While we do identify four other proteins that bind with some specificity to the ESS1 element (Fig. 8), at least two of these proteins (PTB and hnRNP E2) contribute little to the activity of ESS1 (27). Moreover, we find no evidence for altered modification or ESS1 association of any of these four hnRNP proteins between resting and activated conditions (Fig. 3B; see also Fig. S4D in the supplemental material; also data not shown).
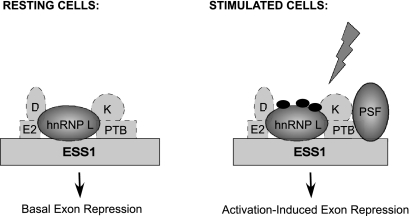
FIG. 8.
Model of the ESS1-binding complex under resting and stimulated conditions. In resting cells, the ESS1 complex is bound by at least five hnRNPs, with hnRNP L acting as the main functional regulator of basal exon silencing. Following cellular activation, the modification state of hnRNP L is altered, correlating to an increase in the silencing activity of the protein. Additionally, PSF is added to the complex and mediates a further increase in the repressive activity of the ESS1 element.
Mechanisms of signal-regulated alternative splicing via posttranslational modifications.
Signal-induced changes in splicing patterns have been described as altering the expression of numerous genes and proteins (30, 31). However, the mechanisms by which specific cellular signaling events influence splicing regulatory proteins and the subsequent effects of these changes on alternative splicing are only beginning to be addressed. Several recent reports have characterized the nucleocytoplasmic regulation of hnRNP A1 in response to osmotic shock, in which the induced phosphorylation of hnRNP A1 leads to its cytoplasmic retention (1, 7, 34). The corresponding decrease in nuclear hnRNP A1 results in alternative splicing of an E1A minigene reporter upon osmotic shock (34). Similar to that of hnRNP A1, the phosphorylation states of the SR proteins SF2/ASF, 9G8, and SRp40 have been shown to be regulated by the kinase Akt in response to stimulation with growth factors or insulin (4, 20). However, in contrast to hnRNP A1, the phosphorylation of SF2/ASF by Akt does not change the localization pattern of SF2/ASF (4), suggesting that the phosphorylation state of this SR protein presumably results in direct changes in its activity.
Signal-induced repression of the CD45 exon 4 is due in part to the regulation of hnRNP L by posttranslational modification, as described for hnRNP A1. However, in this instance, the modification of hnRNP L appears to function more similarly to Akt modification of SF2/ASF, in that we observe no change in the nuclear concentration of this protein or any change in its affinity for ESS1, despite the fact that we do observe an increase in hnRNP L activity. While an acidic shift in pI is often observed following an increase in the phosphorylation state of a protein, the treatment of hnRNP L with phosphatase does not fully shift the acidic population back to the unmodified pI. Therefore we cannot conclusively determine whether the altered migration of hnRNP L upon activation is due to phosphorylation events that are resistant to general phosphatase treatment or due to other modifications such as decreased acetylation. However, given the relatively minor role of the change in hnRNP L activity on the activation-induced alternative splicing of CD45, this differential modification of hnRNP L is unlikely to play a large role in modulating protein function.
Regulated recruitment of PSF to a pre-mRNA substrate is a unique means of controlling splicing.
In contrast to the above-described mechanisms of altered localization or activity, the activation-dependent association of PSF with the ESS1-binding complex provides a unique example of the specific recruitment of a splicing factor to a regulatory sequence in response to signaling events. The mechanisms controlling the binding of PSF to the ESS1-regulatory complex remain unknown; however, the equal levels of nuclear PSF protein in resting and stimulated extract imply that the binding of PSF is not regulated by changes in either protein expression or nuclear-cytoplasmic localization. The mutation data shown in Fig. 6 and hnRNP L knockdown experiments indicate that hnRNP L binding is not a prerequisite for PSF recruitment to the ESS1 complex following stimulation, suggesting that the change in the modification state of hnRNP L is not responsible for PSF binding. Moreover, the differential activities of the Flag-PSF purified from resting and from activated cells indicate that PSF, not other ESS1 binding proteins, is responsible for regulating the function of PSF in ESS1-dependent silencing.
Analysis of PSF by 2D gel electrophoresis shows no evidence for changes in the modification state of PSF itself (data not shown), although this does not rule out the possibility of modifications that do not alter the charge or mobility of the protein. An appealing alternative mechanism for PSF regulation involves signal-induced changes in PSF association with other nuclear proteins. While PSF was initially identified as the binding partner of PTB (21), it has been shown to interact with numerous other nuclear proteins as well as both DNA and RNA (6, 23, 25; reviewed in reference 29). Many of these interactions have been shown to be dynamic and regulated by various cellular events, including altered interaction with PKC in response to phorbol ester treatment and altered subnuclear localization and interactions with other splicing proteins upon apoptosis (24, 28). Thus, PSF may exist in a protein complex in nuclei of resting T cells, which precludes its binding to the ESS1 element. In such a scenario, cellular activation would be predicted to release PSF from sequestration, thereby allowing PSF recruitment to the ESS1-binding complex and an increase in exon skipping. Conversely, PSF may associate with some protein only under an activated condition that either serves as an adaptor to the ESS1 complex or holds PSF in a conformation that is permissive for ESS1 binding. However, further study is still needed to determine if PSF association with the CD45 exon 4, and function in activation-induced silencing, is indeed controlled by such a mechanism.
