Abstract
Vitamin A and its metabolite retinoic acid (RA) are essential elements for normal lung development and the differentiation of lung epithelial cells. We previously showed that RA rapidly activated cyclic AMP response element-binding protein (CREB) in a nonclassical manner in normal human tracheobronchial epithelial (NHTBE) cells. In the present study, we further demonstrated that this nonclassical signaling of RA on the activation of CREB plays a critical role in regulating the expression of airway epithelial cell differentiation markers, the MUC2, MUC5AC, and MUC5B genes. We found that RA rapidly activates the protein kinase Cα isozyme and transmits the activation signal to CREB via the Raf/MEK/extracellular signal-regulated kinase/p90 ribosomal S6 kinase (RSK) pathway. Activated RSK translocated from the cytoplasm to the nucleus, where it phosphorylates CREB. Activated CREB then binds to a cis-acting replication element motif on the promoter (at nucleotides [nt] −878 to −871) of the MUC5AC gene. The depletion of CREB using small interfering RNA abolished not only the RA-induced MUC5AC but also RA-induced MUC2 and MUC5B. Taken together, our findings demonstrate that CREB activation via this nonclassical RA signaling pathway may play an important role in regulating the expression of mucin genes and mediating the early biological effects of RA during normal mucous differentiation in NHTBE cells.
Mucus hyperproduction caused by respiratory disorders such as rhinitis, sinusitis, asthma, cystic fibrosis, and chronic obstructive pulmonary disease is currently incurable. Understanding the mechanisms of mucin gene regulation may lead to a new therapeutic opportunity for treating these disorders. Mucins, the major protein components of airway mucus, are high-molecular-mass glycoproteins produced by the epithelia of the respiratory tract and contribute to the viscoelastic properties of secreted mucus. To date, 18 human mucin genes have been cloned; at least 12 of them are expressed as mRNA in the lower respiratory tract (51). MUC5AC, which is a product of the goblet cells, is the major airway secretory mucin (23). MUC5AC gene expression is known to be regulated by retinoic acids (RAs) (31, 32).
RA, which is a metabolite of vitamin A, has an essential role in the development and maintenance of mucociliary differentiation of the epithelium in the conducting airways (18, 20, 21). When normal human tracheobronchial epithelial (NHTBE) cells are cultured in medium deficient in RAs, they undergo squamous differentiation; the addition of RAs restores mucous differentiation (31, 32) and induces mucin gene expression. We previously showed that RA receptor α (RARα) played an important role in the induction of expression of the mucin genes MUC2, MUC5AC, and MUC5B and mucous cell differentiation of bronchial epithelial cells by RA (31). Unlike this genetic event, an early effect of RA on mucociliary differentiation of bronchial epithelial cells is the nongenomic activation of cyclic AMP (cAMP) response element (CRE)-binding protein (CREB), as we previously demonstrated that RA rapidly activated a CREB transcription factor without using its canonical RAR/retinoid X receptors (RXRs) (1).
The CREB family of transcription factors plays critical roles in controlling cell growth, survival, and cell cycle progression and determining the fate of many cell types (11, 26, 34, 35, 50). Studies have shown that CREB is important for the proliferation and differentiation of neuronal cells (9, 35, 59), vascular smooth muscle cells (28), adipocytes (49), thyrocytes (42), Sertoli cells (53), pancreatic beta cells (27), and hematopoietic cells (47). CREBs recognize the specific DNA sequence 5′-TGACGTCA-3′, known as the CRE, in the transcription regulatory regions of many cAMP-regulated genes (8, 25, 38, 40, 61) with cell type specificity (5). Studies have shown that numerous kinases are involved in the activation of CREB in response to various extracellular stimuli (e.g., growth factors and stress signals) via diverse signaling pathways (28, 35, 55) including protein kinase A (PKA) (7, 15), protein kinase C (PKC) (64), p90 ribosomal S6 kinase 1/2 (RSK1/2) (63), mitogen- and stress-activated protein kinase 1 (10), mitogen-activated protein kinase (MAPK)-activated protein 2 kinase (61), Akt (12), calcium/calmodulin-dependent protein kinase II (58), and calcium/calmodulin-dependent protein kinase IV (37).
PKCs are a family of serine/threonine kinases that assume important physiological functions and are highly activated in certain malignancies (19). They can be categorized into three subfamilies, conventional, novel, and atypical, based on the structural domains that confer their coactivator dependency (41). A novel isoform, PKCδ, has been shown to play an important role in mucin exocytosis stimulated by human neutrophil elastase (45) and mucin gene expression induced by phorbol 12-myristate 13-acetate (62). Conventional PKC (cPKC) isoforms were shown to be directly activated by RA (48). A recent crystallography study showed that RA may exert its effect by binding to the C2 domain of PKC, a structure domain that present only in conventional isoforms (43). However, as PKCs are differentially expressed in different cell types (19), it is not known which isoform of cPKC is involved in RA-induced CREB activation and whether such an activation has a role in mucin gene expression and the normal differentiation of lung epithelial cells. Here, we hypothesized that RA-induced CREB activation has a critical role in mucin gene (especially MUC5AC) regulation and triggers early events in the differentiation of the bronchial epithelia. To test our hypothesis, we performed the study described herein to further determine the role of molecules involved in the nonclassical signaling pathway of RA in the activation of CREB. In addition, we analyzed the transcriptional action of CREB on the promoter region of the MUC5AC gene in NHTBE cells. An exploration of transcriptional regulation demonstrated that the RA-induced up-regulation of the MUC5AC gene in NHTBE cells was mediated by the CRE motif on the promoter. RA transactivates the promoter function of the other secretory mucin genes, including MUC2 and MUC5B, which are also regulated by CREB. Together, these findings give new insight into the role of CREB in mucin gene expression in mucous cell differentiation.
MATERIALS AND METHODS
Antibodies and reagents.
Antibodies against MEK1/2, phospho-MEK1/2 (Ser-217 and Ser-221 phosphorylated), Raf, phospho-Raf (Ser-259 phosphorylated), extracellular signal-regulated kinase (ERK), phospho-ERK (Thr-202 and Thr-204 phosphorylated), RSK, phospho-RSK (Ser-380 phosphorylated), and epidermal growth factor receptor were purchased from Cell Signaling Technology (Beverly, MA). Antibodies against PKCα, phospho-PKCα (Ser-659 phosphorylated), PKCβII, RARα, RARβ, RARγ, and RXRα were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). An antibody against β-actin (clone AC-15) was purchased from Sigma-Aldrich (St. Louis, MO). Antibodies against CREB, phospho-CREB (Ser-133 phosphorylated), phospho-PKCβII (Thr-641 phosphorylated), and cPKCαβγ were purchased from Upstate Biotechnology (Waltham, MA). MUC5AC antibody was purchased from NeoMarkers (Freemont, CA). All-trans RA, 9-cis RA, 13-cis RA, retinol, cycloheximide, and actinomycin D were purchased from Sigma-Aldrich. A pan-RAR antagonist (Ro 61-8431) and a pan-RXR antagonist (Ro 26-5405) were provided by Roche Bioscience (Palo Alto, CA). Other signal transduction inhibitors [H89, wortmannin, PP2, U0126, SB203580, Sp600125, staurosporine, Go6976, GF109203X, rottlerin, U73122, 2′,5′-dideoxyadenosine, calphostin C, EGTA-ethyleneglycol-bis(β-aminoethyl)-N,N,N′,N′-tetraacetoxymethyl ester (EGTA-AM), 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetra(acetoxymethyl) ester (BAPTA-AM), 12-O-tetradecanoyl-phorbol 13-acetate (TPA), and 8-bromo (8Br)-cAMP] were purchased from Calbiochem (San Diego, CA). All of the chemicals were dissolved in dimethyl sulfoxide (DMSO), and all other reagents were of analytical reagent grade or better.
NHTBE cell culture.
NHTBE cells were purchased from Clonetics (San Diego, CA). Second-passage NHTBE cells (1 × 105) were seeded onto a 24-mm Transwell plate (Corning, Acton, MA) and grown in serum-free growth factor-supplemented and hormone-supplemented culture medium as described previously (1, 18, 30, 32). After 7 days under immersed culture conditions, the cell culture was switched to an air-liquid interface system. For signal transduction experiments, NHTBE cells were incubated with bronchial epithelial cell basal medium for 24 h prior to RA treatment. To study the effect of chemical inhibitors on signal transduction pathways, cells were pretreated with each inhibitor 2 h prior to treatment with RA. To study the effect of RA on MUC5AC mRNA expression and secretion, NHTBE cells were grown in RA-deficient medium for 7 days and then further incubated with 1 μM RA for the indicated times or with various concentrations of RA for 24 h. All cells were grown at 37°C in a humidified atmosphere of 5% CO2.
QRT-PCR analysis.
Total RNA was extracted from NHTBE cells and the NCI-H292 (H292) lung cancer cell line using RNeasy Mini kits (QIAGEN, Valencia, CA). Each reverse transcription (RT) reaction was performed using 1 μg of total RNA that was reverse transcribed into cDNA using random hexamer primers (GeneAmp RNA PCR core kit; Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. PCR conditions were an initial reaction at 95°C for 5 min followed by 30 cycles at 95°C, 55°C, and 72°C for 30 s each. PCR analysis of β-actin expression (Ambion, Austin, TX) was performed as an internal control. The primer sequences, which were described previously (24), were as follows: 5′-AGCCTACGTGCCAAAAAAGG-3′ (PKCα forward), 5′-TCTAGGTGTGGAGGCAAATGG-3′ (PKCα reverse), 5′-ACCATGGAATCTGGAGCCGAGAAC-3′ (PKCβI forward), 5′-CTGTAGGAAGGCCTCCTTGAAAGA-3′ (PKCβI reverse), 5′-ACCATGGAATCTGGAGCCGAGAAC-3′ (PKCβII forward), 5′-CTGTAGGAAGGCCTCCTTGAAAGA-3′ (PKCβII reverse), 5′-ACCATGGAATCTGGAGCCGAGAAC-3′ (PKCβγ forward), 5′-CTGTAGGAAGGCCTCCTTGAAAGA-3′ (PKCβγ reverse), 5′-AGCCTACGTGCCAAAAAAGG-3′ (RARβ forward), 5′-TCTAGGTGTGGAGGCAAATGG-3′ (RARβ reverse), 5′-ACCATGGAATCTGGAGCCGAGAAC-3′ (CREB forward), and 5′-CTGTAGGAAGGCCTCCTTGAAAGA-3′ (CREB reverse). PCR products were then separated in a 2% agarose DNA gel and stained with ethidium bromide. To investigate the expression level of MUC5AC, mRNA expression was determined using quantitative RT-PCR (QRT-PCR). MUC5AC primer sequences were as follows: 5′-TGTGGCGGGAAAGACAGC-3′ (forward) and 5′-CCTTCCTATGGCTTAGCTTCAGC-3′ (reverse). MUC5B primer sequences were as follows: 5′-CGATCCCAACAGTGCCTTCT-3′ (forward) and 5′-CCTCGCTCCGCTCACAGT-3′ (reverse). MUC2 primer sequences were as follows: 5′-GCTGGCTGGATTCTGGAAAA-3′ (forward) and 5′-TGGCTCTGCAAGAGATGTTAGC-3′ (reverse). Human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) endogenous control reagents (Applied Biosystems) were used as normal controls. The PCR was performed in a 96-well format using the iCycler Real Time PCR detection system (Bio-Rad, Richmond, CA) by using SYBR green PCR core reagent (Applied Biosystems). Samples were analyzed in triplicate in a PCR program with an initial 3-min 95°C step followed by 40 cycles of 1 min at 95°C and 1 min at 65°C. After each analysis, a melting curve was performed to ensure that no primer dimers or secondary products were formed, and data were then analyzed using the accompanying iCycler version 3.1 software (Bio-Rad).
