Abstract
The transcription repressor Bach1 is a sensor and effector of heme that regulates the expression of heme oxygenase 1 and globin genes. Heme binds to Bach1, inhibiting its DNA binding activity and inducing its nuclear export. We found that hemin further induced the degradation of endogenous Bach1 in NIH 3T3 cells, murine embryonic fibroblasts, and murine erythroleukemia cells. In contrast, succinylacetone, an inhibitor of heme synthesis, caused accumulation of Bach1 in murine embryonic fibroblasts, indicating that physiological levels of heme regulated the Bach1 turnover. Polyubiquitination and rapid degradation of overexpressed Bach1 were induced by hemin treatment. HOIL-1, an ubiquitin-protein ligase which recognizes heme-bound, oxidized iron regulatory protein 2, was found to bind with Bach1 when both were overexpressed in NIH 3T3 cells. HOIL-1 stimulated the polyubiquitination of Bach1 in a purified in vitro ubiquitination system depending on the intact heme binding motifs of Bach1. Expression of dominant-negative HOIL-1 in murine erythroleukemia cells resulted in higher stability of endogenous Bach1, raising the possibility that the heme-regulated degradation involved HOIL-1 in murine erythroleukemia cells. These results suggest that heme within a cell regulates the polyubiquitination and degradation of Bach1.
Heme is essential for life, as it is a prosthetic group consisting of many heme proteins in reactions involving molecular oxygen, electron transfer, and diatomic gases. In addition, evidence that heme plays a regulatory role by conditionally binding to proteins has recently emerged. As a ligand, heme regulates transcription factors (7, 21, 48), sorting of mitochondrial and nuclear proteins (16, 34), protein kinase HRI (5, 6, 26), and the potassium channel (39). Heme also regulates protein degradation in both prokaryotes and eukaryotes. Heme binds to the bacterial iron response regulator (Irr) through its heme regulatory motif (HRM), thus causing a rapid degradation of Irr (24, 25, 44). In mammalian cells, heme binds to iron regulatory protein 2 (IRP2), a regulator of iron homeostasis, through its HRM (11, 42). Upon binding, heme is suggested to cause the oxidation of IRP2, thus leading to its recognition by the E3 ubiquitin-protein ligase HOIL-1, and also resulting in the polyubiquitination and subsequent proteasome-dependent degradation (12, 42). While the polyubiquitination of protein is often regulated by covalent modifications, such as the phosphorylation of a target, the finding of the heme-regulated degradation of IRP2 suggests that heme may constitute another class of molecular signature for recognition by an E3 ubiquitin ligase. However, the generality of this concept has not yet been established, and IRP2 is the only example.
The transcription repressor Bach1 is a sensor and effector of heme (10). Bach1 forms heterodimers with the small Maf proteins (MafF, MafG, and MafK) to bind to MARE (Maf recognition element), thus repressing the expression of target genes (23). As a sensor of heme, Bach1 binds heme through its multiple HRMs (21), thereby losing its activity as a repressor. First, the DNA binding activity of Bach1 dramatically decreases upon heme binding in vitro (21, 33). Second, a heme-regulated nuclear export signal of Bach1 is activated upon heme binding, thus leading to its accumulation in the cytoplasmic region (34). The net effect of heme is a derepression of the Bach1 target genes, constituting a fundamental mechanism in transducing the heme metabolism as an input into gene expression as an output. The target genes of Bach1 in mice include the α- and β-globin genes (3, 32, 36, 37) and the heme oxygenase 1 (HO-1) gene (Hmox-1) (32, 33). These genes possess MARE in their cis regulatory elements to which Bach1 binds, causing repression. When the heme levels increase, the displacement of Bach1 from the enhancers ensues (32). Simultaneously with the derepression by heme, activators such as NF-E2 and Nrf2 are stimulated, thus binding to the vacant enhancers to realize the activation of target genes. In the Bach1-deficient mice, HO-1 is highly expressed in many tissues (33), thus leading to decreased damage of tissue injuries, such as arteriosclerosis (22) and ischemic-reperfusion of the heart (45). We herein demonstrate another layer of the heme-mediated regulation of Bach1 involving its polyubiquitination and degradation. By analyzing the effects of hemin (ferric protoporphyrin IX) upon endogenous Bach1 in NIH 3T3 and murine erythroleukemia (MEL) cells, we found that increased levels of heme induced not only the nuclear export of Bach1 but also its polyubiquitination and degradation. HOIL-1 bound Bach1 in vivo and thus stimulated its polyubiquitination in vitro. These results suggest that heme regulates the polyubiquitination of Bach1 and subsequent degradation and that HOIL-1 may function as an E3 ligase in this process.
MATERIALS AND METHODS
Reagents.
Dulbecco's modified Eagle's medium was from Sigma, while fetal bovine serum was obtained from JRH Biosciences. Restriction endonuclease and other DNA-modifying enzymes were purchased from either New England Biolabs or Takara. Oligonucleotides were synthesized by Invitrogen. Hemin was obtained from Wako, and it was dissolved in dimethyl sulfoxide to make a 10 mM stock solution. The protease inhibitor cocktail was from Roche. All other chemicals were reagent grade.
Immunoblotting analysis.
Isolation of murine embryonic fibroblasts (MEFs) from wild-type or Bach1-deficient mice (33) will be described elsewhere. Total cell extracts were prepared from MEFs or NIH 3T3 cells treated with or without hemin or other reagents as described previously (43). An immunoblotting analysis was carried out as previously described using anti-Bach1 antiserum (A1-5) (32).
SA treatment and determination of heme levels.
MEFs were cultured in the presence or absence of 1 mM succinylacetone (SA), an inhibitor of heme synthesis, for 24 h. Heme contents in these cells were determined by the fluorometric method (28, 49). Pelleted cells were treated with 0.5 ml of 2 M oxalic acid at 100°C for 30 min. The mixtures without heating were used as a blank for measurement of endogenous porphyrins. After the mixtures were cooled, fluorescence was determined in an RF-5300PC spectrofluorometer (Shimadzu Corp., Kyoto, Japan). The excitation wavelength was 400 nm, and the fluorescence emission was determined at 662 nm. Hemin solutions, containing 0, 1.0, 2.5, 5.0, 10, 25, 50, or 100 pmol of hemin, were prepared in 0.5 ml of oxalic acid and used as standards.
Subcellular fractionation.
MEL cells transduced with FLAG- or hemagglutinin (HA)-tagged Bach1-expressing retrovirus (Y. Dohi, submitted for publication) were washed with phosphate-buffered saline (PBS) and fed with serum-free Dulbecco's modified Eagle's medium with or without 10 μM hemin. After the cells were incubated for 3 h, they were washed in PBS. The cells were suspended in buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.3% NP-40, and 1× protease inhibitor cocktail), then kept on ice for 10 min, and vortexed for 10 seconds. After centrifugation in a microcentrifuge, the supernatants were used as cytoplasmic extracts. The pellets were resuspended in buffer C (20 mM HEPES, pH 7.9, 20% glycerol, 400 mM NaCl, 1.0 mM MgCl2, 0.5 mM DTT, 0.3% NP-40, and 0.5 mM PMSF) and then were kept on ice for 30 min. After being centrifuged at 12,000 rpm for 2 min in a microcentrifuge, the supernatants were used as nuclear extracts.
Plasmids.