Combinatorial control of signal-regulated alternative splicing.
Of particular interest in this system is the fact that the overall signal-induced regulation of exon 4 is not controlled by a single mechanism but by the separate activities of hnRNP L and PSF, which combine to yield the total increase in exon skipping. Combinatorial control of signal-induced alternative splicing of CD45 and other genes has been previously suggested by studies demonstrating cross talk and/or redundancy in signaling pathways necessary for a particular splicing phenotype (15, 22, 35), but these studies have not investigated combinatorial changes in the RNA-bound splicing regulatory machinery. The use of multiple small effects to achieve a functionally significant change in splicing, as shown here for CD45, would be predicted to increase the specificity of signal-induced regulation, as only genes controlled by the specific combination of activated factors would be responsive to a particular stimulus. In addition, the requirement for the integration of multiple “hits” to alter splicing prevents accidental changes in splicing due to spurious activation of individual signaling cascades. Therefore, the combinatorial control of signal-induced splicing, such as that identified here for CD45, may prove to be a more global mechanism of achieving specific regulation of alternative splicing in response to precise extracellular cues.
In conclusion, we show that the signal-induced splicing regulation of the CD45 exon 4 can be recapitulated using resting and stimulated nuclear extracts derived from JSL1 cells. These extracts provided a platform for the identification of a combinatorial mechanism of activation-induced repression of exon 4 via hnRNP L and PSF. Future characterization of the changes in hnRNP L and PSF induced upon T-cell activation will provide further insight as to how integration of signaling pathways leads to the unique remodeling of a silencer complex to promote signal-induced changes in alternative splicing.
Supplementary Material
Acknowledgments
We thank the Protein Technology Core at UT Southwestern Medical Center for the mass spectrometry analysis. We thank D. Black (UCLA), R. Andino (UCSF), and I. Mattaj (EMBL, Heidelberg, Germany) for the generous gifts of antibodies.
This work was funded by NIH grant R01 GM067719 and NSF grant MCB0347104 to K.W.L and by the Welch Foundation (I-1634). Alexis Melton is funded by training grant T32 GM07062.
Footnotes
Published ahead of print on 30 July 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Allemand, E., S. Guil, M. Myers, J. Moscat, J. F. Caceres, and A. R. Krainer. 2005. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc. Natl. Acad. Sci. USA 102:3605-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, P., and P. J. Grabowski. 2007. Exon silencing by UAGG motifs in response to neuronal excitation. PLoS Biol. 5:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 4.Blaustein, M., F. Pelisch, T. Tanos, M. J. Munoz, D. Wengier, L. Quadrana, J. R. Sanford, J. P. Muschietti, A. R. Kornblihtt, J. F. Caceres, O. A. Coso, and A. Srebrow. 2005. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat. Struct. Mol. Biol. 12:1037-1044. [DOI] [PubMed] [Google Scholar]
- 5.Dawes, R., B. Hennig, W. Irving, S. Petrova, S. Boxall, V. Ward, D. Wallace, D. C. Macallan, M. Thursz, A. Hill, W. Bodmer, P. C. Beverley, and E. Z. Tchilian. 2006. Altered CD45 expression in C77G carriers influences immune function and outcome of hepatitis C infection. J. Med. Genet. 43:678-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emili, A., M. Shales, S. McCracken, W. Xie, P. W. Tucker, R. Kobayashi, B. J. Blencowe, and C. J. Ingles. 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8:1102-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guil, S., J. C. Long, and J. F. Caceres. 2006. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol. Cell. Biol. 26:5744-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hermiston, M. L., Z. Xu, R. Majeti, and A. Weiss. 2002. Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. J. Clin. Investig. 109:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.House, A. E., and K. W. Lynch. 2006. An exonic splicing silencer represses spliceosome assembly after ATP-dependent exon recognition. Nat. Struct. Mol. Biol. 13:937-944. [DOI] [PubMed] [Google Scholar]
- 10.Hui, J., K. Stangl, W. S. Lane, and A. Bindereif. 2003. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat. Struct. Biol. 10:33-37. [DOI] [PubMed] [Google Scholar]
- 11.Ip, J. Y., A. Tong, Q. Pan, J. D. Topp, B. J. Blencowe, and K. W. Lynch. 2007. Global analysis of alternative splicing during T-cell activation. RNA 13:563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobsen, M., D. Schweer, A. Ziegler, R. Gaber, S. Schock, R. Schwinzer, K. Wonigeit, R. B. Lindert, O. Kantarci, B. Hemmer, et al. 2000. A point mutation in PTPRC is associated with the development of multiple sclerosis. Nat. Genet. 26:495-499. [DOI] [PubMed] [Google Scholar]