PKC activation assay and preparation of cell fractionation.
To investigate PKC activation in NHTBE cells, we measured the translocation of PKCα and PKCβII from cytosolic to membrane fractions as described previously (14). Briefly, the cells were treated with 1 μM RA for the indicated time periods at 37°C and then washed twice with ice-cold phosphate-buffered saline (PBS), scraped, and lysed using Tris-sucrose buffer (10 mM Tris-HCl [pH 7.5], 0.25 M sucrose, 0.2 mM CaCl2, 1 mM EDTA) supplemented with protease inhibitor cocktail tablets (Roche Diagnostics, Indianapolis, IN). The cells were sonicated using a sonic dismembrator (Fisher Scientific, Pittsburgh, PA) three times for 30 s each with 5 min of incubation on ice between sonications. Nuclei and unbroken cells were removed by centrifugation at 1,000 × g for 10 min in a Beckman SW28 rotor (Beckman Coulter, Fullerton, CA) at 4°C. The resulting supernatant was further centrifuged at 100,000 × g for 60 min in a Beckman SW28 rotor at 4°C. The supernatant from this centrifugation (cytosolic fraction) was removed and saved. The pellet (membrane fraction) was resuspended in Tris-sucrose buffer containing 1% Triton X-100 and 10 mM 2-mercaptoethanol. The suspensions were then briefly sonicated and centrifuged (100,000 × g for 60 min at 4°C). Insoluble material was discarded, and the supernatant was collected as a Triton X-100-soluble membrane fraction after the last centrifugation. The protein concentrations of the cytosolic and nuclear fractions were determined according to the Bradford protocol (4).
Western blot analysis.
Western blot analysis was performed as previously described (1). Briefly, whole-cell extracts were prepared using 2× sodium dodecyl sulfate (SDS) Laemmli lysis buffer. Equal amounts of total protein (20 μg) were resolved by 10% SDS-polyacrylamide gel electrophoresis. Proteins that were reactive with primary antibody were visualized with a horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence reagents (Amersham Bioscience, Arlington Heights, IL). Equal protein loading was confirmed by stripping the blots and reprobing them with an anti-β-actin antibody.
RNA interference.
RNA interference was performed with NHTBE cells by using the siIMPORTER small interfering RNA (siRNA) transfection reagent (Upstate Biotechnology) as described previously (1). For target gene silencing, SMARTpool-sequenced siRNAs targeting human PKCα (GenBank accession no. NM_002953), RSK1 (GenBank accession no. NM_002953), RSK2 (GenBank accession no. NM_004586), ERK2 (GenBank accession no. NM_002745), RARα (GenBank accession no. NM_000964), RARβ (GenBank accession no. NM_000965), RARγ (GenBank accession no. NM_000966), RXRα (GenBank accession no. NM_002957), or CREB (GenBank accession no. NM_004379) and a nonspecific control pool (siRNA-negative control; Dharmacon RNA Technologies, Lafayette, CO) were diluted and stored according to the manufacturer's instructions. NHTBE cells at 60% or 70% confluence were transfected with a final concentration of 100 nM SMARTpool siRNA or the nonspecific control pool. Cells were analyzed 72 h after transfection.
Transient transfection and luciferase assays.
Transfections of NHTBE cells were performed as described previously (1). Briefly, NHTBE cells (1 × 104 cells/well) were plated in 12-well plates using bronchial epithelial growth medium in the absence of RA. When the cells reached 60% to 70% confluence, transfections were performed using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) with the CRE promoter-luciferase reporter plasmid (Stratagene, La Jolla, CA), the construct of deletion mutants of MUC5AC or point-mutated CRE sites of MUC5AC, and the β-galactosidase (β-gal) reporter construct (BD Biosciences Clontech, Palo Alto, CA) according to the manufacturer's instructions. Four hours after transfection, cells were treated with RA and cultured for another 48 h. The luciferase activity was measured using a Lumat LB 9507 luminometer (EG&G, Berthold, Germany). β-gal activity was measured using a β-gal enzyme assay system (Promega, Madison, WI) and used to normalize transfection efficiency.
For knockdown by siRNA, 100 nM siRNA of the indicated target gene or nonspecific siRNA was cotransfected with luciferase plasmid and β-gal reporter constructs. The cells were treated with or without RA for another 24 h after 72 h of transfection, when target protein levels had been reduced by 70% to 80% as assessed by Western blot analysis. After transfection, whole-cell lysate was prepared for luciferase assay.
Immunofluorescence analysis.
For immunofluorescence staining, NHTBE cells were grown on coverslips for 7 days in an RA-deficient medium. After treatment with RA, the cells were fixed in a methanol-acetone mixture (1:1, vol/vol), washed with PBS, and blocked with 5% preimmune serum for 30 min. The cells were then incubated with the indicated primary antibodies (1:100 dilution) for 2 h at room temperature. The coverslips were washed with PBS containing 0.1% Tween 20, incubated with an AlexaFluor 488-tagged secondary antibody (Molecular Probes, Eugene, OR) for 1 h at room temperature, and then counterstained for nuclei with 4′,6-diamidino-2-phenylindole (DAPI) for 30 min. After being washed with PBS, slides were mounted using the SlowFade Antifade kit (Molecular Probes). The stained cells were visualized under a fluorescence microscope (Axioskop 40; Carl Zeiss, Thornwood, NY), and the images were captured at a magnification of ×400 and stored using the AxioVision software program (Carl Zeiss) according to the instructions provided by the manufacturer.
Quantification of secreted MUC5AC.
To analyze the production of MUC5AC, accumulating apical secretion fluid with or without RA was collected and analyzed by dot blot analysis (16). Briefly, diluted apical secretion fluid was applied to nitrocellulose membranes, which were incubated with human MUC5AC antibody, followed by a reaction with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG). The signal was detected by chemiluminescence using the ECL kit (Amersham, Little Chalfont, Buckinghamshire, United Kingdom). The blot intensities were analyzed densitometrically using Quantity One 4.6.1 software (Bio-Rad).
Preparation of MUC5AC promoter-luciferase constructs.
Methods to measure MUC5AC promoter activity with luciferase as a reporter were reported previously (17). Fragments ranging in size from 3.7 kb (nt −3752 to +7) to 0.29 kb (nt −296 to +7), which were located immediately adjacent to the 5′ transcriptional start site of human MUC5AC, were generated by digestion with exonuclease (Erase a Base system; Promega Corp., Madison, WI) of the 3.7-kb fragment of the MUC5AC promoter and cloned into the pGL3-Basic luciferase vector (Promega Corp.).
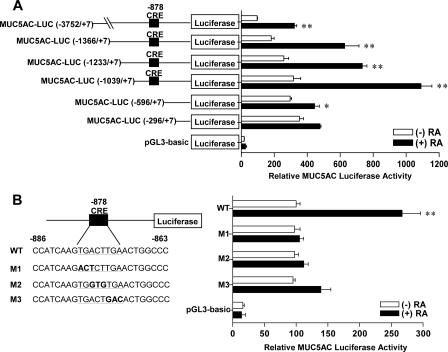
Site-directed mutations of the human MUC5AC promoter were made in the context of MUC5AC-LUC (positions −1366 to +7), using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and confirmed by sequencing. The oligomers used to introduce point mutations included the following: M1 (5′-CCATCAAGACTCTTGAACTGGCCC-3′), M2 (5′-CCATCAAGTGGTGTGAACTGGCCC-3′), and M3 (5′-CCATCAAGTGACTGACACTGGCCC-3′) (mutations are shown in boldface type; underlined type indicates putative CRE sites) (56).
EMSA.
To assess the DNA-binding activity of CREB, we performed an electrophoretic mobility shift analysis (EMSA) as described previously (1), with necessary modifications. Briefly, nuclear proteins from RA-treated or untreated cells were prepared and stored at −80°C. For EMSA, oligonucleotides corresponding to the consensus CRE sequences (5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′ [boldface type indicates CRE {Santa Cruz Biotechnology}]), CRE-specific sequences in the MUC5AC promoter region at nt −878 to −871 (wild type [wt] [5′-CCATCAAGTGACTTGAACTGGCCC-3′ {boldface type indicates putative CRE}]), and the CRE mutant sequence (M1 [5′-CCATCAAGACTCTTGAACTGGCCC-3′], M2 [5′-CCATCAAGTGGTGTGAACTGGCCC-3′], and M3 [5′-CCATCAAGTGACTGACACTGGCCC-3′ [underlining indicates a CRE mutant]) were synthesized, annealed, and end labeled with [γ-32P]ATP using T4 polynucleotide kinase. Nuclear extract (10 μg) was incubated at room temperature for 30 min with the 32P-labeled CRE probe in binding buffer [20% glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM dithiothreitol, 250 mM NaCl, 50 mM Tris-HCl (pH 7.5), and 0.25 mg/ml poly(dI-dC)]. For supershift analysis, nuclear extracts were incubated with antibodies against phospho-CREB (pCREB) or CREB and wild-type CRE-oligonucleotide probe for 30 min at 37°C, and the complex was then analyzed by EMSA. Antibodies against preimmune serum were included as negative controls. The DNA-protein complexes were resolved by electrophoresis in 5% nondenaturing polyacrylamide gels in 0.5× TBE (1× TBE is 25 mM Tris base, 25 mM boric acid, and 0.5 mM EDTA). Gels were dried and autoradiographed at −80°C. For EMSA, all studies were repeated three times; consistent results were obtained.
ChIP assay.
