Abstract
Global mRNA translation is transiently inhibited during cellular division. We demonstrate that mitotic cells contain heavy polysomes, but these are significantly less translationally active than polysomes in cycling cells. Several observations indicate that mitotic translational attenuation occurs during the elongation stage: (i) in cycling nonsynchronized cultures, only mitotic cells fail to assemble stress granules when treated with agents that inhibit translational initiation; (ii) mitotic cells contain fewer free 80S complexes, which are less sensitive to high salt disassembly; (iii) mitotic polysomes are more resistant to enforced disassembly using puromycin; and (iv) ribosome transit time increases during mitosis. Elongation slowdown guarantees that polysomes are retained even if initiation is inhibited at the same time. Stalling translating ribosomes during mitosis may protect mRNAs and allow rapid resumption of translation immediately upon entry into the G1 phase.
Global translation in mammals is subjected to transient inhibition under specific physiological conditions, in keeping with cellular requirements. Studying translational regulation during cellular division is important owing to the pivotal role mRNA translation plays in cellular transformation and tumor progression. More than 35 years ago it was noticed that protein synthesis is inhibited during mitosis (13). Previous studies indicate that translational regulation at the initiation stage is achieved by phosphorylation events that interfere with the formation of active eukaryotic translation initiation factor 4F (eIF4F; the 5′ cap-binding complex) (4, 15, 28) and active ternary complex (eIF2-GTP-Met tRNA) (11). The inhibition of 5′ cap-dependent translation during mitosis leads to global reduction in mRNA translation, while certain internal ribosomal entry site (IRES)-containing mRNAs (such as ornithine decarboxylase and p58/cyclin-dependent kinase/PITSLRE kinase) are efficiently translated via their IRES elements (9, 27). The switch from 5′ cap- to IRES-dependent translation initiation is necessary for the translation of mRNAs whose protein products are required at mitosis (9, 27).
Although a block at the initiation stage provides a mechanistic explanation for the temporal global inhibition of translation observed during mitosis, postinitiation regulation has not been ruled out. Recently, temporary translational arrest at both the stages of initiation and elongation has been noted in yeast under hypoxic conditions (33). Accordingly, arrest at both stages may allow separate control of different classes of mRNA transcripts, enabling those arrested during elongation to rapidly resume translation immediately upon entry into the G1 phase. The current study evaluates the state of the translational machinery during cell division. Our data clearly demonstrate that polysomes remain intact in mitotic cells, yet they become significantly less active in global protein synthesis. Stress granules (SGs), hallmarks of stalled translational initiation (6, 19), are not induced in mitotic cells using diverse stimuli that block initiation by different mechanisms, including arsenite (which increases eIF2α phosphorylation and prevents ternary complex formation), energy starvation (also presumed to impair ternary complex formation), or pateamine A treatment (which binds eIF4A and prevents 48S scanning). The current study clearly indicates that stalled elongation is a major cause of translational attenuation during mitosis.
MATERIALS AND METHODS
Cells, synchronization, and cell cycle analysis.
HeLa S3 or U2OS cells were grown in Dulbecco modified Eagle medium (Gibco), supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (Biological Industries). Cells were synchronized by incubation of freshly plated cells grown to 50% confluence in medium containing 2 mM thymidine (Sigma) for 16 to 17 h, followed by three washes with phosphate-buffered saline (PBS), incubation with regular medium lacking additional thymidine for 8 h, and then an additional incubation for 16 to 17 h with medium containing 2 mM thymidine. The cells were then released from the double thymidine block by three washes with PBS and incubation with regular medium for various time periods. For cell cycle analysis, 5 × 105 cells were analyzed using flow cytometry on a Becton Dickinson FACSort instrument with Cell Quest software, as previously described (14).
Polysomal profile analysis, RNA, and protein extraction from sucrose gradient fractions.
Polysomal profiles were performed according to Johannes and Sarnow (17) with modifications. Specifically, 10 million to 12 million cells were grown in 150-mm tissue culture dishes, incubated for 5 min with 100 μg of cycloheximide (CHX), harvested, and stored at −70°C. For puromycin sensitivity experiments, the incubation with CHX was replaced by a 3-min incubation with 100 μg/μl puromycin (Sigma). Prior to analysis, the cells were resuspended in 0.4 ml of LBA buffer (18 mM Tris, pH 7.5, 50 mM KCl, 10 mM MgCl, 10 mM NaF, 10 mM α-glycerolphosphate, 1.4 μg/ml pepstatin, 2 μg/ml leupeptin, EDTA-free protease inhibitor cocktail [Complete; Roche], 70 μg/ml CHX, 1.25 mM dithiothreitol, and 200 μg/ml heparin), and Triton X-100 and deoxycholate were added to a final concentration of 1.2% each for lysis of 5 min on ice. Twenty optical density units (260 nm) were loaded on each sucrose gradient. Following centrifugation of the sucrose gradients, 0.5-ml fractions were collected. For RNA extraction, 1 ml of cold ethanol was added to each fraction, which was then incubated for 12 h at −20°C and centrifuged for 15 min at 20,000 × g. RNA was extracted from each fraction by guanidium-chloride followed by ethanol precipitations as described previously (2). For protein extraction, each fraction was diluted 1:1 with 20 mM Tris, pH 7.5, followed by the addition of 7 μl of StrataClean resin (Stratagene). Following rotation overnight at 4°C and spinning down, the proteins were eluted from the beads by boiling in Laemmli sample buffer. ImageJ software was used for quantification.
Northern and Western blotting.