Plasmids pMT123HA-Ub (40), pCMV-FLAGMafK (19), pCMVFLAGBach1 and its derivatives (18, 34, 35), and pcDNAHAHOIL-1 and its derivatives (42) have been described previously. The Parkin expression plasmid, pcDNA3.1FLAGParkin, was a kind gift of N. Hattori (Juntendo University). Plasmids that express fragments of mouse Bach1 were constructed after amplifying respective DNA fragments by PCR. For amplifying the C-terminal fragments, PCRs were carried out with 5′ primers containing BamHI or BclI sites and a 3′ primer containing HindIII site (5′-GTTAAGAAGCTTTTACTCGTCAGTAGTGCACTT-3′). The 5′ primers were 5′-GTTAAGTGATCAGCCTCTGTCCAGCCAAAT-3′ (Bach1 252-739), 5′-GTTAAGTGATCAAGCAGCAGCCTTGCATCT-3′ (Bach1 380-739), and 5′-GCAGCAGGATCCGACTCTGAGACGGACACG-3′ (Bach1 506-739). For amplifying the N-terminal fragments, PCRs were carried out with 5′ primers containing the BclI site (5′-GTTAAGAATGATCAATGTCTGTGAGTGAGAGT-3′) and 3′ primers containing the HindIII site. The 3′ primers were 5′-GTTAAGAAGCTTTTACAGGCTAATCACACA-3′ (Bach1 1-503) and 5′-GTTAAGAAGCTTTTAGTCTTCTTTAGGGCC-3′ (Bach1 1-379). The amplified DNAs were digested with restriction enzymes and then were cloned into pcDNAFLAG (18).
In vivo ubiquitination assay.
The transfection of plasmids was done with GeneJuice (Novagen) as suggested by the supplier. NIH 3T3 cells in 6-cm dishes were transfected with 1 μg pMT123HA-Ub, 0.2 μg pCMV-FLAGMafK, and 1 μg pCMVFLAGBach1 or plasmids that express Bach1 derivatives. After 24 to 36 h, cells were washed with PBS, fed with serum-free medium, and then treated with chemical reagents at the indicated concentrations and periods. The cells were scraped into PBS and then were recovered by centrifugation. Total cell extracts were prepared using radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40, 0.5% deoxycholate, 0.02% NaN3, 3 mM dithiothreitol, 1 mM PMSF, and protease inhibitor cocktail). The extracts were diluted with 4× volume of ID(0) buffer (20 mM HEPES, pH 7.9, 20% glycerol, 5 mM MgCl2, 1.3 mg/ml bovine serum albumin, and 1× protease inhibitor cocktail), cleared for precipitates, and reacted with 25 μl slurry (1:1) of anti-FLAG M2 agarose beads (Sigma) at 4°C for 4 h. After the extracts were washed four times with wash buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 2 mM EDTA, 0.1% NP-40), the precipitates were analyzed by SDS-polyacrylamide electrophoresis and immunoblotting with anti-HA antibodies.
In vitro ubiquitination assay.
The purification of recombinant E1, E2 (UbcH5c), and HOIL-1 was described previously (42). The indicated amounts of Bach1 or Bach1mCP1-6 (9, 21) were incubated with 0.5 μg of ubiquitin aldehyde (Affiniti), 5 μg of ubiquitin (Sigma), and ATP and ATP-regenerating system (0.5 mM ATP, 10 mM creatine phosphate, 10 μg of creatine phosphokinase) in 20 μl of 20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, and 2 mM DTT in the presence or absence of 100 ng of E1, 100 ng of E2 (UbcH5c), and 1 μg of HOIL-1 as indicated for 2 h at 37°C. Where indicated, hemin was included at various concentrations. The reactions were stopped by the addition of 4× SDS sample buffer (200 mM Tris-HCl, pH 6.8, 8% SDS, 400 mM DTT, 40% glycerol, 0.2% bromophenol blue) and then resolved on a 6% SDS-polyacrylamide gel, followed by immunoblotting with rabbit anti-Bach1 antibody.
Coimmunoprecipitation assays.
To analyze protein interaction, NIH 3T3 cells in 10-cm dishes were transfected with 3 μg each of pCMVFLAGBach1 and pcDNA3.1HA-HOIL1 or their derivatives. After 24 to 36 h, the total protein extracts were prepared and precipitated with anti-FLAG M2 agarose beads as described previously (43).
MEL cells expressing DN HOIL-1.
The open reading frame fragment of dominant-negative (DN) HOIL-1 (C240 and 243S) (42) was amplified by PCR with a 5′ primer containing a XhoI site (5′-CCGctcgagATGGGCACAGCCACGCCAGAT-3′) and a 3′ primer containing a NotI site (5′-GTTAAgcggccgcTCAGTGGCAGTTCTGACAGCT-3′). Amplified DNA was digested with restriction enzymes and subcloned between the XhoI and NotI sites of pOZ-FH-N (20), resulting in pOZ-HOIL-1DN that directs expression of a bicistronic mRNA encoding the DN form of FLAG- or HA-tagged HOIL-1 (FLAG-HOIL-1 or HA-HOIL-2, respectively) and interleukin 2 receptor α-subunit. 293P packaging cells were transfected with pOZ-HOIL-1DN using GeneJuice (Novagen). Viral supernatants were harvested 2 days after transfection and used to transduce MEL cells. Transduced cells were enriched by repeated cycles of affinity cell sorting using anti-interleukin 2 receptor α-subunit antibody. Nontransduced cells were used as a control.
Half-life determination.
NIH 3T3 cells or MEL cells were incubated with 20 μg/ml cycloheximide in the presence or absence of 10 μM hemin. Band intensities of Western blots were measured by densitometric analysis. MafK or actin bands were used as internal controls to correct for protein loadings.
RESULTS
Heme induces the degradation of Bach1.
To examine the effect of heme upon endogenous Bach1, we first carried out detailed characterization of the anti-Bach1 antibody (A1-5) described previously (32). Total cell extracts were prepared from MEFs isolated from control wild-type and Bach1-deficient mice. The A1-5 antibody detected an antigen with a mobility expected for Bach1 in the extracts from wild-type MEFs (Fig. 1A). The remaining faint band in the extracts of Bach1-deficient MEFs may be due to an unrelated cross-reactive antigen. The identity of the other antigen is not known at present. However, they were judged unrelated to Bach1 because they were present in both extracts from control and Bach1-deficient MEFs. To further confirm these conclusions, extracts from MEL cells were immunoprecipitated with A1-5, and the precipitates were analyzed by immunoblotting with A1-5 (Fig. 1B). Only the presumed Bach1 antigen was enriched by the immunoprecipitation. A1-5 was thus useful to detect endogenous Bach1 in murine cells.
FIG. 1.
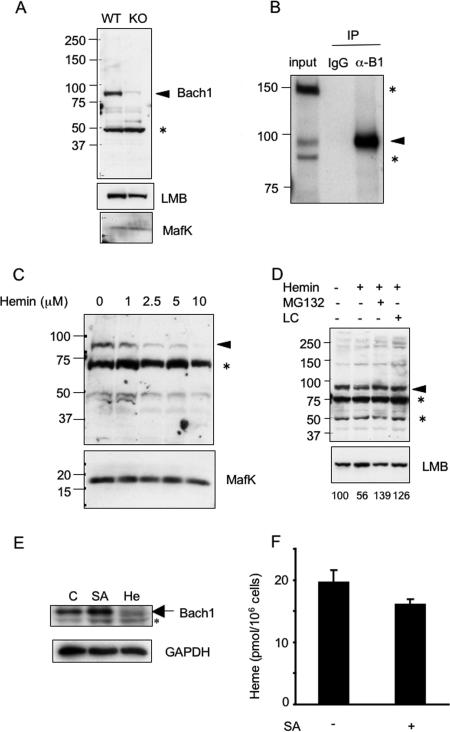
Degradation of Bach1 in response to hemin treatment. (A) Total cell extracts from wild-type (WT) and Bach1-deficient (knockout [KO]) MEFs were immunoblotted with anti-Bach1, anti-lamin B (anti-LMB), and anti-MafK antibodies. The arrowhead points to the position of the Bach1 band. The asterisk indicates an unknown cross-reactive band. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel. (B) Total cell extracts from MEL cells were immunoprecipitated (IP) with control or anti-Bach1 antibodies, and the resulting precipitates were immunoblotted with anti-Bach1 antibody (α-B1). The arrowhead points to the position of Bach1. The asterisks indicate unknown cross-reactive bands. IgG, immunoglobulin G. (C) NIH 3T3 cells were cultured in the presence of the indicated concentrations of hemin for 2 h. Bach1 and MafK (top and bottom panels, respectively) were detected with the corresponding antibodies. The asterisk indicates an unknown cross-reactive band. (D) NIH 3T3 cells were cultured in the presence (+) of the indicated combinations of hemin (2.5 μM), MG132 (10 μM), and lactacystin (LC) (10 μM) for 2 h. Bach1 (top) and lamin B (bottom) were detected with the corresponding antibodies. The asterisks indicate unknown cross-reactive bands. Relative Bach1 levels were determined after correction for LMB levels and shown below the panels. (E) MEF cells were cultured in the absence (control [C]) or presence of 1 mM SA (24 h) or 10 μM hemin (He) (3 h). Total cell extracts were analyzed with antibodies against Bach1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The asterisk indicates a nonspecific band cross-reacting with the Bach1 antibody. (F) Amounts of heme in MEFs treated with SA (+) for 24 h or without SA (−) are shown.