To determine transcription factor binding on the promoter region, we performed chromatin immunoprecipitation (ChIP) assays (34). NHTBE cells were treated with RA for 4 h, cross-linked with 1% formaldehyde for 10 min at 37°C, and then washed with cold PBS. The cell pellet was resuspended in 0.3 ml of lysis buffer (1% SDS, 100 mM NaCl, 50 mM Tris-HCl [pH 8.1], 5 mM EDTA), followed by sonication to an average DNA length of 500 to 1,000 bp, and then rotated at 4°C overnight. After interaction with protein A beads and incubation overnight at 65°C to reverse the cross-links, the DNA was dissolved in Tris-EDTA buffer and then analyzed by PCR. The antibodies anti-CREB, anti-phospho-CREB (Upstate Biotechnology), and normal rabbit IgG were added separately into the reaction solutions. Primers used for PCR were from MUC5AC promoter sequences 5′-TCACTGGGACCTTTCTGTGCT-3′ (forward) and 5′-ACTGAAGTAGCGCCAGCCA-3′ (reverse).
Data analysis.
For all transfection assays and QRT-PCR analyses, values are given as means ± standard errors (SE) of triplicate assays in three independent experiments. Statistical analysis was performed with the Prism program (GraphPad Software, San Diego, CA) using one-way analysis of variance, followed by Dunnett's test for comparing experiment groups against a single control or by Bonferroni's test for paired comparison.
RESULTS
PKCα is a predominant cPKC isozyme expressed in NHTBE cells.
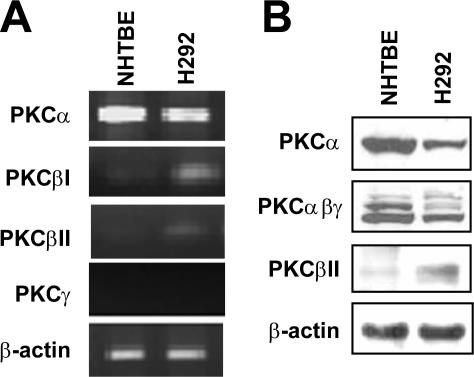
We previously showed that RA induced CREB activation via PKC in NHTBE cells (1). A recent crystallography study indicates that RA activates PKCα by binding to its C2 domain (43). Because this domain is present only in cPKC, to further determine which cPKC isotype mediates the RA activation of CREB, we measured the level of expression of cPKC isozymes in NHTBE cells using RT-PCR and Western blot analysis with isoform-specific primers and antibodies. H292 cells were included as a control for their known high expression level of PKC. As shown in Fig. 1, PKCα was highly expressed in NHTBE and H292 cells. However, PKCβI and PKCβII were barely detectable in NHTBE cells although fairly detectable in H292 cells. PKCγ was not detected in either cell type (Fig. 1A). We further confirmed the predominant expression of PKCα among cPKC isoforms in NHTBE cells using Western blot analysis with antibodies against PKCα, PKCα/β/γ, and PKCβII (Fig. 1B). We used an antibody that recognizes all three types (α/β/γ) to assess the level of PKCβI because a PKCβI-specific antibody was not available. Immunoblotting of PKCγ was not performed because we did not detect mRNA of PKCγ in RT-PCR. This result is in agreement with data reported previously by Park et al. (45) showing that NHTBE cells express PKCα, PKCδ, PKCμ, and PKCζ but not PKCβ, PKCγ, or PKCɛ. These results clearly showed that PKCα is a major cPKC isozyme expressed in NHTBE cells.
FIG. 1.
PKCα is highly expressed in NHTBE and H292 cells. NHTBE cells were cultured for 7 days in RA-deficient medium. (A) RT-PCR products amplified using specific primers for the cPKC isotypes from total RNA. The RT-PCR products were visualized on a 2% agarose gel containing 0.01% (wt/vol) ethidium bromide. (B) Equal amounts of whole-cell lysates were prepared and subjected to Western blot analysis with antibodies against PKCα, PKCαβγ, and PKCβII. β-Actin expression was used as an internal control for RT-PCR and Western blotting. Both figures are representative of three independent experiments.
RA activates PKCα and CREB, and activated PKCα translocates from the cytoplasm to plasma and perinuclear membranes.
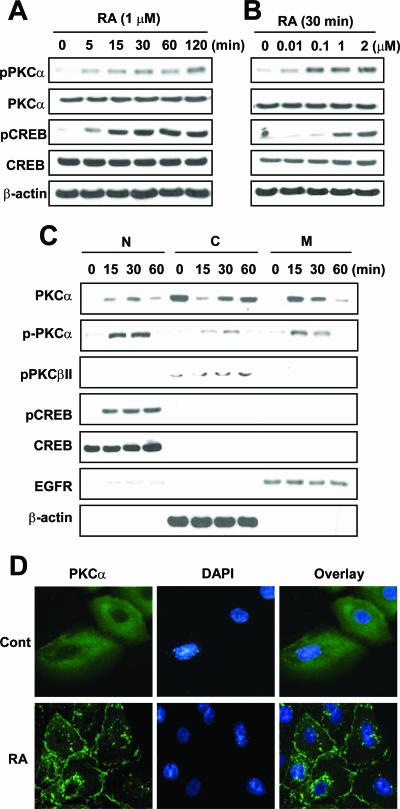
We next determined the effect of RA on the activation status of PKCα. We analyzed the effect of RA on PKCα phosphorylation at Ser-659, which indicates activation of PKCα (3, 44), and PKCα translocation using Western blot and immunofluorescence analysis. RA clearly induced phosphorylation of PKCα and CREB in a time-dependent manner (Fig. 2A). We detected PKCα phosphorylation as early as 5 min after treatment with RA; the level of phosphorylation reached a maximum at 120 min. CREB phosphorylation showed a similar pattern. The levels of expression of PKCα and CREB remained unchanged. We also observed the occurrence of RA-induced phosphorylation of PKCα and CREB in a dose-dependent manner (Fig. 2B). Both PKCα and CREB were phosphorylated with treatment with as little as 10 nM RA for 30 min.
FIG. 2.
Time- and dose-dependent activation of PKCα and CREB by RA. NHTBE cells grown in an RA-deficient medium were treated with 1 μM RA for the indicated periods (A) or RA at the indicated concentrations for 30 min (B). (C) RA-induced PKC activation was determined according to the translocation of PKC. RA-treated NHTBE cells were fractionated to nuclear (N), cytosolic (C), and membrane (M) fractions for the indicated periods as described in Materials and Methods. Equal loading of proteins in each compartment was verified using Western blot analysis with the indicated antibodies. EGFR, EGF receptor. (D) NHTBE cells grown on an RA-deficient medium on coverslips for 7 days were subjected to immunocytofluorescence analysis. NHTBE cells were incubated with a vehicle (Cont) (top) or 1 μM RA (bottom) for 15 min before fixation and stained with a mouse monoclonal anti-PKCα antibody followed by anti-mouse AlexaFluor 488 antibodies (green). Nuclei were stained with DAPI (blue), and the two images were then merged. All figures are representative of three independent experiments.
To determine whether RA relocates PKCα in bronchial epithelial cells, we isolated nuclear, cytoplasmic, and membrane fractions from the NHTBE cells treated with 1 μM RA for 15, 30, or 60 min. As shown in Fig. 2C, the majority of PKCαs were located in the cytoplasm of RA-deficient NHTBE cells. However, upon treatment with RA, more than 80% of the PKCαs were detected in the membrane and nuclear fractions within 15 min after treatment, demonstrating translocation of PKCα by RA. As analyzed using Western blot analysis to detect phosphorylated PKCα, PKCα translocated to nuclear and membrane fractions in a phosphorylated form. In contrast, PKCα residing in the cytoplasm was not phosphorylated. Treatment with RA did not cause the translocation of cytoplasmic PKCβII. These results are consistent with those of immunocytochemical analyses showing that most of the diffusely localized PKCα in the cytoplasm when the cells were in an RA-deficient state was translocated to the plasma and perinuclear membranes within 15 min after treatment with RA (Fig. 2D). Consistent with our previous results, the majority of CREB was localized in nuclear fractions. Phosphorylation of CREB was maximal after 15 min of treatment with RA and continued for another 60 min with no change in the level of CREB expression in nuclear fractions (1).
Activation of PKCα and CREB by RA is independent of RAR and RXR.
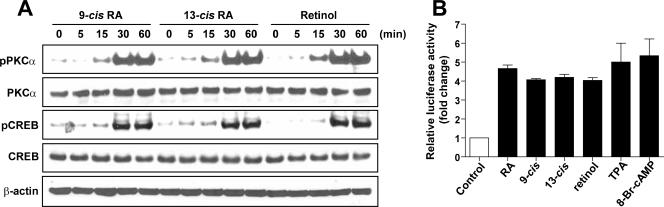
To determine whether RAR and RXR are involved in the activation of PKCα by RA, we initially examined the effect of several retinoids, including 9-cis RA, 13-cis RA, and retinol, on the phosphorylation of PKCα and CREB in NHTBE cells. We found that all these retinoids equipotently induced the phosphorylation of PKCα and CREB (Fig. 3A). Furthermore, all-trans RA, 9-cis RA, 13-cis RA, and retinol induced the transactivation of the CRE-luciferase reporter four- to fivefold, which is similar to the capacity of TPA and 8Br-cAMP, which were used as positive controls for CRE activation (Fig. 3B). These findings clearly demonstrate that all-trans RA is not the only retinoid that activates PKCα and CREB.
FIG. 3.
Activation of PKCα and CREB by retinoids and retinol. (A) NHTBE cells were treated with 1 μM 9-cis RA, 13-cis RA, or retinol for the indicated periods. After the treatment, equal amounts of whole-cell lysates were prepared and subjected to Western blot analysis using the indicated antibodies. (B) NHTBE cells were transiently transfected with a CRE promoter-driven luciferase-containing vector. After transfection, cells were further incubated with RA, 9-cis RA, 13-cis RA, or retinol for 48 h. TPA and 8Br-cAMP were used as positive controls. Cell lysates were then assayed for luciferase activity. The data represent luciferase units normalized according to β-gal in the same cell lysates. Values are the means ± SE of triplicate experiments, and the figures represent three independent experiments.
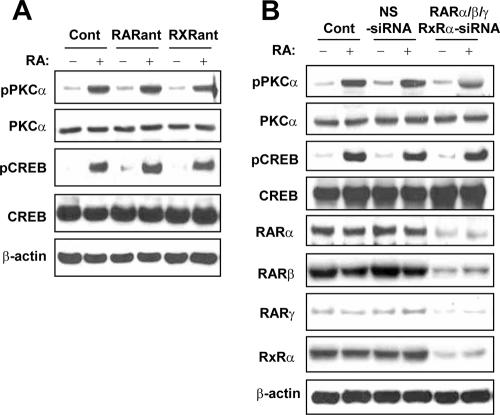
To determine whether RAR and RXR are required for the activation of PKCα by RA, we next examined the effects of pan-RAR and pan-RXR antagonists on the activation of PKC. We incubated NHTBE cells with 10 μM Ro 61-8431 and Ro 26-5405 before treatment with RA. Pretreatment with either antagonist did not block RA-induced activation of PKCα or CREB (Fig. 4A). As we previously reported (1), these antagonists did not affect the RA-induced CRE transactivation either.