Total RNA was separated in 1.2% agarose-formaldehyde gel and blotted onto Hybond-N membrane (Amersham). Digoxigenin (DIG)-labeled β-actin cDNA probe was prepared using a PCR DIG Probe Synthesis Kit (Roche). Anti-DIG-AP and the CDP-Star substrate (Roche) were used for detection. For protein analysis, cells were lysed in lysis buffer containing 10 mM HEPES, pH 7.5, 0.5% NP-40, 100 mM NaCl, 10 mM MgCl, 1 mM sodium orthovanadate, 10 mM NaF, 20 mM β-glycerolphosphate, 1.4 μg/ml pepstatin, 2 μg/ml leupeptin, and EDTA-free protease inhibitor cocktail (Complete; Roche). To detect eukaryotic elongation factor 2 (eEF2) and eEF2 kinase (eEF2K) phosphorylation levels, 0.1 μM microcystin, 1 mM EDTA, and 0.1 mM EGTA were included in the lysis buffer. Equal amounts of protein were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and subjected to Western blot analysis according to standard procedures.
[35S]methionine-cysteine labeling and immunoprecipitation.
A total of 0.5 million to 1 million HeLa cells in a 60-mm plate were labeled for 10 min with 0.5 ml of methionine-cysteine-free Dulbecco modified Eagle medium (Sigma) supplemented with 2 mM l-glutamine, 10% dialyzed fetal calf serum (Sigma), and 100 μCi/ml of l-[35S]methionine and l-[35S]cysteine mix (Easy Tag, item NEG7720; NEN). The cells were harvested following addition of 1 ml of cold PBS containing 100 μg/ml CHX and two washes with cold PBS. The global protein synthesis rate was determined as described before (14). For immunoprecipitation of β-actin, a total of 8 million to 10 million cells in 150-mm plates were labeled for 15 min with 5 ml of labeling medium. Four milligrams of total protein extracted as described above was incubated overnight at 4°C with 5 μl of polyclonal anti-β-actin antibody (Cell Signaling), followed by immunoprecipitation using protein A-Sepharose beads (Santa Cruz). Monoclonal antibody specific for β-actin (clone C4; MP Biomedical) was used for Western blot analysis of the immunoprecipitated protein.
Antibodies.
Rabbit polyclonal antibodies against human β-actin, eEF2, eEF2K, and the phosphorylated forms of eEF2 and eEF2K (at Ser366) were obtained from Cell Signaling Technology. Monoclonal antibody specific for β-actin was from MP Biomedical (clone C4). Anti-eIF3b antibody (sc-16377) and anti-Hedls (sc-8418) used for immunofluorescence were obtained from Santa Cruz Biotechnology. Anti-DCP1a used to detect P bodies was a kind gift from Jens Lykke-Andersen. Anti-RCK to detect P bodies was obtained from Bethyl Labs (BL2142 A300-461A). Secondary antibodies for immunofluorescence were all multilabeling grade from Jackson ImmunoResearch Laboratories, Inc.
Immunofluorescence of SGs and PBs.
Immunostaining was performed as described previously. Briefly, cells were plated on coverslips and allowed to recover for 24 to 48 h. Cells were exposed to stresses including sodium arsenite (500 mM for 45 min; Sigma), DMDA-pateamine A (50 nM for 1 h) (a kind gift from Jun O. Liu, John's Hopkins University), clotrimazole (used at 20 mM in serum-free medium for 1 h; Sigma), or heat shock (45 min at 44°C). The cells were then washed in PBS, fixed for 15 min at room temperature in 4% paraformaldehyde in PBS, and then postfixed and permeabilized by a 5-min incubation in −29°C methanol. Cells were washed with PBS and blocked with 5% horse serum in PBS prior to a 1-h incubation in primary antibodies. Cells were then washed twice (for 5 min with PBS), incubated with secondary antibodies (multilabeling grade; Jackson ImmunoResearch Laboratories) in blocking buffer supplemented with 50 ng/ml Hoechst dye (33258; Sigma) for 1 h, and then washed. Mounting was performed using a home-made polyvinyl-based mounting medium. Images used in Fig. 1 were obtained using a wide-field Nikon Eclipse E800 microscope and were photographed using a charge-coupled-device SPOT RT camera. All images were compiled using Adobe Photoshop, version 7.0. Quantification of SGs and processing bodies (PBs) shown in Tables 1 and 2 was obtained using eIF3 as the SG marker, DCP1a as a PB marker, and Hoechst dye to visualize DNA and determine the mitotic state of the cells. Cells were treated with the indicated drugs for 1 h prior to staining and scoring.
FIG. 1.
Cells in mitosis do not contain SGs. U2OS cells in log-phase growth were unstressed (A and D), exposed to sodium arsenite for 1 h (B and E), or exposed to DMDA-pateamine A for 1 h (C and F) prior to fixation and staining for markers of PBs (green; DCP1a) and for SGs (red; eIF3). DNA is stained blue with Hoechst dye. Arrows indicate mitotic cells.
TABLE 1.
SG distribution in mitotic cells
| Treatment | Distribution (%) of SGs in cells at the indicated phase (n)a
|
|||
|---|---|---|---|---|
| Interphase | Prophase | Metaphase | Anaphase/telophase | |
| No drug | 0 (>200) | 0 (>200) | 0 (>200) | 0 (>200) |
| Arsenite | 100 (>200) | 73 (14) | 0 (32) | 7.6 (13) |
| Pateamine A | 100 (>200) | 46 (13) | 0 (27) | 0 (33) |
eIF3 was used as an SG marker. Hoechst dye was used to visualize DNA and determine the mitotic state of the cells. n, number of cells scored.
TABLE 2.