We treated NIH 3T3 cells with various concentrations of hemin for 2 h and compared Bach1 levels (Fig. 1C). Hemin at concentrations as low as 2.5 μM caused significant reduction of Bach1. In contrast to Bach1, the amounts of MafK remained unchanged after hemin treatment (Fig. 1C). Thus, we regarded MafK as an internal loading control. When treated with proteasome inhibitorsMG132 or lactacystin, the hemin-induced Bach1 reduction was negated (Fig. 1D, top panel), suggesting that hemin induced proteasome-dependent degradation of Bach1. These reagents did not affect the levels of lamin B (Fig. 1D, bottom panel).
To investigate whether endogenous levels of heme affect Bach1 protein levels, we examined the effect of succinylacetone, an inhibitor of δ-aminolevulinic acid dehydratase, the second enzyme involved in the heme synthesis pathway, upon the levels of Bach1 in MEFs. SA caused an increase of Bach1 level (Fig. 1E) and a reduction of heme level (Fig. 1F). Since most of heme is expected to bind proteins, such as cytochrome c, tightly, the observed reduction of heme may reflect changes in the levels of “uncommitted” heme pool which could be a ligand for Bach1 (27). Taken together, these results suggest that physiological levels of heme can function as a trigger of Bach1 degradation. The effect of heme upon endogenous Bach1 was also observed in MEL cells in which even 0.5 μM of hemin in the medium caused a reduction of Bach1 in 2 h (Fig. 2A).
FIG. 2.
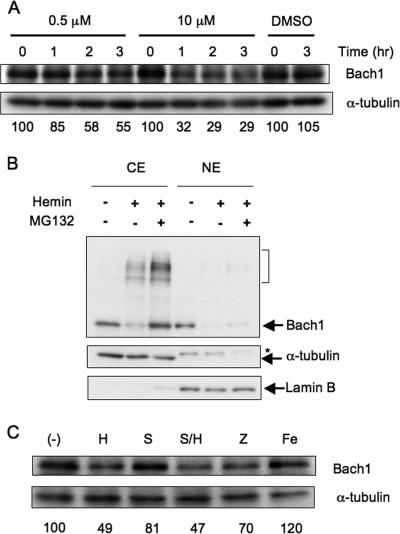
Degradation of Bach1 in MEL cells. (A) MEL cells were cultured in the presence of the indicated concentrations of hemin for 1 to 3 h. Immunoblots for Bach1 and α-tubulin are shown. Relative Bach1 levels were determined after correction for α-tubulin levels and shown below the panels. The effect of dimethyl sulfoxide (DMSO), which was used to prepare the hemin solution, was also examined. (B) MEL cells expressing FLAG-Bach1 were treated with 10 μM hemin (+) for 3 h or without hemin (−). Where indicated, 5 μM MG132 was added (+) 20 min before hemin treatment. Cytoplasmic and nuclear fractions (cytoplasmic extract [CE] and nuclear extract [NE]) were immunoblotted with antibodies against Bach1, α-tubulin, and lamin B. The bracket indicates lower-mobility bands containing Bach1. The asterisk indicates a cross-reactive band. Partitioning of α-tubulin and lamin B in the cytoplasmic and nuclear fractions, respectively, indicates successful fractionation. (C) MEL cells were cultured in the presence of 0.5 μM hemin (H), SnPP (S), ZnPP (Z), or ammonium iron(III) citrate (Fe) for 3 h. Cell extracts were analyzed with antibodies against Bach1 and α-tubulin. Relative Bach1 levels were determined by correcting for α-tubulin levels, and mean values from two experiments are shown below the panel. (−), negative control.
Heme stimulates the nuclear export of Bach1 when it is overexpressed in several cell lines (34). To investigate the relationship between the heme-induced reduction and nuclear export of Bach1, we examined the effect of hemin on the subcellular distribution of overexpressed Bach1 in MEL cells. An immunoblot analysis using anti-Bach1 antibody revealed that Bach1 localized in both the cytoplasmic and nuclear fractions under normal culture conditions (Fig. 2B). We then treated MEL cells with 10 μM hemin for 3 h. Not only the nuclear but also the cytoplasmic levels of Bach1 significantly decreased (Fig. 2B). Proteasome inhibitor MG132 negated the disappearance of Bach1 from the cytoplasmic fraction but not in the nuclear fraction. Hemin induced an appearance of lower-mobility bands in the cytoplasmic fraction, which was augmented by MG132 treatment. These bands may represent polyubiquitinated Bach1 (see below). Thus, heme appears to induce proteasome-dependent degradation of Bach1 in the cytoplasmic region following its nuclear export, although we cannot exclude the possibility that Bach1 is also degraded in the nuclei.
Because hemin induces expression of HO-1 very rapidly, it is highly likely that hemin added was degraded by HO-1 in cells. Thus, it is possible that products of heme degradation, in particular iron, were responsible for the Bach1 decay. For example, it has been reported recently that inhibition of IRP1 activity by heme is mediated by the iron produced by HO-1 (30). To address this possibility, we utilized tin protoporphyrin IX (SnPP), an inhibitor of HO. As in Fig. 2A, 0.5 μM hemin caused Bach1 decay (Fig. 2C). SnPP itself did not show this effect. Importantly, SnPP did not inhibit the hemin-induced Bach1 degradation (Fig. 2C). When cells were treated with ammonium iron(III) citrate to increase intracellular iron levels (11), Bach1 remained the same (Fig. 2C). These results strongly suggest that heme itself was responsible for the Bach1 degradation. Zinc protoporphyrin IX (ZnPP) also caused degradation (Fig. 2C) which became more clear at higher concentrations of ZnPP (data not shown). Because ZnPP is unable to bind to a sixth ligand, it does not interact with molecular oxygen when bound to a protein in a five-coordinated manner (2). Therefore, ZnPP bound to Bach1 may not produce reactive oxygen species. A simple interpretation would be that heme-regulated degradation of Bach1 takes place without protein oxidation (see Discussion).