FIG. 4.
RAR/RXR-independent activation of PKCα by RA. RA-induced PKC activation does not involve classical retinoid receptors. (A) To examine the role of RAR and RXR in RA-induced PKC activation, NHTBE cells were preincubated with 10 μM Ro 61-8431 and Ro 26-5405 for 2 h and further incubated with or without RA for 30 min. Equal amounts of whole-cell lysates were subjected to Western blot analysis. Cont, control. (B) NHTBE cells were transfected with siRNA of RARα, RARβ, RARγ, and RXRα combined (RARαβγ/RXRα-siRNA) or a nonspecific control pool (NS-siRNA) as described in Materials and Methods. Control cells were not transfected. Three days after transfection, the cells were treated with or without RA for 30 min. After the treatment, equal amounts of whole-cell lysates were prepared and subjected to Western blot analysis. All figures are representative of three independent experiments.
To further rule out the involvement of RAR and RXR in this rapid activation of PKCα by RA, we silenced the expression of RARα, RARβ, RARγ, and RXRα using siRNA; we silenced only the α type of RXR because it is known to be a predominant RXR isotype in eliciting the classical action of RA. At 72 h after transfection of siRNAs targeting RARs and RXRα, we confirmed the maximal silencing of target gene expression (data not shown). Western blot analysis of RA-induced active PKCα levels after transfection with pooled synthetic siRNAs for RARs and RXRα revealed that the depletion of RARs and RXRα in NHTBE cells did not affect RA-induced rapid phosphorylation of either PKCα or CREB (Fig. 4B). Silencing of RARs, RXRα, and a nonspecific control pool (negative control for siRNA) did not affect the activation status of PKCα or CREB in the vehicle-treated control. We also examined the effect of RARs and RXRα knockdown on CRE-luciferase activity and found no effect of such a knockdown on the RA-induced CRE transcriptional activity (data not shown). Taken together, these results confirmed that the RA-induced rapid activation of PKCα and CREB does not require RAR or RXR function in NHTBE cells.
cPKC and ERK are critically involved in RA-induced activation of CREB.
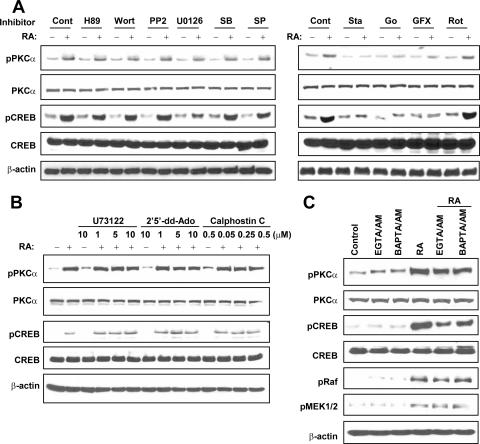
To investigate the signaling pathway by which RA induces the activation of PKCα and CREB, we treated NHTBE cells with various signaling inhibitors alone or in combination with RA and determined the activation status of PKCα and CREB. We found that of the signal transduction inhibitors used, only staurosporine (general PKC inhibitor), Go6976 (cPKC inhibitor), and GF109203X (cPKC and novel PKC inhibitor) substantially blocked PKCα and CREB activation induced by RA (Fig. 5A). Other inhibitors, including H89 (PKA inhibitor), wortmannin (phosphoinositide 3-kinase inhibitor), PP2 (Src tyrosine kinase inhibitor), SB203580 (p38 MAPK inhibitor), Sp600125 (c-Jun N-terminal kinase inhibitor), and rottlerin (novel PKC inhibitor), did not block RA activation of PKCα or CREB. Also, U0126 (MEK1/2 inhibitor) did not inhibit the RA activation of PKCα but did inhibit the activation of CREB. These results reconfirmed that cPKC isotypes and ERK1/2 are involved in the RA activation of CREB.
FIG. 5.
RA induces CREB activation via the PKCα/MEK/ERK pathway but not PKA or other MAPKs or tyrosine kinase pathways. (A) NHTBE cells were preincubated with various signal transduction inhibitors (10 μM) for 2 h and then further treated with or without 1 μM RA for 30 min. Cont, control; Wort, wortmannin; SB, SB203580; SP, SP600125; Sta, staurosporine; Go, Go6976; GFX, GF109203X; Rot, rottlerin. (B) NHTBE cells were preincubated with the PLC inhibitor U73122, the AC inhibitor 2′,5′-dideoxyadenosine (2′5′-dd-Ado), or the PKC inhibitor calphostin C at the indicated concentrations for 2 h before treatment with RA. (C) NHTBE cells were incubated with membrane-permeable specific calcium chelators (20 μM EGTA-AM or 50 μM BAPTA-AM) for 2 h before treatment with RA. Cells treated as controls received equivalent amounts of solvent (DMSO) (Cont). Whole-cell lysates were prepared, and equal amounts of proteins were subjected to Western blot analysis using the indicated antibodies. All figures are representative of three independent experiments.
To determine whether the adenylate cyclase (AC)-cAMP-PKA pathway is involved in the RA activation of PKCα and CREB, we inhibited endogenous AC using the AC inhibitor 2′,5′-dideoxyadenosine in NHTBE cells before treatment with RA. As shown in Fig. 5B, 2′,5′-dideoxyadenosine did not inhibit the RA-induced activation of PKCα or CREB. We also sought to determine whether phospholipase C (PLC) is involved in the RA activation of PKCα and CREB by treating NHTBE cells with U73122 (an inhibitor of PLC) before treatment with RA. Treatment with 10 μM U73122 did not affect the activation of PKCα or CREB (Fig. 5B).
To further determine whether diacylglycerol and Ca2+ are required for RA-induced PKCα and CREB activation, we treated NHTBE cells with the indicated concentrations of calphostin C (diacylglycerol inhibitor), EGTA-AM, or BAPTA-AM (an intracellular Ca2+ chelator) before treatment with RA. As shown in Fig. 5B, treatment with calphostin C did not block RA-induced activation. However, treatment with inhibitors that specifically chelated intracellular calcium did not inhibit the phosphorylation of PKCα but slightly blocked the phosphorylation of CREB (Fig. 5C).
RA activation of CREB is mediated by the PKCα/Raf/MEK/ERK/RSK signaling pathway.
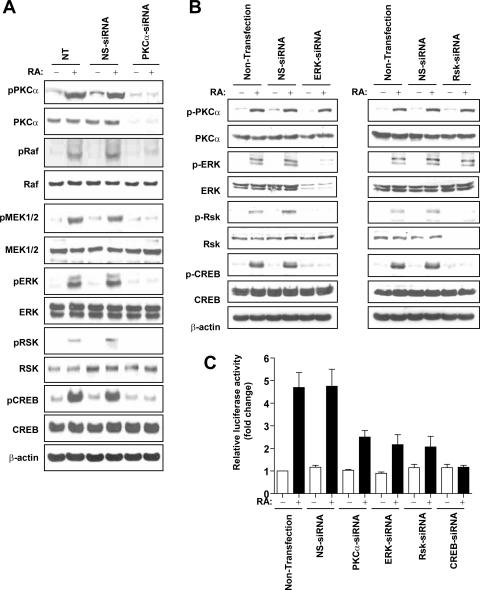
To determine whether RA-activated PKCα uses the linear signal transduction of the Raf/MEK/ERK/RSK signaling pathway for CREB activation, we examined the effect of RA on the PKCα/Raf/MEK/ERK/RSK/CREB signaling pathway after the knockdown of the expression of PKCα with an siRNA targeting PKCα. We verified the successful knockdown of PKCα expression in NHTBE cells using Western blot and RT-PCR analyses (Fig. 6A). Treatment with RA resulted in the activation of the PKCα/Raf/MEK/ERK/RSK/CREB signaling cascade in control transfections with a nonspecific siRNA. However, RA activation of this signaling pathway was completely blocked in PKCα-silenced NHTBE cells (Fig. 6A). This result clearly demonstrated that PKCα is indispensable for RA activation of the CREB signaling cascade.
FIG. 6.
RA-induced PKC-CREB activation is mediated by the Raf/MEK/ERK/RSK signaling pathways. NHTBE cells were transfected with PKCα-siRNA (A), ERK-siRNA (B, left), RSK-siRNA (B, right), or a nonspecific control pool (NS-siRNA) as described in Materials and Methods. Three days after transfection, the cells were further treated with or without RA for 30 min. After the treatment, equal amounts of whole-cell lysates were prepared and subjected to Western blot analysis using the indicated antibodies. All figures are representative of three independent experiments. β-Actin expression was used as an internal control. NT, nontransfection. (C) To determine the effects of siRNA on CRE transactivation, cells were cotransfected with a CRE-luciferase vector and siRNA of PKCα, ERK, RSK, or CREB. After transfection, cells were incubated with or without RA for 48 h. Cell lysates were then assayed for luciferase activity. The data represent luciferase units normalized according to β-gal in the same cell lysates. Values are means ± SE of triplicate experiments, and the figures are representative of three independent experiments.
We further examined the roles of ERK and RSK in the RA activation of CREB signaling using a knockdown approach. We transfected NHTBE cells with small interfering ERK1/2 (siERK1/2) or small interfering RSK1/2 and treated them with RA for 3 days after transfection. Compared with the control, transfection of NHTBE cells with siERK1/2 dramatically blocked ERK, RSK, and CREB activation, whereas the activation of PKCα (which is upstream of ERK) remained intact (Fig. 6B, left). The knockdown of RSK1/2 blocked the activation of CREB (downstream of RSK) only, without affecting upstream PKCα or ERK (Fig. 6B, right). To determine whether the inhibition of this signaling cascade inhibits the transcriptional activity of CREB, we transfected NHTBE cells with a CRE-luciferase reporter vector alone or with small interfering PKCα, siERK1/2, or small interfering CREB (siCREB). As shown in Fig. 6C, the knockdown of individual signaling mediators significantly blocked the RA-induced transcriptional activity of CREB.
ERK and RSK are activated by RA, and RSK but not ERK is translocated to the nucleus.