PB distribution in mitotic cells
| Treatment | Distribution (%) of PBs in mitotic cells at the indicated phase (n)a
|
|||
|---|---|---|---|---|
| Interphase | Prophase | Metaphase | Anaphase/telophase | |
| No drug | 49.1 (166) | 100 (10) | 4.2 (47) | 11.1 (27) |
| Arsenite | 100 (>200) | 100 (14) | 15.6 (32) | 15.4 (13) |
| Pateamine A | 41.8 (134) | 76.9 (13) | 40.7 (27) | 21.2 (33) |
DCP1a was used as a PB marker. Hoechst dye was used to visualize DNA and determine the mitotic state of the cells. n, number of cells scored.
Measurement of ribosome half-transit time.
Ribosome half-transit time measurements were performed as previously described (31) with the following modifications: 20 million mitotic or nonsynchronized cells were suspended in 5 ml of labeling medium containing 12 μCi/ml l-[35S]methionine, l-[35S]cysteine mix (Easy Tag, item NEG7720; NEN). At 2, 4, 6, 8, and 10 min after labeling, 1-ml aliquots were transferred to Eppendorf tubes containing 0.5 ml of ice-cold PBS with 100 μg/ml CHX.
RESULTS
Mitotic cells are immune to SG formation.
Drugs and environmental conditions (pateamine A, energy starvation, heat shock, or viral infection) that prevent translational initiation induce the rapid formation of cytoplasmic SGs, which contain mRNA released from disassembling polysomes. Drugs that inhibit translational elongation and stabilize polysomes (e.g., CHX or emetine) prevent the formation of SGs. Similarly, PBs are smaller mRNA-containing structures whose assembly is also dependent on mRNA release from polysomes and is blocked by CHX treatment (6, 19). If translational arrest in mitotic cells is due to blocked translational initiation that results in polysome disassembly, mitotic cells should display SGs. Alternatively, if the translational block in mitotic cells is due to stalled elongation and polysome stabilization rather than abortive initiation, mitotic cells should not be able to form SGs even when exposed to conditions that normally prevent initiation and result in SG assembly. As shown in Fig. 1, in the absence of stress (Fig. 1A and D), the smooth distribution of eIF3 (red) in actively growing U2OS cells indicates the absence of SGs in both mitotic (indicated by arrows) and nonmitotic cells. Treatment of cells with the oxidative stressor sodium arsenite (Fig. 1B and E) induces prominent SGs (red foci) in nonmitotic cells, but SGs are completely absent in mitotic cells. Similarly, arsenite treatment induces PB assembly (green foci) in 100% of interphase cells, whereas only 15.6% of mitotic cells display PBs. The data are quantified in Tables 1 and 2, which show that resistance to SG assembly begins in prophase and is complete in metaphase cells. As arsenite potently induces eIF2α phosphorylation leading to ternary complex deficiency and polysome disassembly, the failure of mitotic cells to assemble SGs or PBs suggests that they contain mRNA unavailable for SG/PB assembly, i.e., that mitotic cells contain stabilized polysomes. An alternative possibility is that phosphorylation of eIF2α is blocked in mitotic cells, preventing SG assembly. This seems less likely since eIF2α phosphorylation increases at the G2/M boundary (11), and the effects of arsenite on PB assembly are independent of eIF2α phosphorylation (19).
However, the eIF4E inhibitor pateamine A (Fig. 1C and F) promotes SG assembly by acting downstream of ternary complex formation and can induce SGs even in cells that cannot phosphorylate eIF2α, as it targets the helicase eIF4A-1 required for mRNA scanning prior to large ribosomal joining (5, 10). We therefore treated cells with a derivative of pateamine A in order to induce SGs by a mechanism distinct from that of arsenite. As shown in Fig. 1C and F, pateamine A treatment induced SG assembly in virtually 100% of interphase cells, whereas metaphase cells were completely devoid of SGs. Unlike arsenite, pateamine A does not induce PB assembly (10), yet it seemingly increased the percentage of mitotic cells with PBs relative to untreated cells (Table 2). As pateamine A also inhibits the helicase eIF4AIII (involved in nonsense-mediated decay) as well as eIF4AI involved in scanning, it is possible that the pateamine A-induced increase in PBs is due to effects on mRNA decay rather than stalled translation and that these PBs contain mRNA from nonpolysome sources (e.g., error-containing mRNA destined for nonsense-mediated decay). Future work will be required to address this point. A variety of other SG-inducing treatments all failed to induce SGs in mitotic cells, including energy starvation, heat shock, thapsigargin, and mitochondrial toxins. Similar results were also obtained using other markers of SGs, such as eIF4G, G3BP-1, FXR1, and TIA-1, and using other cell lines (HeLa, mouse embryonic fibroblasts, DU145, and COS) (data not shown). The complete lack of SGs in mitotic cells leads us to conclude that mitotic cells are generally resistant to SG assembly, consistent with the idea that polysome disassembly is prevented in mitotic cells.
Reduced translation during mitosis is not accompanied by disassembly of heavy polysomes.