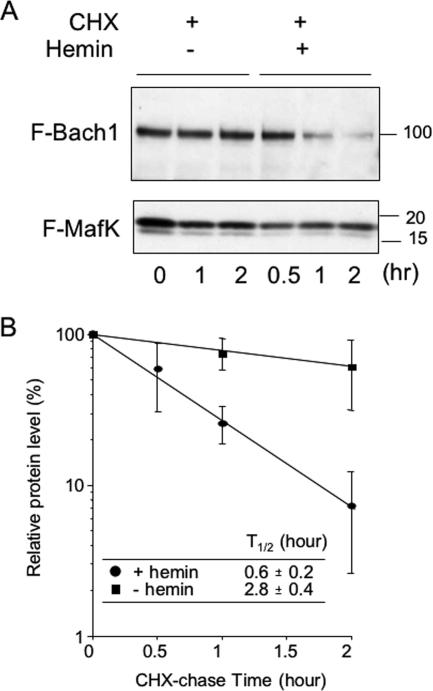
To confirm that hemin induced Bach1 degradation, we expressed FLAG-tagged Bach1 (FLAG-Bach1) and FLAG-MafK in NIH 3T3 cells, treated the cells with cycloheximide, and then cultured them in the absence or presence of hemin in the medium. Upon immunoblotting with anti-FLAG antibody, FLAG-Bach1 and FLAG-MafK were identified based on their mobilities. As shown in Fig. 3A, FLAG-Bach1 was rather stable in the absence of hemin, but it disappeared more rapidly in the presence of hemin. Its half-life was shortened less than 1 h by the presence of hemin (Fig. 3B). We conclude that heme induces degradation of Bach1 in vivo.
FIG. 3.
Hemin-induced degradation of Bach1. (A) NIH 3T3 cells were transfected with FLAG-Bach1 (F-Bach1), FLAG-MafK (F-MafK), and HA-ubiquitin expression plasmids. The cells were treated with 10 μM hemin (+) or without hemin (−) for the indicated periods (in hours). Cycloheximide (CHX) (25 μg/ml) was added (+) to the medium 10 min before hemin treatment. Anti-Bach1 and anti-MafK antibodies were used to detect each protein. The positions of molecular mass markers (in kilodaltons) are indicated to the right of the gel. (B) The intensities of bands in panel A were quantified by densitometry, and the relative mean intensities ± standard deviations (error bars) were calculated from three experiments and plotted. The band intensity of Bach1 from untreated sample (0 h) was set at 100%. Symbols: ovals, treated with 10 μM hemin; squares, not treated with hemin.
Heme induces ubiquitination of Bach1.
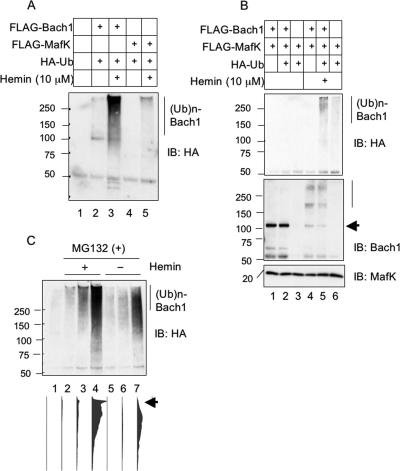
Since the proteasome-dependent degradation of proteins involves their modification by polyubiquitination, we examined whether heme induced ubiquitination of Bach1. Various combinations of FLAG-Bach1, FLAG-MafK, and HA-tagged ubiquitin (HA-ubiquitin) were expressed in NIH 3T3 cells by transfecting the respective expression plasmids. After treating cells with or without hemin, whole-cell extracts were prepared from the cells and were subjected to immunoprecipitation of FLAG-Bach1 or FLAG-MafK with anti-FLAG antibody. An immunoblotting analysis of the precipitates with an anti-HA antibody revealed that polyubiquitination of FLAG-Bach1 or FLAG-MafK was not apparent under normal conditions (Fig. 4A and B). In contrast, it significantly increased when the cells expressing FLAG-Bach1 (Fig. 4A, lane 3) or FLAG-Bach1 and FLAG-MafK (Fig. 4B, anti-HA blot, lane 5) were treated with hemin. Concomitantly, the amounts of FLAG-Bach1 decreased (Fig. 4B, anti-Bach1 blot). In contrast, when FLAG-MafK alone was expressed, it did not show a significant level of ubiquitination (Fig. 4A, lane 5, and B, lane 6), and it did not decrease in response to hemin (Fig. 4B, anti-MafK blot). An immunoblot analysis with anti-Bach1 antibody confirmed that the polyubiquitinated bands contained Bach1 (Fig. 4B, anti-Bach1 blot). The time course of hemin-induced ubiquitination of FLAG-Bach1 was examined (Fig. 4C). In this experiment, MG132 was added to inhibit degradation and thus to compare the levels of polyubiquitinated FLAG-Bach1. Ubiquitination of FLAG-Bach1 was induced as early as 1 h after hemin treatment (Fig. 4C). Even though ubiquitination of FLAG-Bach1 was detected in the absence of hemin after 4 h, densitometry analysis revealed that the levels were much less (Fig. 4C, lower panel). The extent of polyubiquitination as judged by the mobility was also less. The observed polyubiquitination without added hemin may be due to endogenous levels of heme or due to a misfolding of a portion of overexpressed Bach1. Nonetheless, these results established that hemin enhanced the polyubiquitination of Bach1.
FIG. 4.
Hemin-induced ubiquitination of Bach1. (A) NIH 3T3 cells were transfected (+) with the indicated combinations of FLAG-Bach1, FLAG-MafK, and HA-ubiquitin (HA-Ub) expression plasmids. Cells were treated without or with 10 μM hemin for 2 h (+). Immunoprecipitation was carried out using FLAG antibody. Ubiquitin conjugation was examined with anti-HA antibodies. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel. (Ub)n-Bach1, polyubiquitinated Bach1; IB: HA, immunoblotting with anti-HA antibody. (B) Ubiquitination of FLAG-Bach1 and FLAG-MafK were compared as described above for panel A (IB: HA). The same membranes were probed with anti-Bach1 and anti-MafK antibodies (immunoblotting with anti-Bach1 antibody [IB: Bach1]; immunoblotting with anti-MafK antibody [IB: MafK]). The arrow points to the position of Bach1. (C) NIH 3T3 cells were transfected as described above for panel A. Cells were treated (+) with hemin and MG132 for 1, 2, or 4 h (lanes 2 to 4). Cells were treated only with MG132 for the same periods (lanes 5 to 7). Lane 1 contains transfected cells without hemin/MG132 treatment. Cell extracts were analyzed as described above for panel A. Results of densitometric scanning of the entire lanes are shown below. The arrow indicates highly ubiquitinated bands.
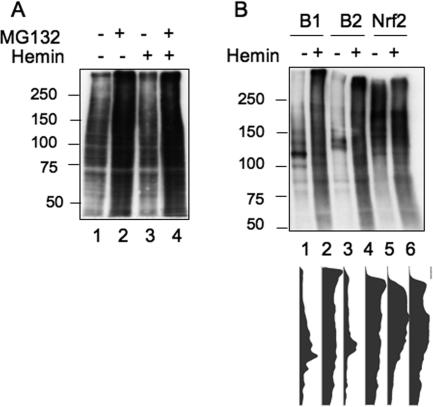
To rule out the possibility that the increased polyubiquitination of Bach1 is not a specific event, we compared the ubiquitination of bulk proteins with direct immunoblotting of whole-cell extracts. As shown in Fig. 5A, the total ubiquitination levels increased by treating cells with MG132 presumably due to the accumulation of ubiquitinated proteins. Hemin treatment did not increase the levels of ubiquitinated proteins with or without MG132 treatment. We thus concluded that the heme-induced polyubiquitination was specific to Bach1 among the many proteins that are modified by ubiquitination. We next compared Bach2 and Nrf2 and found that Bach2 was also polyubiquitinated in response to hemin treatment (Fig. 5B). Nrf2 was polyubiquitinated under normal culture conditions as reported previously (15, 17, 46), which was not obviously affected by hemin treatment (Fig. 5B). These results suggest that heme-induced polyubiquitination is thus a shared feature between Bach1 and Bach2.
FIG. 5.