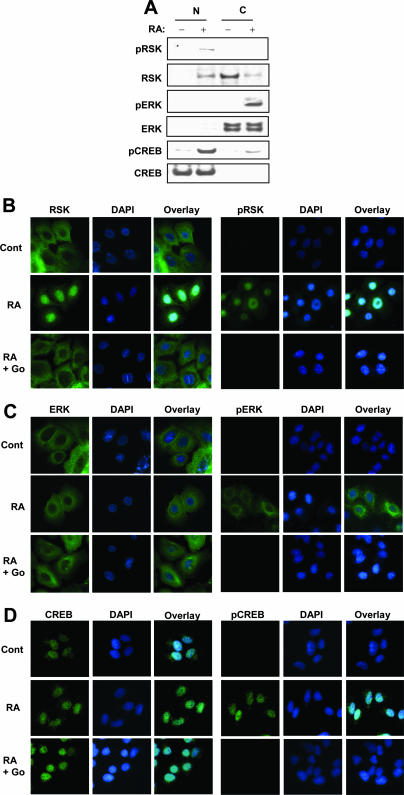
To determine which molecule enters the nucleus upon stimulation with RA, we examined the localization of activated RSK and ERK in NHTBE cells using Western blot analysis and immunocytofluorescence analysis. As shown in Fig. 7A, the majority of RSKs and ERKs were located in cytoplasmic fractions in the absence of RA. Treatment with RA (1 μM for 30 min) reduced the amount of RSK in the cytoplasm while increasing the amount of RSK in the nuclear fractions. We detected phosphorylated RSKs only in the nuclear fractions. However, we detected ERK only in the cytoplasmic fractions, and treatment with RA induced only the activation of ERK but not the translocation of it.
FIG. 7.
ERK and RSK are activated by RA, and RSK but not ERK is translocated to the nucleus. (A) NHTBE cells were preincubated with 10 μM Go6976 (selective PKCα inhibitor) for 2 h and then further treated with or without 1 μM RA for 30 min. Nuclear (N) and cytosolic (C) extracts were prepared as described in Materials and Methods. Equal amounts of proteins from each fraction were examined using Western blot analysis with the indicated antibodies. (B to D) Immunocytofluorescence analysis of the activation and translocation of RSK, ERK, and CREB. NHTBE cells grown in an RA-deficient medium on coverslips for 7 days were subjected to immunocytofluorescence analysis. Cells were preincubated with or without 10 μM Go6976 (Go) and then further incubated with a vehicle (Cont) (top) or 1 μM RA (middle) for 30 min before fixation. After fixation, the cells were stained with the indicated antibodies followed by anti-mouse AlexaFluor 488 antibodies (green). Nuclei were stained with DAPI (blue) and then merged. All figures are representative of three independent experiments.
To further examine the location of RSK and ERK in NHTBE cells, we performed immunocytochemical fluorescence analysis of these cells. As shown in Fig. 7B, unphosphorylated RSK resided in the cytoplasm, and treatment with RA induced the relocalization of RSK to the nuclei. We detected phosphorylated RSK only in the nuclei of RA-treated NHTBE cells. Also, the cPKC inhibitor Go6976 completely blocked the RA-induced activation and translocation of RSK. In contrast, RA phosphorylated ERK, but ERK was not translocated into the nuclei, although RA-induced phosphorylated ERK showed a tendency to migrate to the perinuclear membrane (Fig. 7C). The cPKC inhibitor Go6976 completely blocked the activation of ERK as expected. These results clearly demonstrated the translocation of active RSK but not ERK from the cytoplasm to the nucleus. Consistently, CREB resided in nuclear fractions in the absence of RA, and RA induced the phosphorylation of nuclear CREB (Fig. 7D). Again, the cPKC inhibitor Go6976 completely abolished the phosphorylation of CREB.
Mapping of RA-induced transactivation regions within the MUC5AC promoter.
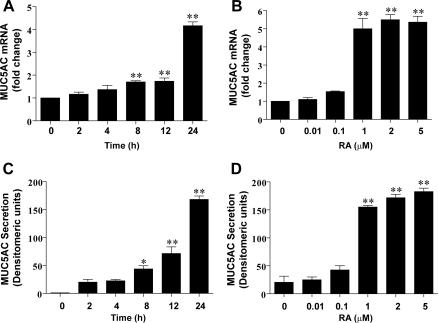
To determine whether RA induces the upregulation of MUC5AC, we treated the NHTBE cells with 1 μM RA for different periods of time or with different concentrations of RA for 24 h and measured MUC5AC gene expression and secretion. We found that RA increased both the MUC5AC mRNA level and protein secretion in a time- and dose-dependent manner (Fig. 8).
FIG. 8.
RA induced MUC5AC mRNA expression and secretion. NHTBE cells were grown in RA-deficient medium for 7 days and then further incubated with 1 μM RA for the indicated times (A and C) or with the indicated concentrations of RA for 24 h (B and D). MUC5AC mRNA expression and MUC5AC protein secretion were measured by QRT-PCR (A and B) and dot blot analysis (C and D) as described in Materials and Methods. Values shown are means ± SE of three independent experiments, each performed with triplicate cultures. *, P < 0.05; **, P < 0.01 (compared with RA-untreated control).
Next, to determine the promoter regions of the MUC5AC gene responsible for the RA-induced transcriptional activity, luciferase reporter vectors with progressive 5′ deletion mutants of the MUC5AC promoter were constructed and analyzed in NHTBE cells (Fig. 9A). Results of the transient transfection study showed that RA increased the luciferase activity of the construct with the region from nt −3752 to +7 of the MUC5AC promoter. Deletion of the MUC5AC 5′-flanking sequence from nt −3752 to −1039 had no apparent effect on the promoter activity induced by RA. However, this activation was completely abolished when the deletion proceeded from nt −1039 to −596. This result suggests that the region from nt −1039 to −596 is the core promoter of MUC5AC that responds to RA and that positive regulatory elements might locate between nt −1039 and −596. By comparing this core promoter sequence with the TFsearch database (http://mbs.cbrc.jp/research/db/TFSEARCH.html) (22), we found the putative CRE at nt −878 using a threshold score of 78.0. The native sequence of the putative CRE site in the sense strand is 5′-TGACTTGA-3′. To determine whether this cis-acting element is critical for RA-induced MUC5AC regulation, we mutated the putative CREB binding site by site-directed mutagenesis (Fig. 9B). Results of the transient transfection study indicated that mutant constructs (M1, M2, and M3) abolished the RA responsiveness of the wt MUC5AC promoter construct. These results strongly suggested that the CRE motif located between nt −878 and −871 on the MUC5AC promoter was critical for the up-regulation by RA.
FIG. 9.
The CRE motif is required for RA-induced MUC5AC promoter activity. NHTBE cells were transiently transfected with a luciferase expression vector that contained various 5′-deleted MUC5AC promoter constructs (A) or with the region from nt −1366 to +7 of the MUC5AC promoter construct containing various mutated CRE sites (B). After transfection, cells were treated with 1 μM RA or vehicle control (DMSO) for 48 h, and luciferase activity was the measured. The data were normalized with β-gal activity and expressed relative to the basal activity of the MUC5AC promoter (MUC5AC-LUC [nt −3752 to +7] = 100% [A]; MUC5AC-LUC [nt −1366 to +7] = 100% [B]). Values shown are means ± SE of three independent experiments, each performed with triplicate cultures. *, P < 0.05; *, P < 0.01 (compared in pairs between RA-treated and RA-untreated cultures).
RA induced the binding of CREB to the CRE motif on the MUC5AC promoter both in vitro and in vivo.
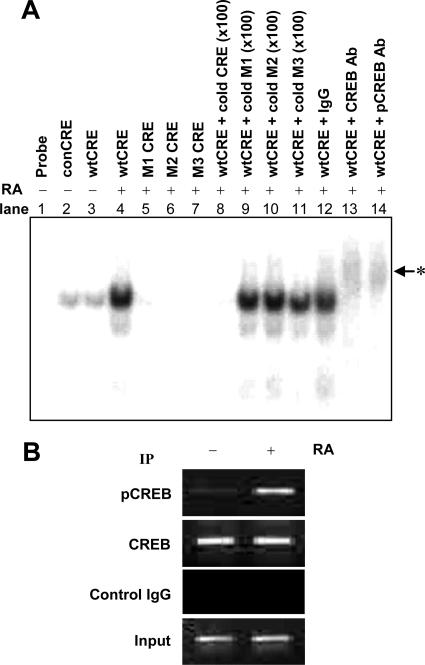
Because promoter analysis experiments showed that the CRE element was involved in RA-induced MUC5AC gene transcription, we examined the RA-induced CREB-CRE interaction in vitro using EMSA. We found that the binding of the radiolabeled MUC5AC CRE oligonucleotides (wtCRE) to nuclear protein was increased by RA treatment (Fig. 10A). The wtCRE band was abolished by a 100-fold excess of cold wtCRE (Fig. 10A, lane 8). However, unlabeled CRE mutants (cold M1, M2, and M3) (Fig. 10A, lanes 9 to 11) had no effect, indicating that the binding is specific. To further confirm the binding of CREB to the putative CRE motif after RA treatment, supershift assays were performed with anti-CREB or anti-pCREB antibodies (Fig. 10A, lanes 13 and 14). After incubation with either of these antibodies, a further retention of the labeled wtCRE band was observed (Fig. 10A, lanes 13 and 14). However, normal rabbit IgG did not cause such a retention (Fig. 10A, lane 12). These results indicated that CREB was the component that binds to the putative CRE site in response to RA stimulation. To confirm the interactions of CREB with putative CRE elements in vivo, a ChIP assay was performed using anti-CREB and anti-pCREB antibodies (Fig. 10B). ChIP analysis showed that a single PCR band (273 bp), which represents a fragment of the MUC5AC promoter, was detected in preimmunoprecipitation chromatin from both RA-treated and untreated NHTBE cells. Such a PCR signal was observed only in RA-treated NHTBE cells when immunoprecipitation was performed with anti-pCREB antibody, but no PCR signal was observed in controls with normal rabbit serum. Interestingly, in vivo bindings of pCREB to this fragment occurred as early as 30 min (data not shown) and were sustained until 4 h after RA treatment. Taken together, these results suggest that the binding of CREB to the CRE motif is critical for the RA-induced transactivation of MUC5AC.
FIG. 10.