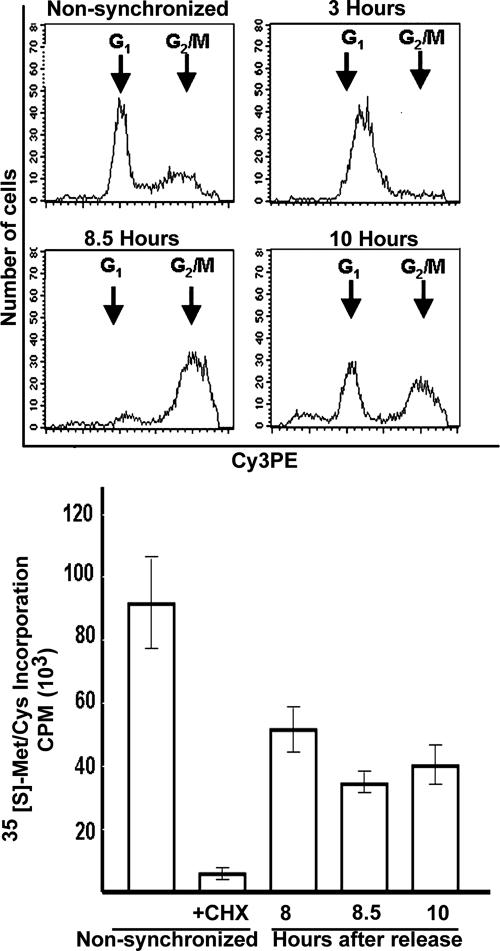
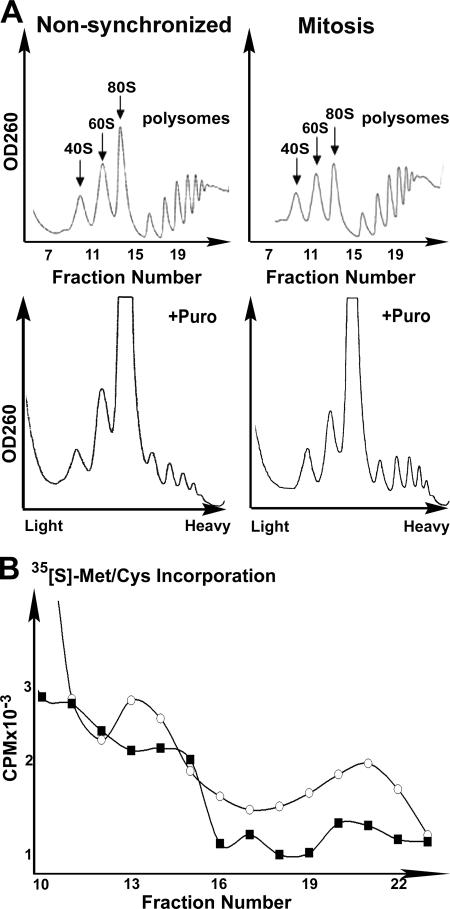
To enrich HeLa cell culture for mitotic cells, we used a double thymidine block followed by release of the arrested synchronized cells to allow them to reach mitosis. Figure 2 demonstrates that 8.5 h after the release from the block, most of the cells were at the G2/M phase of the cell cycle, as confirmed by fluorescence-activated cell sorting analysis (Fig. 2, upper panel). The appearance of mitotic condensed chromosomes detected by fluorescence microscopy confirmed that the culture is highly enriched in mitotic cells (not shown). [35S]methionine-cysteine incorporation showed that global protein synthesis was decreased by 30 to 40% during mitosis compared to translation rate in nonsynchronized cells (Fig. 2, lower panel). However, in contrast to what is expected under conditions of global protein synthesis inhibition, polysomal profile analyses clearly showed that heavy polysomes did not disassemble during mitosis (Fig. 3A). Since the polysomes remained the same size, the initiation may also have been defective, as was previously noted (4, 11, 15, 28). However, our data suggest that a major regulatory point during normal mitosis occurs downstream of initiation, possibly at the elongation and/or termination stages. To assess if this phenomenon is due to attenuation of the rate of elongation, we checked the sensitivity of the polysomes to puromycin, which leads to polysome disassembly by causing premature termination of translocating ribosomes. Due to these effects, puromycin leads the collapse of translating heavy polysomes (3). Figure 3A clearly shows that heavy polysomes of mitotic cells are more resistant to puromycin than heavy polysomes of nonsynchronized cells, suggesting that the rate of translational elongation is slowed during mitosis. To confirm that the heavy polysomes observed during mitosis are indeed less active in protein synthesis, the cells were pulse-labeled with [35S]methionine-cysteine just before their lysis and fractionation. Since free [35S]methionine and [35S]cysteine sediment at a light fraction of the gradient, the association of the [35S]Met-Cys label with the heavy fractions represents its incorporation into nascent polypeptide chains. Figure 3B demonstrates the decreased association of the [35S]Met-Cys label with the 80S and with the heavy polysome peaks of mitotic cells. This indicates decreased formation of initiation complexes as well as decreased active translation by heavy polysomes, respectively, during mitosis.
FIG. 2.
Protein synthesis is reduced during mitosis. HeLa cells were synchronized by a double thymidine block and released with fresh medium lacking thymidine for 3, 8.5, or 10 h. At the indicated time points, cell cycle analysis was performed by flow cytometry (upper panel), or the global protein synthesis rate was measured by [35S]methionine-cysteine incorporation during 10-min labeling (lower panel). For the CHX sample, the cells were treated with 100 μg/ml CHX at 30 min prior to labeling in the presence of CHX. The radioactivity at each time point is presented as the mean ± standard error of the mean of four independent measurements.
FIG. 3.
Translational arrest during mitosis is beyond the initiation stage. (A) Cycling nonsynchronized or mitotic (8.5 h after block release) HeLa cells were used for polysomal profile analysis before (upper panel) or after (lower panel) an additional 3-min incubation in the presence of 100 μg/μl of puromycin (+puro). The 40S, 60S, 80S, and polysomes peaks are indicated. (B) Cycling nonsynchronized (circles) or mitotic (squares) HeLa cells were labeled with 100 μCi/ml [35S]methionine-cysteine for the last 10 min prior to their harvest for the polysomal profile analysis presented in panel A. The counts per minute of fractions 10 to 23 as counted in a Beckman scintillation counter are shown. The data shown represent one of three independent similar experiments.
Mitotic cells exhibit increased ribosome transit time and decreased sensitivity of 80S particles to high salt concentrations.