Specificity of hemin-induced polyubiquitination. (A) NIH 3T3 cells were transfected with HA-ubiquitin expression plasmid and treated (+) with the indicated reagents for 2 h. Total cell extracts were analyzed by immunoblotting with anti-HA antibodies. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel. (B) NIH 3T3 cells were transfected with FLAG-Bach1 (B1), FLAG-Bach2 (B2), or FLAG-Nrf2 together with HA-ubiquitin expression plasmids. The ubiquitination of each FLAG-tagged protein was analyzed as described in the legend to Fig. 4. Results of densitometric scanning of the entire lanes are shown below.
HOIL-1 binds to Bach1 and mediates its ubiquitination.
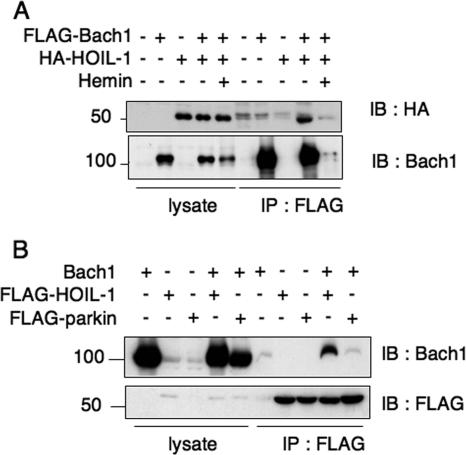
HOIL-1 is the only E3 ubiquitin ligase whose target recognition is reported to be regulated by heme (42). In the case of IRP2, the binding of heme to IRP2 results in its polyubiquitination by HOIL-1. To investigate whether HOIL-1 is involved in the regulation of Bach1, we first examined their binding after overexpressing them in NIH 3T3 cells. As shown in Fig. 6A, HOIL-1 was coimmunoprecipitated along with Bach1. In a reverse experiment in which FLAG-HOIL1 was immunoprecipitated, the coexpressed Bach1 was also precipitated (Fig. 6B). The Bach1-HOIL1 interaction was judged to be specific, because Bach1 was not coimmunoprecipitated with another E3 ligase Parkin as efficiently as with HOIL-1 (Fig. 6B). Quantification of the bands showed that the efficiency of Bach1 recovery with HOIL-1 was at least 10-fold more than that with Parkin. The interaction of Bach1 and HOIL-1 was lost after hemin treatment due to the degradation of Bach1 (Fig. 6A). We failed to detect interaction of endogenous Bach1 and HOIL-1 proteins in NIH 3T3 cells in coimmunoprecipitation assay even in the presence of MG132 (data not shown). Their interaction may be transient and unstable as expected for an enzyme-substrate interaction. Further study is necessary to understand in vivo interaction between Bach1 and HOIL-1.
FIG. 6.
Interaction of HOIL-1 with Bach1. (A) Lysates from NIH 3T3 cells expressing (+) the indicated combinations of proteins were immunoprecipitated with anti-FLAG agarose beads (IP : FLAG) and analyzed by immunoblotting with anti-HA (IB: HA) to detect HOIL-1 or anti-Bach1 (IB: Bach1). The total lysates were also compared to document equal protein input. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel. (B) Lysates from NIH 3T3 cells expressing the indicated combinations of proteins that were immunoprecipitated with anti-FLAG agarose beads and then analyzed by immunoblotting with anti-FLAG to detect HOIL-1 or Parkin and anti-Bach1.
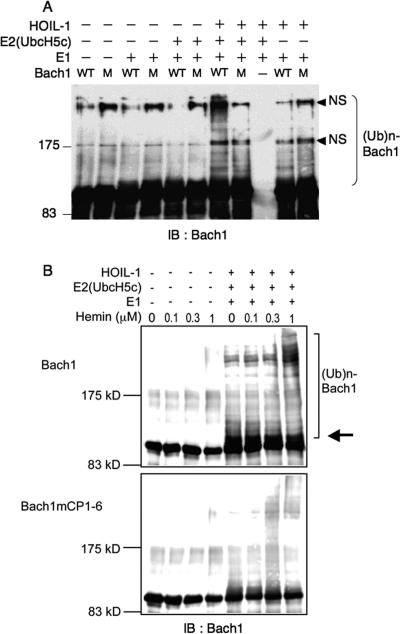
To investigate whether HOIL-1 is an E3 ligase of Bach1, we carried out an in vitro ubiquitination assay using purified proteins. As shown in Fig. 7A, HOIL-1 stimulated polyubiquitination of Bach1 only in the presence of both E1 and E2 enzymes. Omission of either E1 or E2 abolished Bach1 polyubiquitination. Bach1 carries six HRM-like sequences each containing cysteine-proline (CP) motifs (21). Among them, CP motif 3 (CP3), CP4, CP5, and CP6 have been shown to bind to heme in vitro (21, 34). Mutations in all of the CP motifs significantly reduced the HOIL-1-mediated ubiquitination, thus suggesting an involvement of some of the HRMs/CP motifs in the HOIL-1-mediated polyubiquitination. Consistent with this interpretation, the addition of hemin in the reaction mixtures strongly increased the levels of polyubiquitination of Bach1 but not Bach1 with mutations in the CP motifs (Fig. 7B). This set of experiments also revealed that HOIL-1 together with E1 and E2 enzymes efficiently monoubiquitinated Bach1. In contrast, monoubiquitination of Bach1 with CP mutations was less efficient. These results strongly suggest that HOIL-1 is one of the E3 ligases that mediate the heme-regulated polyubiquitination of Bach1.
FIG. 7.
Polybiquitination of Bach1 in vitro. (A) Recombinant Bach1 (wild type [WT]) or Bach1mCP1-6 containing mutations in the six CP motifs (M) were incubated with ubiquitin aldehyde, ubiquitin, ATP and the ATP-regenerating system in the presence (+) or absence (−) of combinations of E1, E2 (UbcH5c), and HOIL-1 as indicated. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel. IB : Bach1, immunoblotting with anti-Bach1 antibody; (Ub)n-Bach1, polyubiquitinated Bach1; NS, nonspecific. (B) Ubiquitination reactions of wild-type and Bach1mCP1-6 (0.5 μg) were carried out as described above for panel A in the presence of the indicated concentrations of hemin. HOIL-1 together with E1 and E2 enzymes efficiently monoubiquitinated Bach1 (indicated by the arrow). The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
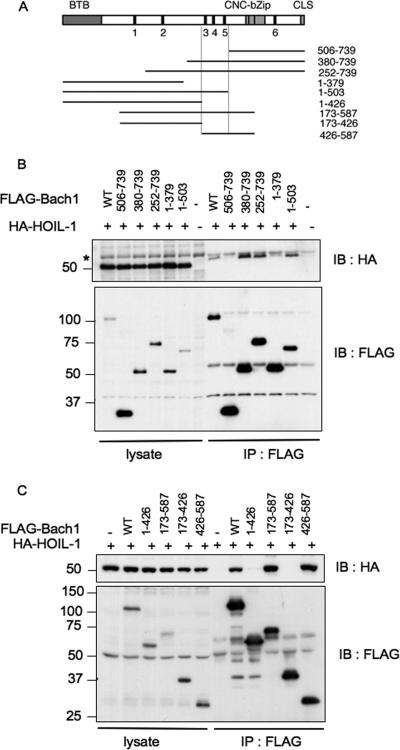
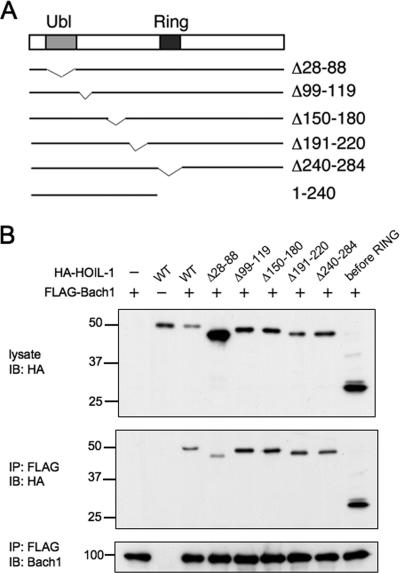
To map a region of Bach1 that is involved in the HOIL-1 binding, we expressed various FLAG-tagged fragments of Bach1 (Fig. 8A) together with HA-HOIL-1 in NIH 3T3 cells, immunoprecipitated with anti-FLAG antibody, and looked for the presence of HOIL-1 in the immunoprecipitates. The results shown in Fig. 8B and C can be interpreted as follows. Both the N- and C-terminal regions of Bach1 were not involved in HOIL-1 binding, and a central region (amino acids 426 to 503) containing the CP3, CP4, and CP5 motifs was involved specifically in HOIL-1 binding.