RA induced a specific interaction between CREB and CRE sites on the MUC5AC promoter. NHTBE cells were cultured for 7 days with or without RA. (A) Nuclear extracts were prepared, and DNA-protein binding activity was measured by EMSA using various [γ-32P]ATP-labeled oligonucleotides (as indicated in Fig. 9B) that corresponded to nt −886 to nt −863 in the MUC5AC promoter. The DNA-binding reaction was performed in the absence or presence of a 100-fold excess (×100) of unlabeled oligonucleotides (as an unlabeled competitor). Supershift assays were performed using 2 μg of the anti-CREB and anti-pCREB antibodies (Ab), as indicated above each lane. The labeled nucleotide and nuclear protein complexes were separated by electrophoresis on 5% polyacrylamide gels and subjected to autoradiography. The arrowhead with the asterisk indicates shifted bands. (B) ChIP analysis. NHTBE cells were treated with 1 μM RA for 4 h, and ChIP assays were then performed using anti-CREB and anti-pCREB to precipitate MUC5AC promoter regions (nt −980 to nt −708) as described in Materials and Methods. As a positive control, 1% of the chromatin was assayed to verify equal loading (Input). Nonspecific IgG (Control IgG) was assayed under otherwise identical conditions as a negative control. Results shown are representative of three independent experiments. IP, immunoprecipitation.
Silencing of CREB decreases RA-induced mucin gene expression and secretion.
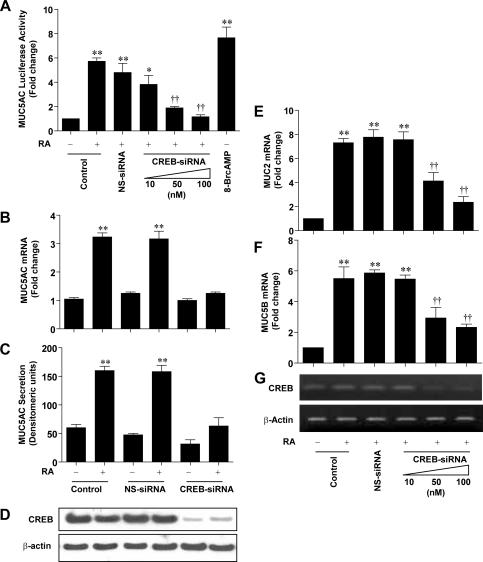
To further assess the role of CREB in RA-induced MUC5AC expression, we used the siRNA approach to silence CREB expression. As shown in Fig. 11A, RA induced MUC5AC promoter activities in siRNA-untransfected control and nonspecific siRNA-transfected samples by 5.8-fold and 4.3-fold, respectively. However, such a promoter activity was substantially reduced by an increasing concentration of the CREB-siRNA transfection (10 nM siCREB, 3.9-fold; 50 nM siCREB, 2.1-fold; 100 nM siCREB, 1.3-fold). 8Br-cAMP was used as a positive control for CREB activation. To determine the functional consequence of CREB silencing on MUC5AC gene regulation, we then transiently transfected siCREB and measured MUC5AC mRNA gene expression (Fig. 11B) and secretion (Fig. 11C). RA induced the up-regulation of MUC5AC mRNA expression and secretion. This up-regulation was completely abolished when the cells were transfected with siRNA of CREB (Fig. 11B and C). However, CREB knockdown alone did not show any effect on the mRNA expression level and secretion of MUC5AC compared with RA-untreated control cells. The efficient CREB knockdown by siRNA was verified by reduced CREB protein expression using Western blot analysis (Fig. 11D). These results demonstrate a stimulatory effect of RA on MUC5AC gene transcription and also indicated the critical role of CREB in MUC5AC transcriptional regulation by RA.
FIG. 11.
CREB is required for RA-induced expression of MUC5AC, MUC2, and MUC5B. (A) NHTBE cells were transiently cotransfected with a luciferase expression vector that contained the region from nt −1366 to +7 of the MUC5AC promoter construct and various concentrations of CREB siRNA (CREB-siRNA) (as indicated) or 100 nM nonspecific control pool (NS-siRNA). After transfection, cells were treated with 1 μM RA or vehicle control (DMSO) for 72 h, and luciferase activity was then measured. 8-Br-cAMP (1 μM) was used as a positive control for the CREB activator. The data were normalized according to β-gal activity. (B and C) NHTBE cells were transiently transfected with 100 nM CREB siRNA or nonspecific control siRNA. Three days after transfection, the cells were incubated with or without 1 μM RA or vehicle control (DMSO) for another 24 h, and MUC5AC mRNA expression and MUC5AC protein secretion were then analyzed. (D) CREB protein expression was measured as a control for the siRNA experiments. After treatment, MUC2 (E) and MUC5B (G) mRNA expression levels were analyzed. (G) CREB mRNA expression was measured to determine the siRNA transfection efficiency. Values are means ± SE of triplicate independent experiments, each performed with triplicate cultures. *, P < 0.05; **, P < 0.01 (compared with RA-untreated control). †, P < 0.05; ††, P < 0.01 (compared with the nonspecific control siRNA-transfected group).
Next, to determine whether CREB is required for the RA-induced transactivation of MUC2 and MUC5B, we transiently transfected various concentrations of siCREB and then measured MUC2 and MUC5B gene expression. As shown in Fig. 11E and F, RA induced MUC2 and MUC5B gene expression by 7.4-fold and 5.6-fold, respectively. However, increasing siCREB transfection decreased RA-upregulated MUC2 and MUC5B expression in a dose-dependent manner. The effective knockdown of CREB was confirmed by CREB mRNA expression using RT-PCR analysis (Fig. 11G). Taken together, these data strongly indicate that CREB has an essential role in RA-regulated mucin expression through CRE transactivation.
DISCUSSION
The major goals of our studies were to identify signaling components involved in the activation of CREB by the nonclassical action of RA and to understand the role of CREB and its signaling in the RA-induced regulation of mucin gene expression and mucous cell differentiation. In the present study, we showed that RA, 9-cis RA, 13-cis RA, and retinol rapidly activated the α isoform of cPKC in primary bronchial epithelial cells and that RA-activated PKCα transmitted the signal via the Raf/MEK/ERK/RSK pathway to induce the transcriptional activity of CREB. We further confirmed the critical role of CREB in the RA-induced expression of mucin genes, including MUC5AC, MUC5B, and MUC2.
Translocation of PKC from the cytoplasm to the membrane is a key event in classical PKC activation (2, 39). We showed that the majority of PKCαs were rapidly phosphorylated and translocated from the cytoplasm to the plasma and perinuclear membranes after treatment with RA (Fig. 2D). A recent study (13) showed that the calcium-binding loop and β-strands 3 and 4 in the C2 domain of PKCα are essential for the specific translocation of PKCα to the plasma membrane in response to Ca2+. Because the RA-binding site has been suggested to be the region of R239-V271 (48) that encompasses the third loop of the calcium-binding loop, RA may act like Ca2+ or enhance the effect of Ca2+ in coordinating the active structure of PKCα. In fact, our study showed that the deprivation of cytosolic Ca2+ by a calcium chelator did not block the RA-induced activation of PKCα but slightly reduced downstream CREB activation (Fig. 5C), indicating a cooperative effect between Ca2+ and RA, and that Ca2+ may be needed for the full activity of RA-activated PKCα. Interestingly, unlike Ca2+-induced activation, which translocates PKC only to the inner leaflet of the plasma membrane (13), RA stimulation induced the translocation of PKCα to both the plasma membrane and the perinuclear membrane. It has been suggested that nucleus-translocated PKC may play certain roles in transcriptional regulation (36). However, we did not observe a bypassing of the intermediate pathway in terms of RA-induced CREB activation. The role of nuclear PKC remains to be elucidated.
It has been shown that PKC isoforms are differentially expressed according to the cell types and the differentiational stages of the same cell linage (19). Other than cPKCs, a novel PKCδ has been reported to play an important role in mucin secretion in airway epithelial cells (45, 62). However, in our preliminary study, we did not detect any RA-induced phosphorylation of PKCδ in NHTBE cells (data not shown). Thereafter, we found that rottlerin, a novel PKC-selective inhibitor, was not able to inhibit RA-induced PKC phosphorylation and subsequent CREB activation (Fig. 5B). In contrast, the cPKC-selective inhibitors GF109203X and Go6976 completely abolished RA-induced PKC activation and its downstream signal transduction (Fig. 5A and Fig. 7B to D), indicating a cPKC-selective activation of RA. Moreover, treatment with calphostin C, a PKC inhibitor that competes with diacylglycerol for binding to the C1 domain of PKCs, did not block the aforementioned activation, which further supported the model that RA directly activates PKCα by binding to its C2 domain (43).
It is well known that RA activity is mediated mainly through RAR/RXR. However, our results showed that the activation of PKCα and CREB by RA does not require RAR/RXR. The functional nullification of RAR/RXR by treatment with pan-RAR and pan-RXR antagonists or by transfection of siRNAs targeting RARs and RXRα did not block RA-induced activation of PKCα or CREB (Fig. 4). Also, the treatment did not affect the transactivation activity of CREB in NHTBE cells. These results clearly demonstrated a nonclassical effect of RA on the activation of PKCα and CREB. We sought to explore the role of other conventional pathways that are known to activate PKC or CREB and found that neither PLC (upstream activator of PKC) nor PKA (upstream activator of CREB) is required for the RA-induced activation of PKCα and CREB (Fig. 5A).
Our previous study and other previous reports showed that the downstream signaling of PKC to CREB can be mediated through the Raf/MEK/ERK/RSK/CREB pathway (1, 29, 46, 52). Our results clearly showed that the RA activation of this signaling pathway was completely blocked in PKCα-silenced NHTBE cells (Fig. 6A), which indicates that PKCα is indispensable in this signaling cascade. Our previous studies and the current one also showed that the RA activation of CREB requires RSK (CREB kinase) and ERK (RSK kinase); the knockdown of either ERK or RSK abolished the RA-induced phosphorylation and transactivation activity of CREB but not the activation of PKCα (Fig. 6B). In addition, our results clearly showed that RA induced the translocation of RSK but not ERK from the cytoplasm to the nucleus, implying that CREB remained in the nucleus and is activated by the translocated active RSK (Fig. 7). Our results confirmed that RA-activated PKCα activates the signaling cascade of Raf/MEK/ERK/RSK/CREB during the mucociliary differentiation of primary bronchial epithelial cells.