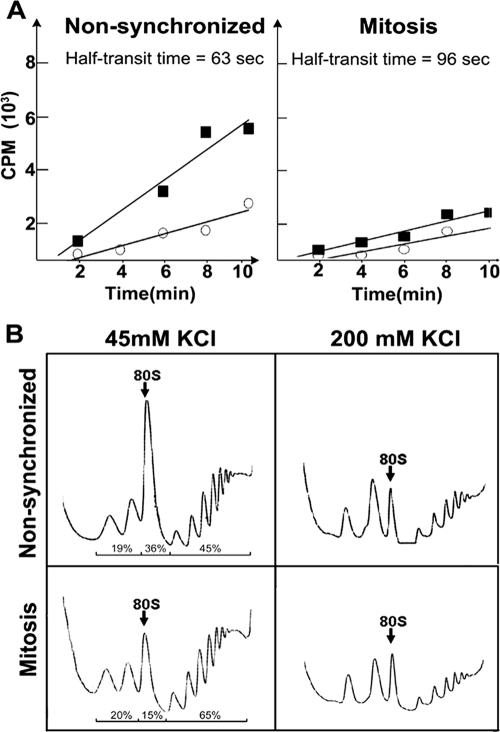
To examine whether the decreased protein synthesis in mitotic cells results from decelerated peptide chain elongation, we determined the ribosome half-transit time (13). This was performed by measuring the kinetics of [35S]Met-Cys incorporation into total protein chains and into completed polypeptides released from the ribosomes. Ribosome transit time refers to the time required for a ribosome, after it has initiated, to traverse an average-sized mRNA and release the complete polypeptide (26). The average half-transit time was determined from the displacement in time between the two lines corresponding to the total and released protein data plotted as a function of time (Fig. 4A). The ribosome transit time was calculated to be 2.1 and 3.4 min for cycling nonsynchronized and mitotic cells, respectively. The 1.62-fold increase in the transit time of the mitotic ribosomes reflects a decrease in the elongation rate during mitosis.
FIG. 4.
Increased ribosome half-transit time and increased resistance of 80S particles to 200 mM KCl. (A) The ribosome half-transit time was obtained as described in Materials and Methods. Incorporation of [35S]methionine-cysteine into total proteins (postmitochondrial supernatant; squares) or into completed peptides released from ribosomes (postribosomal supernatant; circles) is shown. The radioactivity at each time point is presented as a mean of three measurements. The transit time was determined from the displacement in time between the two lines, which were obtained by linear regression analysis. (B) Cycling nonsynchronized or mitotic (8.5 h after block release) HeLa cells were used for polysomal profile analysis in the presence of 45 mM (left) or 200 mM (right) KCl. The 80S peak is marked. The data shown represent one of three independent similar experiments.
Quantification of ribosomal profiles of nonsynchronized versus mitotic cells reveals that 19% or 20% of the total migrates as free ribosomal subunits, 36% or 15% migrates as 80S particles, and 45% or 65% migrates as heavy polysomes, respectively (Fig. 4B, left panel). The decreased proportion of 80S particles is accompanied by a reciprocal, increased proportion of heavy polysomes in mitotic cells, consistent with a decreased elongation rate. An early report showed that the 80S peak of yeast polysomal profiles represents a mixed population of empty complexes lacking mRNA and genuine translation initiation complexes. The empty complexes were distinguished by their elevated sensitivity to high salt concentrations (22). Figure 4B exhibits the significantly decreased salt sensitivity of the 80S peak of mitotic cells compared to that of nonsynchronized cells exposed to 200 mM KCl in the sucrose gradient, indicating that mitotic cells contain fewer ribosomes capable of forming empty complexes. Taken together with the increased ribosome transit time (Fig. 4A) and the observation that the poorly translating heavy polysomes in mitotic cells do not collapse and are more resistant to puromycin (Fig. 3), the data strongly suggest that translation elongation during mitosis results from ribosomal stalling on mRNA.
β-Actin mRNA translation is inhibited during mitosis, and yet it is associated with heavy polysomes.
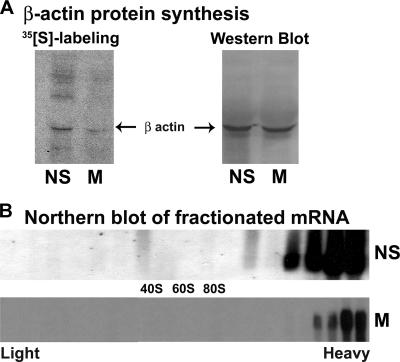
To further confirm that mitotic heavy polysomes are not engaged in efficient mRNA translation, we specifically examined the translational efficiency of β-actin mRNA as well as its association with ribosomes during interphase and mitosis. Pulse-labeling of the cells with [35S]Met-Cys followed by immunoprecipitation of the β-actin protein clearly showed that β-actin mRNA is translated much less efficiently during mitosis than during interphase (Fig. 5A). Cycling (nonsynchronized) or mitotic cells were used for polysomal profile analysis on sucrose gradients, followed by RNA extraction from each of the 23 gradient fractions for Northern blot analysis using DIG-labeled probe specific for β-actin cDNA. Figure 5B shows that despite its inefficient translation during mitosis, β-actin mRNA is associated with heavy polysomes just as it is during active cell growth. This observation is in agreement with the idea that the translation of β-actin mRNA during mitosis is attenuated at the stage of elongation.
FIG. 5.
β-Actin mRNA remains associated with heavy polysomes during mitosis despite its translation attenuation. (A) Cycling nonsynchronized (NS) or mitotic (8.5 h after block release; M) HeLa cells were labeled for 20 min with [35S]methionine-cysteine; β-actin was immunoprecipitated and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto a nitrocellulose membrane. The autoradiogram shows the labeled proteins (left). The same membrane was probed by Western blot analysis using antibody specific for β-actin (right). (B) Cycling nonsynchronized (NS) or mitotic (M) cells were used for polysomal analysis on sucrose gradients. Total RNA extracted from each of the 23 gradient fractions was subjected to Northern blot analysis using DIG-labeled probe specific for β-actin cDNA. The data shown represent one of three independent similar experiments.