FIG. 8.
Mapping of the HOIL-1 binding region of Bach1. (A) Schematic representation of Bach1 and its deletion derivatives. CP motifs are indicated with filled boxes and numbers. BTB/POZ domain, cnc homology-basic region-leucine zipper (bZip), and CLS are also shown with boxes. The boundaries of the HOIL-1-binding region determined in panels B and C are shown by the vertical lines. (B and C) HA-tagged HOIL-1 and the indicated fragments of Bach1 with FLAG tag (wild type [WT]) were expressed. Bach1 fragments were purified with anti-FLAG agarose beads, and copurification of HOIL-1 was determined by immunoblotting with anti-HA antibodies (IB : HA) (top panels). The total lysates were also compared to document equal input of proteins (left portions of gels). The asterisk in panel B indicates a cross-reactive band whose identity is not known. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
We next determined a region of HOIL-1 that mediated Bach1 binding by coexpressing HA-tagged HOIL-1 fragments (Fig. 9A) and FLAG-Bach1. Among the fragments examined, Δ28-88 lacking the ubiquitin-like domain showed less binding to FLAG-Bach1 (Fig. 9B). This seemed to be significant especially when taking into account the fact that Δ28-88 was expressed more abundantly than other HOIL-1 fragments. Consistent with this, an N-terminal fragment containing the ubiquitin-like domain was coprecipitated along with Bach1. Considering that the ubiquitin-like domain is involved in the recognition of IRP2 (42), we concluded that the N-terminal region containing the ubiquitin-like domain was therefore also involved in Bach1 binding. There may be several, redundant Bach1-binding sites within this region.
FIG. 9.
Mapping of Bach1 binding region of HOIL-1. (A) Schematic representation of HOIL-1 and its deletion derivatives. (B) Coimmunoprecipitation assays were carried out as described in the legend to Fig. 8 with the indicated combinations of expression plasmids. The total lysates and anti-FLAG immunoprecipitates were analyzed with anti-HA antibodies for HOIL-1 (top and middle panels) or with anti-FLAG for Bach1. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel. WT, wild type; IB: HA, immunoblotting with anti-HA antibody; IP: FLAG, immunoprecipitation with anti-FLAG antibody.
Bach1 is a substrate of HOIL-1 in vivo.
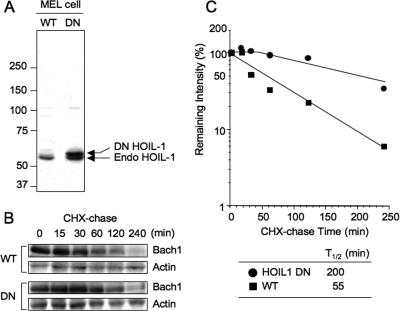
To examine whether HOIL-1 is the E3 ligase that facilitates the degradation of Bach1 in vivo, we expressed stably in MEL cells the dominant-negative form of FLAG/HA-HOIL-1 that carried two point mutations (C240 and 243S) in the RING finger domain and was shown previously to sequester its substrate IRP2 in cells (42). Because Bach1 bound to the N-terminal region of HOIL-1 (Fig. 9), we anticipated that this DN form would sequester Bach1 as well. Compared to endogenous HOIL-1, DN HOIL-1 was expressed at two- to threefold-higher levels (Fig. 10A). To examine the kinetics of Bach1 degradation, we treated the control and DN HOIL-1-expressing cells with cycloheximide. As shown in Fig. 10B and C, endogenous Bach1 in MEL cells underwent degradation under normal culture conditions with a half-life of approximately 55 min. This relatively short half-life may be due to degradation of Bach1 driven by endogenous levels of heme as in MEF cells (Fig. 1E). This basal-level degradation of Bach1 was delayed substantially by expressing DN HOIL-1 (Fig. 10A and B), suggesting that HOIL-1 was indeed involved in the basal, heme-regulated degradation of Bach1 in MEL cells.
FIG. 10.
HOIL-1-mediated degradation of Bach1 in vivo. (A) Expression of HOIL-1 in wild-type (WT) and DN HOIL-1-expressing MEL cells were examined using anti-HOIL-1 antibody. The arrows indicate the positions of endogenous (Endo) and DN HOIL-1. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel. (B) Control (wild type [WT]) and DN HOIL-1-expressing cells were cultured in the presence of 10 μg/ml cycloheximide (CHX) for 0 to 240 min. Total cell extracts were analyzed with antibodies against Bach1 and actin. (C) Determination of the degradation half-life of Bach1. The band intensities in panel A were measured by densitometric analysis. The relative mean values (Bach1/Actin) against CHX chase time from two independent experiments are shown; the average values are shown below the plot.
DISCUSSION
Our results provide strong evidence that heme regulates polyubiquitination and subsequent proteasome-dependent degradation of Bach1, thus extending the role of heme as a signaling molecule. Heme is a critical prosthetic group for diverse life forms, and it is synthesized and/or present in virtually every cell. The cellular heme levels can change due to the balance of its synthesis and degradation. Heme can also be taken up from outside of the cell via specific transport systems upon disease conditions, such as hemolysis (31, 38). Taken together with our previous observations (21, 33, 34), our findings suggest that heme inhibits the activity of Bach1 through multiple mechanisms, including inhibition of DNA binding, induction of nuclear export, and degradation to achieve derepression of target genes.
We found that endogenous levels of heme regulated the turnover of Bach1 in vivo (Fig. 1E). Although the physiological relevance of the heme-regulated degradation of Bach1 remains to be evaluated, we can envisage two meanings for the heme-induced proteolytic dismantling of Bach1. First, it allows for the timely inactivation of a Bach1 repressor complex on a target enhancer by reducing the overall levels of Bach1 and/or by promoting in situ ubiquitination of DNA-bound Bach1. It should also preclude the subsequent assembly of a Bach1 repressor complex on the vacant enhancer after any inadvertent reactivation of this critical regulator. Second, because Bach1 and its competitors, such as Nrf2, share small Maf proteins as their obligate heterodimer partners (13, 23), reduced levels of Bach1 are thus expected to shift the equilibrium toward formation of the activating Nrf2/small Maf heterodimers. The level of Nrf2 in a cell is kept low under normal conditions by a proteasome-dependent degradation system in which Keap1, a substrate recognition adaptor protein for Cullin 3-based E3 ligase, directs the polyubiquitination of Nrf2 (8, 14, 15, 47). The fact that both the repressor and activator are regulated by distinct polyubiquitination systems seems very important because the regulated protein stability can effectively widen the range of protein levels in vivo. Such an increased range of regulatory proteins has been shown to be associated with a robust operation of the genetic circuits (4). Furthermore, because degradation of Bach1 and Nrf2 is regulated by distinct mechanisms, combinations of their upstream regulatory signals (e.g., heme for Bach1 and thiol-reactive electrophiles for Nrf2) can thus widen the ratio between the two protein concentrations in vivo. When Bach1 is degraded in a timely manner, target genes are expected to be expressed at higher levels due to a higher activator/repressor ratio. When Bach1 persists, Nrf2 may be inefficient in target gene activation. The combination of Bach1 and Nrf2 can thus make their target gene circuits more robust and stable.