- 13.Lee, J. A., Y. Xing, D. Nguyen, J. Xie, C. J. Lee, and D. L. Black. 2007. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 5:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch, K. W. 2004. Consequences of regulated pre-mRNA splicing in the immune system. Nat. Rev. Immunol. 4:931-940. [DOI] [PubMed] [Google Scholar]
- 15.Lynch, K. W., and A. Weiss. 2000. A model system for the activation-induced alternative-splicing of CD45 implicates protein kinase C and Ras. Mol. Cell. Biol. 20:70-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matlin, A. J., F. Clark, and C. W. Smith. 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6:386-398. [DOI] [PubMed] [Google Scholar]
- 17.Matter, N., P. Herrlich, and H. Konig. 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420:691-695. [DOI] [PubMed] [Google Scholar]
- 18.Matter, N., and H. Konig. 2005. Targeted “knockdown” of spliceosome function in mammalian cells. Nucleic Acids Res. 33:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel, N. A., C. E. Chalfant, J. E. Watson, J. R. Wyatt, N. M. Dean, D. C. Eichler, and D. R. Cooper. 2001. Insulin regulates alternative splicing of protein kinase C beta II through a phosphatidylinositol 3-kinase-dependent pathway involving the nuclear serine/arginine-rich splicing factor, SRp40, in skeletal muscle cells. J. Biol. Chem. 276:22648-22654. [DOI] [PubMed] [Google Scholar]
- 20.Patel, N. A., S. Kaneko, H. S. Apostolatos, S. S. Bae, J. E. Watson, K. Davidowitz, D. S. Chappell, M. J. Birnbaum, J. Q. Cheng, and D. R. Cooper. 2005. Molecular and genetic studies imply Akt-mediated signaling promotes protein kinase CbetaII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. J. Biol. Chem. 280:14302-14309. [DOI] [PubMed] [Google Scholar]
- 21.Patton, J. G., E. B. Porro, J. Galceran, P. Tempst, and B. Nadal-Ginard. 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7:393-406. [DOI] [PubMed] [Google Scholar]
- 22.Pelisch, F., M. Blaustein, A. R. Kornblihtt, and A. Srebrow. 2005. Cross-talk between signaling pathways regulates alternative splicing: a novel role for JNK. J. Biol. Chem. 280:25461-25469. [DOI] [PubMed] [Google Scholar]
- 23.Peng, R., I. Hawkins, A. J. Link, and J. G. Patton. 2006. The splicing factor PSF is part of a large complex that assembles in the absence of pre-mRNA and contains all 5 snRNPs. RNA Biol. 3:69-76. [DOI] [PubMed] [Google Scholar]
- 24.Rosenberger, U., I. Lehmann, C. Weise, P. Franke, F. Hucho, and K. Buchner. 2002. Identification of PSF as a protein kinase C alpha-binding protein in the cell nucleus. J. Cell Biochem. 86:394-402. [DOI] [PubMed] [Google Scholar]
- 25.Rosonina, E., J. Y. Ip, J. A. Calarco, M. A. Bakowski, A. Emili, S. McCracken, P. Tucker, C. J. Ingles, and B. J. Blencowe. 2005. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol. Cell. Biol. 25:6734-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothrock, C., B. Cannon, B. Hahm, and K. W. Lynch. 2003. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell 12:1317-1324. [DOI] [PubMed] [Google Scholar]
- 27.Rothrock, C. R., A. E. House, and K. W. Lynch. 2005. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 24:2792-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shav-Tal, Y., M. Cohen, S. Lapter, B. Dye, J. G. Patton, J. Vandekerckhove, and D. Zipori. 2001. Nuclear relocalization of the pre-mRNA splicing factor PSF during apoptosis involves hyperphosphorylation, masking of antigenic epitopes, and changes in protein interactions. Mol. Biol. Cell 12:2328-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shav-Tal, Y., and D. Zipori. 2002. PSF and p54(nrb)/NonO—multi-functional nuclear proteins. FEBS Lett. 531:109-114. [DOI] [PubMed] [Google Scholar]
- 30.Shin, C., and J. L. Manley. 2004. Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell Biol. 5:727-738. [DOI] [PubMed] [Google Scholar]
- 31.Stamm, S. 2002. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum. Mol. Genet. 11:2409-2416. [DOI] [PubMed] [Google Scholar]
- 32.Tackenberg, B., M. Nitschke, N. Willcox, A. Ziegler, S. Nessler, F. Schumm, W. H. Oertel, B. Hemmer, and N. Sommer. 2003. CD45 isoform expression in autoimmune myasthenia gravis. Autoimmunity 36:117-121. [DOI] [PubMed] [Google Scholar]
- 33.Tchilian, E. Z., D. L. Wallace, R. Dawes, N. Imami, C. Burton, F. Gotch, and P. C. Beverley. 2001. A point mutation in CD45 may be associated with an increased risk of HIV-1 infection. AIDS 15:1892-1894. [DOI] [PubMed] [Google Scholar]
- 34.van der Houven van Oordt, W., M. T. Diaz-Meco, J. Lozano, A. R. Krainer, J. Moscat, and J. F. Caceres. 2000. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 149:307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weg-Remers, S., H. Ponta, P. Herrlich, and H. Konig. 2001. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 20:4194-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.