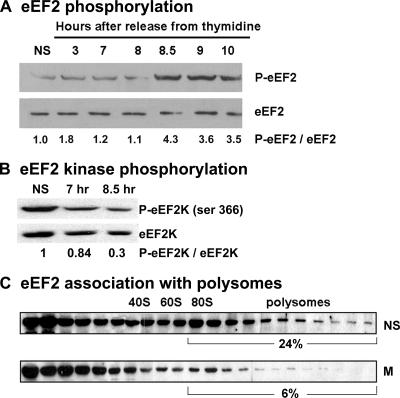
During mitosis eEF2 phosphorylation increases while its association with polysomes decreases.
eEF2 is essential for translation elongation. It has been previously demonstrated that its phosphorylation by the Ca2+/calmodulin-dependent kinase III, known as eEF2K, down-regulates eEF2 activity (32). To further establish that translational elongation is attenuated during mitosis, we assessed eEF2 phosphorylation levels throughout the cell cycle. Figure 6A shows that the eEF2 phosphorylation level is 4.3-fold higher in mitotic cells than in cycling cells. To further support this observation, we assessed the phosphorylation level of eEF2K, whose activity is regulated by multiple phosphorylation sites. Phosphorylation at Ser366 is known to inactivate the kinase, contributing to the dephosphorylation of eEF2 (35). As expected, we found that only 30% of eEF2K was phosphorylated at Ser366 in mitotic cells relative to cycling cells (Fig. 6B), consistent with its increased activity and elevated levels of phosphorylated eEF2 (Fig. 6A). Previous kinetic analysis using purified eEF2 in an eEF2-dependent in vitro translation system showed that eEF2 phosphorylation reduces its affinity for ribosomes (7) and, consequently, its activity in translation (30). To further elucidate the role of eEF2 phosphorylation during mitosis, cells harvested either at log phase or during mitosis were used for polysomal profile analysis on sucrose gradients, followed by protein extraction from each of the 23 gradient fractions for Western blot analysis using antibodies specific for eEF2. This experiment shows that eEF2 association with heavy polysomes decreases from 24% in cycling nonsynchronized cells to 6% in mitotic cells (Fig. 6C), concurrent with its increased phosphorylation. These data indicate a decreased translation elongation rate during mitosis.
FIG. 6.
eEF2 is phosphorylated during mitosis and dissociates from heavy polysomes. Equal amounts of total protein extracted from cycling nonsynchronized (NS) HeLa cells or cells that were released from a double thymidine block at the indicated times were subjected to immunoblot analysis using antibodies specific for phosphorylated eEF2 (p-eEF2) and for total eEF2 (A) or specific for eEF2K phosphorylated at Ser 366 [p-eEF2K(Ser366)] and for total eEF2K (B). Cycling nonsynchronized (NS) or mitotic (8.5 h after block release; M) cells were used for polysomal analysis. Total protein extracted from each of the 23 sucrose gradient fractions was subjected to immunoblot analysis using antibodies specific for eEF2 (C). The data shown represent one of two independent similar experiments. Band intensities were quantified by densitometry using ImageJ software.
DISCUSSION
The inhibition of the 5′ cap-dependent translational initiation during mitosis has been well documented (4, 11, 15, 28). However, the current study provides evidence that translational regulation at the initiation stage is not the exclusive mechanism by which global translation is attenuated in mitotic mammalian cells. We used the double thymidine block protocol to arrest HeLa cells at S phase, followed by release from the block to permit the synchronized cells to enter mitosis 8 to 9 h later under normal conditions, thus avoiding possible complications inherent in using drugs that affect cytoskeleton integrity, as might have occurred in previous studies (29). Under our milder conditions, we observed that polysomes are stabilized in mitotic cells (Fig. 3 to 6). Moreover, their stability reflects their reduced translational activity during mitosis, as indicated by measuring the global translation rate and by [35S]Met-Cys incorporation into each polysomal fraction (Fig. 2B and 3B). Exclusive inhibition at the initiation stage should result in ribosome “runoff” and eventually in the breakdown of polysomes, leading to increased levels of free ribosomal subunits. On the other hand, exclusive inhibition at the elongation stage should result in increased polysomal size. The similar size of polysomes detected in nonsynchronized and mitotic cells indicates that both initiation and postinitiation stages must have been affected during cellular division. Several observations in this study indicate that translational attenuation occurs after initiation in mitotic cells: (i) polysomes remain intact during mitosis (Fig. 3A, upper panel); (ii) there are fewer free 80S complexes in mitotic cells, implying that ribosomes remain associated with the polysomes (Fig. 4B, left panel); (iii) ribosome transit in mitotic cells is slower than in interphase cells (Fig. 4A); (iv) interphase 80S complexes are more sensitive to high salt concentrations, indicating that they lack mRNA (22), relative to the more abundant empty complexes in interphase cells (Fig. 4B, right panel); (v) mitotic polysomes are more resistant to enforced disassembly using puromycin (3), indicating that they are less active in elongation (Fig. 3A, lower panel); and (vi) mitotic cells are immune to SG assembly induced by agents which act at different stages of translational initiation (Fig. 1), as are cells treated with pharmacological agents that stabilize polysomes such as emetine and CHX (1, 18, 19, 10). All of the above observations demonstrate that translation is blocked at the stage of elongation, resulting from ribosomal stalling on the mRNAs. To verify that the heavy polysomes are not actively translating during mitosis, we examined the translational status of β-actin mRNA. Indeed, even though β-actin mRNA was preferably associated with heavy polysomes during mitosis, it was poorly translated (Fig. 5). This phenomenon contradicts the widely accepted perception that heavy polysomes are equivalent to actively translating ribosomes. This insight must be considered when interpreting microarray analyses based on changes in polysomal association of mRNAs as criteria for its translational status under various conditions.