When combined with other mechanisms, such as heme-induced nuclear export, the heme-induced degradation of Bach1 may help to transduce heme levels into target gene expression precisely in magnitude and/or duration. It should be noted that the heme-binding affinity of Bach1 is relatively weak in comparison to proteins such as globins that utilize heme as a prosthetic group (21). Although the relatively low affinity conforms to the regulatory role of heme, such a weak interaction can potentially lead to a leaky regulation. We surmise that the three modes of regulation by heme (i.e., inhibition of DNA binding, nuclear export, and degradation) show a cooperative effect at a system level, thus achieving a sensitive monitoring of heme levels within a narrow fluctuation range. Consistent with this idea, we observe that endogenous levels of heme substantially affected the amount of Bach1 in MEFs (Fig. 1E). As such, Bach1 may be able to transduce a modest change in the heme levels under physiological as well as pathological conditions into gene expression.
We found that HOIL-1 can recognize Bach1, functioning as an E3 ligase in vitro and in vivo. This finding extends the role of HOIL-1 beyond IRP2 regulation. The observed biochemical function of HOIL-1 as an E3 ligase of Bach1 can be integrated nicely in the control of iron homeostasis. When heme is present in excessive amounts, heme inactivates Bach1 partly through HOIL-1-mediated ubiquitination and degradation, thus resulting in HO-1 induction. By the same token, HOIL-1-mediated degradation of IRP2 results in the expression of ferritin (42). HO-1 degrades heme, releasing precious but hazardous iron, which is then chelated by ferritin. As a result, HOIL-1 may regulate both liberation of free iron and its storage by targeting two hitherto unrelated proteins, Bach1 and IRP2. Because free iron can generate reactive oxygen species, the coordination of HO-1 and ferritin expression seems very reasonable. HOIL-1 recognizes IRP2 when IRP2 binds heme through its HRM (11). The oxidation of IRP2 catalyzed by heme appears to play a critical role in its recognition by HOIL-1 (42). Because Bach1 contains multiple HRMs (21), a mechanism similar to IRP2 may thus be involved in the ubiquitination of Bach1. This hypothesis is supported by the results of in vitro ubiquitination assay in which the CP motifs were found to play a critical role (Fig. 7). However, ZnPP induced degradation of Bach1 in cells even though it does not activate molecular oxygen (Fig. 2C). At this moment, we hypothesize two possibilities: namely, the oxidation of Bach1 is not involved in the recognition by HOIL-1, or it is actually involved for the effect of heme but not for ZnPP. ZnPP may induce the degradation of Bach1 through an alternative pathway of ubiquitination in vivo which is independent of HOIL-1 and can be regulated by not only heme but also ZnPP. While we showed that the DN HOIL-1 inhibited the Bach1 degradation in MEL cells (Fig. 10), this observation should be interpreted carefully. The DN HOIL-1 could inhibit other E3 ligases as well. A detailed comparative analysis of Bach1 and IRP2 will thus greatly help to address this issue.
Many genes in higher eukaryotes are regulated by a competitive interplay among repressors and activators as typified by the Myc-Mad-Max (1), E2F (41), and AP-1 (29) systems. In such systems involving both activators and repressors, not only the activation of activators but also the inactivation of repressors may be important to achieve the induction of target genes. The multilayered regulation of Bach1 activity by heme suggests this is indeed the case. Further investigations of such repressors will thus be important for obtaining a better understanding of how gene circuits actually operate in vivo.
Acknowledgments
We thank K. Furuyama and A. Kobayashi (Tohoku University) for help in determining heme levels and reagents, respectively. We also thank M. Yoshida (RIKEN) for providing anti-lamin B antibody and for various suggestions, N. Hattori (Juntendo University) for the Parkin plasmids, D. Bohmann (University of Rochester) for the HA-Ub plasmid, J. Sun and S. Tashiro (Hiroshima University) for help in the initial phase of the project, M. Ikeda-Saito (Tohoku University) for comments on the manuscript, and Ryoko Tsuneishi and Ayako Igarashi for administrative help.
This work was supported by grants-in-aid from the Ministry of Education, Science, Sport, and Culture of Japan and grants from the Uehara Foundation and Yamanouchi Foundation for Research on Metabolic Disorders.
Footnotes
Published ahead of print on 6 August 2007.
REFERENCES
- 1.Baudino, T. A., and J. L. Cleveland. 2001. The Max network gone Mad. Mol. Cell. Biol. 21:691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blough, N. V., H. Zemel, B. M. Hoffman, T. L. K. Lee, and Q. H. Gibson. 1980. Kinetics of carbon monoxide binding to manganese, zinc and cobalt hemoglobins. J. Am. Chem. Soc. 102:5683-5685. [Google Scholar]
- 3.Brand, M., J. A. Ranish, N. T. Kummer, J. Hamilton, K. Igarashi, C. Francastel, T. H. Chi, G. R. Crabtree, R. Aebersold, and M. Groudine. 2004. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat. Struct. Mol. Biol. 11:73-80. [DOI] [PubMed] [Google Scholar]
- 4.Buchler, N. E., U. Gerland, and T. Hwa. 2005. Nonlinear protein degradation and the function of genetic circuits. Proc. Natl. Acad. Sci. USA 102:9559-9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J. J., M. S. Throop, L. Gehrke, I. Kuo, J. K. Pal, M. Brodsky, and I. M. London. 1991. Cloning of the cDNA of the heme-regulated eukaryotic initiation factor 2 alpha (eIF-2 alpha) kinase of rabbit reticulocytes: homology to yeast GCN2 protein kinase and human double-stranded-RNA-dependent eIF-2 alpha kinase. Proc. Natl. Acad. Sci. USA 88:7729-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crosby, J. S., P. J. Chefalo, I. Yeh, S. Ying, I. M. London, P. Leboulch, and J. J. Chen. 2000. Regulation of hemoglobin synthesis and proliferation of differentiating erythroid cells by heme-regulated eIF-2alpha kinase. Blood 96:3241-3248. [PubMed] [Google Scholar]
- 7.Dioum, E. M., J. Rutter, J. R. Tuckerman, G. Gonzalez, M. Gilles-Gonzalez, and S. L. McKnight. 2002. NPAS2: a gas-responsive transcription factor. Science 298:2385-2387. [DOI] [PubMed] [Google Scholar]
- 8.Furukawa, M., and Y. Xiong. 2005. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 25:162-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igarashi, K., H. Hoshino, A. Muto, N. Suwabe, S. Nishikawa, H. Nakauchi, and M. Yamamoto. 1998. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for β-globin locus control region complex. J. Biol. Chem. 273:11783-11790. [DOI] [PubMed] [Google Scholar]
- 10.Igarashi, K., and J. Sun. 2006. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid. Redox Signal. 8:107-118. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa, H., M. Kato, H. Hori, K. Ishimori, T. Kirisako, F. Tokunaga, and K. Iwai. 2005. Involvement of heme regulatory motif in heme-mediated ubiquitination and degradation of IRP2. Mol. Cell 19:171-181. [DOI] [PubMed] [Google Scholar]