Although we have previously shown that RA induces MUC5AC gene expression (32), it is not clear whether cis-regulatory and/or trans-acting factors control the expression of MUC5AC gene in response to RA. In this study, on the basis of the promoter reporter assay, we determined that the region from nt −1039 to nt −596 of the MUC5AC promoter is essential for the response to RA. We found that the CRE motif in the region at nt −878 of the MUC5AC promoter is critical for the RA-induced transcriptional activity of MUC5AC (Fig. 9). In addition, mutation of the CRE motif sequence abolished the responsiveness of the MUC5AC promoter reporter to RA. Further studies employing EMSA and ChIP analyses verified that RA induced the binding of activated CREB to this putative CRE motif of the MUC5AC promoter both in vitro and in vivo. The crucial role of CREB in the transactivation of MUC5AC was further demonstrated by the silencing of CREB with siRNA, which abolished the RA-induced transactivation of the MUC5AC promoter (Fig. 10A). The RA-induced promoter activity of MUC5AC was decreased as the concentration of transfected CREB siRNA increased, indicating a specific control of MUC5AC expression by CREB. However, CREB knockdown abolished not only RA-induced MUC5AC expression/protein secretion but also the expression of MUC2 and MUC5B. Using the TFsearch database, we found corresponding putative CRE sites in the MUC2 and MUC5B promoter regions (Fig. 12B). Taken together, these results strongly suggest that CREB is a potent transcriptional regulator of several mucin genes, including MUC2, MUC5AC, and MUC5B, in human primary airway epithelial cells.
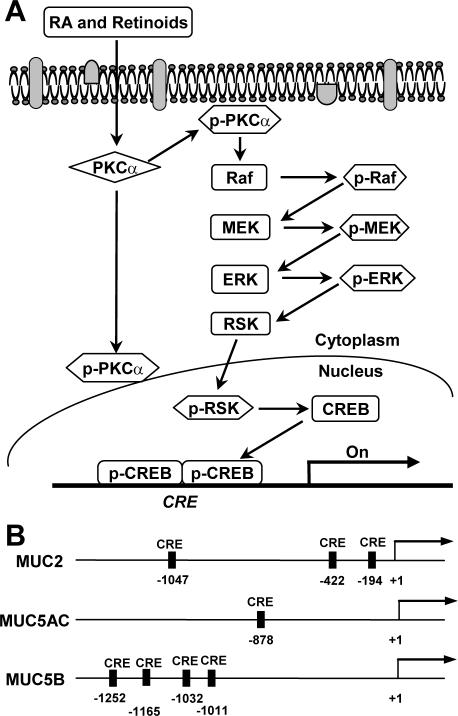
FIG. 12.
Model of the signal transduction pathway of mucin gene regulation by RA-inducible CREB activation. (A) Nonclassical action of RA on the activation of CREB via PKCα/Raf/MEK/ERK/RSK in primary bronchial epithelial cells. (B) Relative positions of the putative CRE sites on the human MUC2, MUC5AC, and MUC5B promoters. Putative CRE sites are indicated by black bars, as determined by comparisons of these sequences with those in the TFsearch database, using threshold scores of 78.0 for MUC5AC and 80.0 for MUC2 and MUC5B.
In agreement with our findings in the current study, it was previously shown that CRE is critical for interleukin-1β- and tumor necrosis factor alpha-induced MUC5AC upregulation in normal human nasal epithelial cells and the NCI-H292 human mucoepidermoid cell line and that the ERK/RSK/CREB signaling pathway is responsible for mediating the resultant MUC5AC promoter activation (56). Correspondingly, CREB has also been shown to regulate MUC8 gene expression induced by prostaglandin E2 and interleukin-1β via the same signaling pathway (6, 57). In addition, the Raf/MEK/ERK pathway has been reported to plays an important role in regulating lipopolysaccharide- and epidermal growth factor (EGF)-induced mucin expression in airway epithelia, especially in the situation of mucin overproduction (33, 54, 60). These results, in concert, suggest that the CRE motif and CREB signaling pathway activated by inflammatory mediators are important in the up-regulation of secretory mucins. It is conceivable that the cross talk among signaling pathways activated by RA, inflammatory mediators, and growth factors is critical for modulating mucin expression under physiological and pathological conditions. Moreover, in the absence of RA, high concentrations of EGF cause squamous metaplasia instead of mucin overproduction (our unpublished observation). This indispensableness of RA in mucous differentiation suggests that the rapid activation of CREB by RA via the PKCα/Raf/MEK/ERK/RSK signaling pathway has a priming effect on MUC5AC production and mucous differentiation in primary bronchial epithelial cells.
In summary, we demonstrated a nonclassical signaling pathway by which RA activates CREB. We showed that RA activates the PKCα isozyme and that RA-activated PKCα immediately translocates from the cytoplasm to the plasma and perinuclear membranes and transmits activation signals to CREB via the activation of a linear signaling cascade, Raf/MEK/ERK/RSK, without using conventional RAR and RXR in primary bronchial epithelial cells. Activated RSK was then translocated from the cytoplasm to the nucleus, where it activates CREB and where the activated CREB transactivates the MUC5AC gene. On the basis of our current and previous findings, we propose a new paradigm for RA-induced mucociliary differentiation of bronchial epithelial cells in which the nonclassical action (mediated through CREB) and the conventional action (mediated through RAR and RXR) of RA each assume important physiological roles and cooperatively facilitate the differentiation of bronchial epithelia.
Acknowledgments
This work was supported by National Heart, Lung, and Blood Institute grant R01-HL-077556 (to J.S.K.) and National Cancer Institute Cancer Center support grant CA-16672 (to The University of Texas M. D. Anderson Cancer Center).
Footnotes
Published ahead of print on 23 July 2007.
REFERENCES
- 1.Aggarwal, S., S. W. Kim, K. Cheon, F. H. Tabassam, J. H. Yoon, and J. S. Koo. 2006. Nonclassical action of retinoic acid on the activation of the cAMP response element-binding protein in normal human bronchial epithelial cells. Mol. Biol. Cell 17:566-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almholt, K., P. O. Arkhammar, O. Thastrup, and S. Tullin. 1999. Simultaneous visualization of the translocation of protein kinase Cα-green fluorescent protein hybrids and intracellular calcium concentrations. Biochem. J. 337:211-218. [PMC free article] [PubMed] [Google Scholar]
- 3.Bornancin, F., and P. J. Parker. 1997. Phosphorylation of protein kinase C-alpha on serine 657 controls the accumulation of active enzyme and contributes to its phosphatase-resistant state. J. Biol. Chem. 272:3544-3549. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Cha-Molstad, H., D. M. Keller, G. S. Yochum, S. Impey, and R. H. Goodman. 2004. Cell-type-specific binding of the transcription factor CREB to the cAMP-response element. Proc. Natl. Acad. Sci. USA 101:13572-13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, K. N., J. Y. Choi, C. H. Kim, S. J. Baek, K. C. Chung, U. Y. Moon, K. S. Kim, W. J. Lee, J. S. Koo, and J. H. Yoon. 2005. Prostaglandin E2 induces MUC8 gene expression via a mechanism involving ERK MAPK/RSK1/cAMP response element binding protein activation in human airway epithelial cells. J. Biol. Chem. 280:6676-6681. [DOI] [PubMed] [Google Scholar]
- 7.Colbran, J. L., P. J. Roach, C. J. Fiol, J. E. Dixon, O. M. Andrisani, and J. D. Corbin. 1992. cAMP-dependent protein kinase, but not the cGMP-dependent enzyme, rapidly phosphorylates delta-CREB, and a synthetic delta-CREB peptide. Biochem. Cell Biol. 70:1277-1282. [DOI] [PubMed] [Google Scholar]
- 8.Conkright, M. D., E. Guzman, L. Flechner, A. I. Su, J. B. Hogenesch, and M. Montminy. 2003. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell 11:1101-1108. [DOI] [PubMed] [Google Scholar]
- 9.Dawson, T. M., and D. D. Ginty. 2002. CREB family transcription factors inhibit neuronal suicide. Nat. Med. 8:450-451. [DOI] [PubMed] [Google Scholar]
- 10.Deak, M., A. D. Clifton, L. M. Lucocq, and D. R. Alessi. 1998. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17:4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desdouets, C., G. Matesic, C. A. Molina, N. S. Foulkes, P. Sassone-Corsi, C. Brechot, and J. Sobczak-Thepot. 1995. Cell cycle regulation of cyclin A gene expression by the cyclic AMP-responsive transcription factors CREB and CREM. Mol. Cell. Biol. 15:3301-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du, K., and M. Montminy. 1998. CREB is a regulatory target for the protein kinase Akt/PKB. J. Biol. Chem. 273:32377-32379. [DOI] [PubMed] [Google Scholar]
- 13.Evans, J. H., D. Murray, C. C. Leslie, and J. J. Falke. 2006. Specific translocation of protein kinase Cα to the plasma membrane requires both Ca2+ and PIP2 recognition by its C2 domain. Mol. Biol. Cell 17:56-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouassier, L., M. T. Nichols, E. Gidey, R. R. McWilliams, H. Robin, C. Finnigan, K. E. Howell, C. Housset, and R. B. Doctor. 2005. Protein kinase C regulates the phosphorylation and oligomerization of ERM binding phosphoprotein 50. Exp. Cell Res. 306:264-273. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, G. A., K. K. Yamamoto, W. H. Fischer, D. Karr, P. Menzel, W. Biggs III, W. W. Vale, and M. R. Montminy. 1989. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature 337:749-752. [DOI] [PubMed] [Google Scholar]
- 16.Gray, T., J. S. Koo, and P. Nettesheim. 2001. Regulation of mucous differentiation and mucin gene expression in the tracheobronchial epithelium. Toxicology 160:35-46. [DOI] [PubMed] [Google Scholar]
- 17.Gray, T., P. Nettesheim, C. Basbaum, and J. Koo. 2001. Regulation of mucin gene expression in human tracheobronchial epithelial cells by thyroid hormone. Biochem. J. 353:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray, T. E., K. Guzman, C. W. Davis, L. H. Abdullah, and P. Nettesheim. 1996. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 14:104-112. [DOI] [PubMed] [Google Scholar]
- 19.Griner, E. M., and M. G. Kazanietz. 2007. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer 7:281-294. [DOI] [PubMed] [Google Scholar]
- 20.Gudas, L. J. 1990. Molecular mechanisms of retinoid action. Am. J. Respir. Cell Mol. Biol. 2:319-320. [DOI] [PubMed] [Google Scholar]
- 21.Guzman, K., T. E. Gray, J. H. Yoon, and P. Nettesheim. 1996. Quantitation of mucin RNA by PCR reveals induction of both MUC2 and MUC5AC mRNA levels by retinoids. Am. J. Physiol. 271:L1023-L1028. [DOI] [PubMed] [Google Scholar]