Since protein synthesis is expensive in terms of metabolic energy, it is temporarily inhibited when increased demands for cellular energy are required for completion of specific cellular tasks. Such temporary translational inhibition allows energy to be diverted to the cellular process with higher priority. When translation rates are decreased, elongation arrest guarantees that polysomes are retained and mRNAs are protected, even if initiation is inhibited at the same time. Elongation arrest also allows translation to be more rapidly resumed upon its release. Cellular division into two daughter cells may be one such case requiring temporary inhibition of translation at both the initiation and elongation stages. Stalling of translating ribosomes should protect mRNAs during mitosis and allow rapid resumption of translation immediately upon entry into the G1 phase of the cell cycle.
The process of peptide chain elongation requires two factors: eEF1 to recruit the amino acyl-tRNAs to the A site of the ribosome and eEF2 to mediate the translocation of the ribosome to the next codon. eEF1 is composed of eEF1A and its guanine nucleotide exchange factor eEF1B (34). The first evidence implicating elongation control during the cell cycle was the discovery that eEF1B is a physiological target of maturation-promoting factor (cyclin-dependent kinase 1/cyclin B, the universal regulator of M phase) during amphibian oocyte maturation (16, 25); this was subsequently observed during early development of sea urchin (23, 24). Studies in sea urchin suggest an essential role eEF1B in the control of gene expression, particularly during the cell cycle (reviewed in reference 21). eEF1A is also known to be involved in several cellular processes including embryogenesis, senescence, oncogenic transformation, cell proliferation, and organization of cytoskeleton (reviewed in reference 20). Characterization of eEF1 posttranslational modifications and their role in controlling translation elongation during mitosis in mammalian systems remains to be determined.
Phosphorylation of eEF2 prevents its binding to ribosomes and thus inhibits its translocational activity, as shown by direct measurements of dissociation constants using purified ribosomal complexes (7). In the present study, we show that eEF2 is phosphorylated during mitosis (Fig. 6A), in agreement with a previous report (8), and in correlation with decreased phosphorylation of eEF2K at Ser366 (Fig. 6B). The kinase p90RSK1, which lies directly downstream of extracellular signal-regulated kinase in the classical mitogen-activated protein kinase pathway, phosphorylates eEF2K at Ser366 and down-regulates eEF2K activity (35). The current study implies that eEF2K activity is up-regulated during mitosis and further demonstrates that phosphorylated eEF2 exhibits decreased association with heavy polysomes (Fig. 6C). Further work will be required to identify the mechanism(s) used during mitosis to arrest translation elongation. At this point, attenuation of translational termination cannot be excluded and should be considered as well.
Differential translational regulation widely attributed to occur at the initiation stage is mediated by specific cis-regulatory elements, which allow initiation when global initiation is repressed. A classical example is the switch from 5′ cap-dependent to IRES-mediated initiation under certain stress conditions, apoptosis, and differentiation, as well as during cellular division (12). Global inhibition at the elongation stage poses new questions as to how such specific mRNAs remain efficiently translated during mitosis. Their as yet unknown cis-regulatory elements remain to be discovered.
Acknowledgments
We thank Dalia Pinchasi for technical assistance and Tyler Hickman for help with the confocal microscopy.
The study was supported by Charles H. Revson grant from the Israel Science Foundation to O.E.-S. and NIH grant AI33600 (N.L.K.).
Footnotes
Published ahead of print on 30 July 2007.
REFERENCES
- 1.Anderson, P., and N. Kedersha. 2002. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arava, Y., F. E. Boas, P. O. Brown, and D. Herschlag. 2005. Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic Acids Res. 33:2421-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blobel, G., and D. Sabatini. 1971. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc. Natl. Acad. Sci. USA 68:390-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonneau, A. M., and N. Sonenberg. 1987. Involvement of the 24-kDa cap-binding protein in regulation of protein synthesis in mitosis. J. Biol. Chem. 262:11134-11139. [PubMed] [Google Scholar]
- 5.Bordeleau, M. E., R. Cencic, L. Lindqvist, M. Oberer, P. Northcote, G. Wagner, and J. Pelletier. 2006. RNA-mediated sequestration of the RNA helicase eIF4A by pateamine A inhibits translation initiation. Chem. Biol. 13:1287-1295. [DOI] [PubMed] [Google Scholar]
- 6.Brengues, M., D. Teixeira, and R. Parker. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlberg, U., A. Nilsson, and O. Nygard. 1990. Functional properties of phosphorylated elongation factor 2. Eur. J. Biochem. 191:639-645. [DOI] [PubMed] [Google Scholar]
- 8.Celis, J. E., P. Madsen, and A. G. Ryazanov. 1990. Increased phosphorylation of elongation factor 2 during mitosis in transformed human amnion cells correlates with a decreased rate of protein synthesis. Proc. Natl. Acad. Sci. USA 87:4231-4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, S., Y. Bruynooghe, G. Denecker, S. Van Huffel, S. Tinton, and R. Beyaert. 2000. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell 5:597-605. [DOI] [PubMed] [Google Scholar]
- 10.Dang, Y., N. Kedersha, W. K. Low, D. Romo, M. Gorospe, R. Kaufman, P. Anderson, and J. O. Liu. 2006. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281:32870-32878. [DOI] [PubMed] [Google Scholar]
- 11.Datta, B., R. Datta, S. Mukherjee, and Z. Zhang. 1999. Increased phosphorylation of eukaryotic initiation factor 2α at the G2/M boundary in human osteosarcoma cells correlates with deglycosylation of p67 and a decreased rate of protein synthesis. Exp. Cell Res. 250:223-230. [DOI] [PubMed] [Google Scholar]
- 12.Elroy-Stein, O., and W. C. Merrick. 2007. Translation initiation via cellular internal ribosome entry sites, p. 155-172. In M. B. Mathews, N. Sonenberg, J. W. B Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Fan, H., and S. Penman. 1970. Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of initiation during mitosis. J. Mol. Biol. 50:655-670. [DOI] [PubMed] [Google Scholar]
- 14.Gerlitz, G., R. Jagus, and O. Elroy-Stein. 2002. Phosphorylation of initiation factor-2 alpha is required for activation of internal translation initiation during cell differentiation. Eur. J. Biochem. 269:2810-2819. [DOI] [PubMed] [Google Scholar]
- 15.Heesom, K. J., A. Gampel, H. Mellor, and R. M. Denton. 2001. Cell cycle-dependent phosphorylation of the translational repressor eIF-4E binding protein-1 (4E-BP1). Curr. Biol. 11:1374-1379. [DOI] [PubMed] [Google Scholar]
- 16.Janssen, G. M., J. Morales, A. Schipper, J. C. Labbe, O. Mulner-Lorillon, R. Belle, and W. Moller. 1991. A major substrate of maturation promoting factor identified as elongation factor 1 beta gamma delta in Xenopus laevis. J. Biol. Chem. 266:14885-14888. [PubMed] [Google Scholar]
- 17.Johannes, G., and P. Sarnow. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4:1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kedersha, N., M. R. Cho, W. Li, P. W. Yacono, S. Chen, N. Gilks, D. E. Golan, and P. Anderson. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M. J. Fritzler, D. Scheuner, R. J. Kaufman, D. E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamberti, A., M. Caraglia, O. Longo, M. Marra, A. Abbruzzese, and P. Arcari. 2004. The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: review article. Amino Acids 26:443-448. [DOI] [PubMed] [Google Scholar]
- 21.Le Sourd, F., S. Boulben, R. Le Bouffant, P. Cormier, J. Morales, R. Belle, and O. Mulner-Lorillon. 2006. eEF1B: at the dawn of the 21st century. Biochim. Biophys. Acta 1759:13-31. [DOI] [PubMed] [Google Scholar]
- 22.Martin, T. E., and L. H. Hartwell. 1970. Resistance of active yeast ribosomes to dissociation by KCl. J. Biol. Chem. 245:1504-1506. [PubMed] [Google Scholar]
- 23.Monnier, A., R. Belle, J. Morales, P. Cormier, S. Boulben, and O. Mulner-Lorillon. 2001. Evidence for regulation of protein synthesis at the elongation step by CDK1/cyclin B phosphorylation. Nucleic Acids Res. 29:1453-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monnier, A., J. Morales, P. Cormier, S. Boulben, R. Belle, and O. Mulner-Lorillon. 2001. Protein translation during early cell divisions of sea urchin embryos regulated at the level of polypeptide chain elongation and highly sensitive to natural polyamines. Zygote 9:229-236. [DOI] [PubMed] [Google Scholar]
- 25.Mulner-Lorillon, O., P. Cormier, J. C. Cavadore, J. Morales, R. Poulhe, and R. Belle. 1992. Phosphorylation of Xenopus elongation factor-1 gamma by cdc2 protein kinase: identification of the phosphorylation site. Exp. Cell Res. 202:549-551. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen, P. J., and E. H. McConkey. 1980. Evidence for control of protein synthesis in HeLa cells via the elongation rate. J. Cell Physiol. 104:269-281. [DOI] [PubMed] [Google Scholar]
- 27.Pyronnet, S., L. Pradayrol, and N. Sonenberg. 2000. A cell cycle-dependent internal ribosome entry site. Mol. Cell 5:607-616. [DOI] [PubMed] [Google Scholar]
- 28.Pyronnet, S., and N. Sonenberg. 2001. Cell-cycle-dependent translational control. Curr. Opin. Genet. Dev. 11:13-18. [DOI] [PubMed] [Google Scholar]
- 29.Qin, X., and P. Sarnow. 2004. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 279:13721-13728. [DOI] [PubMed] [Google Scholar]
- 30.Redpath, N. T., N. T. Price, K. V. Severinov, and C. G. Proud. 1993. Regulation of elongation factor-2 by multisite phosphorylation. Eur. J. Biochem. 213:689-699. [DOI] [PubMed] [Google Scholar]
- 31.Ruvinsky, I., N. Sharon, T. Lerer, H. Cohen, M. Stolovich-Rain, T. Nir, Y. Dor, P. Zisman, and O. Meyuhas. 2005. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 19:2199-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryazanov, A. G., E. A. Shestakova, and P. G. Natapov. 1988. Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature 334:170-173. [DOI] [PubMed] [Google Scholar]
- 33.Shenton, D., J. B. Smirnova, J. N. Selley, K. Carroll, S. J. Hubbard, G. D. Pavitt, M. P. Ashe, and C. M. Grant. 2006. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 281:29011-29021. [DOI] [PubMed] [Google Scholar]
- 34.Taylor, D. J., J. Frank, and T. G. Kinzy. 2007. Structure and function of the eukaryotic ribosome and elongation factors, p. 59-85. In M. B. Mathews, N. Sonenberg, J. W. B. Hershey (ed.), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 35.Wang, X., W. Li, M. Williams, N. Terada, D. R. Alessi, and C. G. Proud. 2001. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 20:4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]