- 22.Heinemeyer, T., E. Wingender, I. Reuter, H. Hermjakob, A. E. Kel, O. V. Kel, E. V. Ignatieva, E. A. Ananko, O. A. Podkolodnaya, F. A. Kolpakov, N. L. Podkolodny, and N. A. Kolchanov. 1998. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 26:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hovenberg, H. W., J. R. Davies, and I. Carlstedt. 1996. Different mucins are produced by the surface epithelium and the submucosa in human trachea: identification of MUC5AC as a major mucin from the goblet cells. Biochem. J. 318:319-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh, Y. S., S. F. Yang, H. L. Chiou, and D. Y. Kuo. 2005. Transcriptional involvement of protein kinase C-alpha isozyme in amphetamine-mediated appetite suppression. Eur. J. Neurosci. 22:715-723. [DOI] [PubMed] [Google Scholar]
- 25.Impey, S., S. R. McCorkle, H. Cha-Molstad, J. M. Dwyer, G. S. Yochum, J. M. Boss, S. McWeeney, J. J. Dunn, G. Mandel, and R. H. Goodman. 2004. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119:1041-1054. [DOI] [PubMed] [Google Scholar]
- 26.Ionescu, A. M., E. M. Schwarz, C. Vinson, J. E. Puzas, R. Rosier, P. R. Reynolds, and R. J. O'Keefe. 2001. PTHrP modulates chondrocyte differentiation through AP-1 and CREB signaling. J. Biol. Chem. 276:11639-11647. [DOI] [PubMed] [Google Scholar]
- 27.Jhala, U. S., G. Canettieri, R. A. Screaton, R. N. Kulkarni, S. Krajewski, J. Reed, J. Walker, X. Lin, M. White, and M. Montminy. 2003. cAMP promotes pancreatic beta-cell survival via CREB-mediated induction of IRS2. Genes Dev. 17:1575-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klemm, D. J., P. A. Watson, M. G. Frid, E. C. Dempsey, J. Schaack, L. A. Colton, A. Nesterova, K. R. Stenmark, and J. E. Reusch. 2001. cAMP response element-binding protein content is a molecular determinant of smooth muscle cell proliferation and migration. J. Biol. Chem. 276:46132-46141. [DOI] [PubMed] [Google Scholar]
- 29.Kolch, W., G. Heidecker, G. Kochs, R. Hummel, H. Vahidi, H. Mischak, G. Finkenzeller, D. Marme, and U. R. Rapp. 1993. Protein kinase C alpha activates RAF-1 by direct phosphorylation. Nature 364:249-252. [DOI] [PubMed] [Google Scholar]
- 30.Kolodziejski, P. J., A. Musial, J. S. Koo, and N. T. Eissa. 2002. Ubiquitination of inducible nitric oxide synthase is required for its degradation. Proc. Natl. Acad. Sci. USA 99:12315-12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koo, J. S., A. M. Jetten, P. Belloni, J. H. Yoon, Y. D. Kim, and P. Nettesheim. 1999. Role of retinoid receptors in the regulation of mucin gene expression by retinoic acid in human tracheobronchial epithelial cells. Biochem. J. 338:351-357. [PMC free article] [PubMed] [Google Scholar]
- 32.Koo, J. S., J. H. Yoon, T. Gray, D. Norford, A. M. Jetten, and P. Nettesheim. 1999. Restoration of the mucous phenotype by retinoic acid in retinoid-deficient human bronchial cell cultures: changes in mucin gene expression. Am. J. Respir. Cell Mol. Biol. 20:43-52. [DOI] [PubMed] [Google Scholar]
- 33.Li, J. D., W. Feng, M. Gallup, J. H. Kim, J. Gum, Y. Kim, and C. Basbaum. 1998. Activation of NF-kappaB via a Src-dependent Ras-MAPK-pp90rsk pathway is required for Pseudomonas aeruginosa-induced mucin overproduction in epithelial cells. Proc. Natl. Acad. Sci. USA 95:5718-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Long, F., E. Schipani, H. Asahara, H. Kronenberg, and M. Montminy. 2001. The CREB family of activators is required for endochondral bone development. Development 128:541-550. [DOI] [PubMed] [Google Scholar]
- 35.Lonze, B. E., and D. D. Ginty. 2002. Function and regulation of CREB family transcription factors in the nervous system. Neuron 35:605-623. [DOI] [PubMed] [Google Scholar]
- 36.Martelli, A. M., C. Evangelisti, M. Nyakern, and F. A. Manzoli. 2006. Nuclear protein kinase C. Biochim. Biophys. Acta 1761:542-551. [DOI] [PubMed] [Google Scholar]
- 37.Matthews, R. P., C. R. Guthrie, L. M. Wailes, X. Zhao, A. R. Means, and G. S. McKnight. 1994. Calcium/calmodulin-dependent protein kinase types II and IV differentially regulate CREB-dependent gene expression. Mol. Cell. Biol. 14:6107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayr, B., and M. Montminy. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2:599-609. [DOI] [PubMed] [Google Scholar]
- 39.Mochly-Rosen, D., C. J. Henrich, L. Cheever, H. Khaner, and P. C. Simpson. 1990. A protein kinase C isozyme is translocated to cytoskeletal elements on activation. Cell Regul. 1:693-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montminy, M. R., K. A. Sevarino, J. A. Wagner, G. Mandel, and R. H. Goodman. 1986. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc. Natl. Acad. Sci. USA 83:6682-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newton, A. C. 2001. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem. Rev. 101:2353-2364. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen, L. Q., P. Kopp, F. Martinson, K. Stanfield, S. I. Roth, and J. L. Jameson. 2000. A dominant negative CREB (cAMP response element-binding protein) isoform inhibits thyrocyte growth, thyroid-specific gene expression, differentiation, and function. Mol. Endocrinol. 14:1448-1461. [DOI] [PubMed] [Google Scholar]
- 43.Ochoa, W. F., A. Torrecillas, I. Fita, N. Verdaguer, S. Corbalan-Garcia, and J. C. Gomez-Fernandez. 2003. Retinoic acid binds to the C2-domain of protein kinase C(alpha). Biochemistry 42:8774-8779. [DOI] [PubMed] [Google Scholar]
- 44.Parekh, D. B., W. Ziegler, and P. J. Parker. 2000. Multiple pathways control protein kinase C phosphorylation. EMBO J. 19:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, J. A., F. He, L. D. Martin, Y. Li, B. N. Chorley, and K. B. Adler. 2005. Human neutrophil elastase induces hypersecretion of mucin from well-differentiated human bronchial epithelial cells in vitro via a protein kinase Cδ-mediated mechanism. Am. J. Pathol. 167:651-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson, G., F. Robinson, T. B. Gibson, B. E. Xu, M. Karandikar, K. Berman, and M. H. Cobb. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22:153-183. [DOI] [PubMed] [Google Scholar]
- 47.Persengiev, S. P., L. R. Devireddy, and M. R. Green. 2002. Inhibition of apoptosis by ATFx: a novel role for a member of the ATF/CREB family of mammalian bZIP transcription factors. Genes Dev. 16:1806-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radominska-Pandya, A., G. Chen, P. J. Czernik, J. M. Little, V. M. Samokyszyn, C. A. Carter, and G. Nowak. 2000. Direct interaction of all-trans-retinoic acid with protein kinase C (PKC). Implications for PKC signaling and cancer therapy. J. Biol. Chem. 275:22324-22330. [DOI] [PubMed] [Google Scholar]
- 49.Reusch, J. E., L. A. Colton, and D. J. Klemm. 2000. CREB activation induces adipogenesis in 3T3-L1 cells. Mol. Cell. Biol. 20:1008-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riccio, A., S. Ahn, C. M. Davenport, J. A. Blendy, and D. D. Ginty. 1999. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science 286:2358-2361. [DOI] [PubMed] [Google Scholar]
- 51.Rose, M. C., and J. A. Voynow. 2006. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol. Rev. 86:245-278. [DOI] [PubMed] [Google Scholar]
- 52.Roux, P. P., and J. Blenis. 2004. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68:320-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scobey, M., S. Bertera, J. Somers, S. Watkins, A. Zeleznik, and W. Walker. 2001. Delivery of a cyclic adenosine 3′,5′-monophosphate response element-binding protein (creb) mutant to seminiferous tubules results in impaired spermatogenesis. Endocrinology 142:948-954. [DOI] [PubMed] [Google Scholar]
- 54.Shao, M. X., I. F. Ueki, and J. A. Nadel. 2003. Tumor necrosis factor alpha-converting enzyme mediates MUC5AC mucin expression in cultured human airway epithelial cells. Proc. Natl. Acad. Sci. USA 100:11618-11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaywitz, A. J., and M. E. Greenberg. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu. Rev. Biochem. 68:821-861. [DOI] [PubMed] [Google Scholar]
- 56.Song, K. S., W. J. Lee, K. C. Chung, J. S. Koo, E. J. Yang, J. Y. Choi, and J. H. Yoon. 2003. Interleukin-1 beta and tumor necrosis factor-alpha induce MUC5AC overexpression through a mechanism involving ERK/p38 mitogen-activated protein kinases-MSK1-CREB activation in human airway epithelial cells. J. Biol. Chem. 278:23243-23250. [DOI] [PubMed] [Google Scholar]
- 57.Song, K. S., J. K. Seong, K. C. Chung, W. J. Lee, C. H. Kim, K. N. Cho, C. D. Kang, J. S. Koo, and J. H. Yoon. 2003. Induction of MUC8 gene expression by interleukin-1 beta is mediated by a sequential ERK MAPK/RSK1/CREB cascade pathway in human airway epithelial cells. J. Biol. Chem. 278:34890-34896. [DOI] [PubMed] [Google Scholar]
- 58.Sun, P., H. Enslen, P. S. Myung, and R. A. Maurer. 1994. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 8:2527-2539. [DOI] [PubMed] [Google Scholar]
- 59.Sung, J. Y., S. W. Shin, Y. S. Ahn, and K. C. Chung. 2001. Basic fibroblast growth factor-induced activation of novel CREB kinase during the differentiation of immortalized hippocampal cells. J. Biol. Chem. 276:13858-13866. [DOI] [PubMed] [Google Scholar]
- 60.Takeyama, K., K. Dabbagh, H. M. Lee, C. Agusti, J. A. Lausier, I. F. Ueki, K. M. Grattan, and J. A. Nadel. 1999. Epidermal growth factor system regulates mucin production in airways. Proc. Natl. Acad. Sci. USA 96:3081-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan, Y., J. Rouse, A. Zhang, S. Cariati, P. Cohen, and M. J. Comb. 1996. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15:4629-4642. [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, D. Y.-C., R. Wu, S. P. Reddy, Y. C. Lee, and M. M. Chang. 2007. Distinctive epidermal growth factor receptor/extracellular regulated kinase-independent and -dependent signaling pathways in the induction of airway mucin 5B and mucin 5AC expression by phorbol 12-myristate 13-acetate. Am. J. Pathol. 170:20-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xing, J., D. D. Ginty, and M. E. Greenberg. 1996. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 273:959-963. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto, K. K., G. A. Gonzalez, W. H. Biggs III, and M. R. Montminy. 1988. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature 334:494-498. [DOI] [PubMed] [Google Scholar]