- 12.Iwai, K., S. K. Drake, N. B. Wehr, A. M. Weissman, T. LaVaute, N. Minato, R. D. Klausner, R. L. Levine, and T. Rouault. 1998. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc. Natl. Acad. Sci. USA 95:4924-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katsuoka, F., H. Motohashi, T. Ishii, H. Aburatani, J. D. Engel, and M. Yamamoto. 2005. Genetic evidence that small Maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol. Cell. Biol. 25:8044-8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi, A., M. I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi, and M. Yamamoto. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi, A., M. I. Kang, Y. Watai, K. I. Tong, T. Shibata, K. Uchida, and M. Yamamoto. 2006. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell. Biol. 26:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lathrop, J. T., and M. P. Timko. 1993. Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science 259:522-525. [DOI] [PubMed] [Google Scholar]
- 17.McMahon, M., K. Itoh, M. Yamamoto, and J. D. Hayes. 2003. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 278:21592-21600. [DOI] [PubMed] [Google Scholar]
- 18.Muto, A., S. Tashiro, H. Tsuchiya, A. Kume, M. Kanno, E. Ito, M. Yamamoto, and K. Igarashi. 2002. Activation of Maf/AP-1 repressor Bach2 promotes apoptosis and its interaction with PML nuclear bodies. J. Biol. Chem. 277:20724-20733. [DOI] [PubMed] [Google Scholar]
- 19.Nagai, T., K. Igarashi, J. Akasaka, K. Furuyama, H. Fujita, N. Hayashi, M. Yamamoto, and S. Sassa. 1998. Regulation of NF-E2 activity in erythroleukemia cell differentiation. J. Biol. Chem. 273:5358-5365. [DOI] [PubMed] [Google Scholar]
- 20.Nakatani, Y., and V. Ogryzko. 2003. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 370:430-444. [DOI] [PubMed] [Google Scholar]
- 21.Ogawa, K., J. Sun, S. Taketani, O. Nakajima, C. Nishitani, S. Sassa, N. Hayashi, M. Yamamoto, S. Shibahara, H. Fujita, and K. Igarashi. 2001. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 20:2835-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Omura, S., H. Suzuki, M. Toyofuku, R. Ozono, N. Kohno, and K. Igarashi. 2005. Effect of genetic ablation of Bach1 upon smooth muscle cell proliferation and atherosclerosis after cuff injury. Genes Cells 10:277-285. [DOI] [PubMed] [Google Scholar]
- 23.Oyake, T., K. Itoh, H. Motohashi, N. Hayashi, H. Hoshino, M. Nishizawa, M. Yamamoto, and K. Igarashi. 1996. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 16:6083-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi, Z., I. Hamza, and M. R. O'Brian. 1999. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. USA 96:13056-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi, Z., and M. R. O'Brian. 2002. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol. Cell 9:155-162. [DOI] [PubMed] [Google Scholar]
- 26.Rafie-Kolpin, M., P. J. Chefalo, Z. Hussain, J. Hahn, S. Uma, R. L. Matts, and J. J. Chen. 2000. Two heme-binding domains of heme-regulated eukaryotic initiation factor-2alpha kinase. N terminus and kinase insertion. J. Biol. Chem. 275:5171-5178. [DOI] [PubMed] [Google Scholar]
- 27.Sassa, S. 2004. Why heme needs to be degraded to iron, biliverdin IXalpha, and carbon monoxide? Antioxid. Redox Signal. 6:819-824. [DOI] [PubMed] [Google Scholar]
- 28.Sassa, S., and A. Kappas. 1977. Induction of aminolevulinate synthase and porphyrins in cultured liver cells maintained in chemically defined medium. Permissive effects of hormones on induction process. J. Biol. Chem. 252:2428-2436. [PubMed] [Google Scholar]
- 29.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 30.Sheftel, A. D., S. F. Kim, and P. Ponka. 2007. Non-heme induction of heme oxygenase-1 does not alter cellular iron metabolism. J. Biol. Chem. 282:10480-10486. [DOI] [PubMed] [Google Scholar]
- 31.Shibahara, S. 2003. The heme oxygenase dilemma in cellular homeostasis: new insights for the feedback regulation of heme catabolism. Tohoku J. Exp. Med. 200:167-186. [DOI] [PubMed] [Google Scholar]
- 32.Sun, J., M. Brand, Y. Zenke, S. Tashiro, M. Groudine, and K. Igarashi. 2004. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc. Natl. Acad. Sci. USA 101:1461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, J., H. Hoshino, K. Takaku, O. Nakajima, A. Muto, H. Suzuki, S. Tashiro, S. Takahashi, S. Shibahara, J. Alam, M. M. Taketo, M. Yamamoto, and K. Igarashi. 2002. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 21:5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, H., S. Tashiro, S. Hira, J. Sun, C. Yamazaki, Y. Zenke, M. Ikeda-Saito, M. Yoshida, and K. Igarashi. 2004. Heme regulates gene expression by triggering Crm1-dependent nuclear export of Bach1. EMBO J. 23:2544-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki, H., S. Tashiro, J. Sun, H. Doi, S. Satomi, and K. Igarashi. 2003. Cadmium induces nuclear export of Bach1, a transcriptional repressor of heme oxygenase-1 gene. J. Biol. Chem. 278:49246-49253. [DOI] [PubMed] [Google Scholar]
- 36.Tahara, T., J. Sun, K. Igarashi, and S. Taketani. 2004. Heme-dependent up-regulation of the alpha-globin gene expression by transcriptional repressor Bach1 in erythroid cells. Biochem. Biophys. Res. Commun. 324:77-85. [DOI] [PubMed] [Google Scholar]
- 37.Tahara, T., J. Sun, K. Nakanishi, M. Yamamoto, H. Mori, T. Saito, H. Fujita, K. Igarashi, and S. Taketani. 2004. Heme positively regulates the expression of beta-globin at the locus control region via the transcriptional factor Bach1 in erythroid cells. J. Biol. Chem. 279:5480-5487. [DOI] [PubMed] [Google Scholar]
- 38.Taketani, S. 2005. Acquisition, mobilization and utilization of cellular iron and heme: endless findings and growing evidence of tight regulation. Tohoku J. Exp. Med. 205:297-318. [DOI] [PubMed] [Google Scholar]
- 39.Tang, X. D., R. Xu, M. F. Reynolds, M. L. Garcia, S. H. Heinemann, and T. Hoshi. 2003. Haem can bind to and inhibit mammalian calcium-dependent Slo1 BK channels. Nature 425:531-535. [DOI] [PubMed] [Google Scholar]
- 40.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 41.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 42.Yamanaka, K., H. Ishikawa, Y. Megumi, F. Tokunaga, M. Kanie, T. Rouault, I. Morishima, N. Minato, K. Ishimori, and K. Iwai. 2003. Identification of the ubiquitin-protein ligase that recognizes oxidized IRP2. Nat. Cell Biol. 5:336-340. [DOI] [PubMed] [Google Scholar]
- 43.Yamasaki, C., S. Tashiro, Y. Nishito, T. Sueda, and K. Igarashi. 2005. Dynamic cytoplasmic anchoring of the transcription factor Bach1 by intracellular hyaluronic acid binding protein IHABP. J. Biochem. (Tokyo) 137:287-296. [DOI] [PubMed] [Google Scholar]
- 44.Yang, J. M., K. Ishimori, and M. R. O'Brian. 2005. Two heme binding sites are involved in the regulated degradation of the bacterial iron response regulator (Irr) protein. J. Biol. Chem. 280:7671-7676. [DOI] [PubMed] [Google Scholar]
- 45.Yano, Y., R. Ozono, Y. Oishi, M. Kambe, M. Yoshizumi, T. Ishida, S. Omura, T. Oshima, and K. Igarashi. 2006. Genetic ablation of the transcription repressor Bach1 leads to myocardial protection against ischemia/reperfusion in mice. Genes Cells 11:791-803. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, D. D., and M. Hannink. 2003. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23:8137-8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, D. D., S. C. Lo, J. V. Cross, D. J. Templeton, and M. Hannink. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24:10941-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, L., and L. Guarente. 1995. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 14:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Y., K. Furuyama, K. Kaneko, Y. Ding, K. Ogawa, M. Yoshizawa, M. Kawamura, K. Takeda, T. Yoshida, and S. Shibahara. 2006. Hypoxia reduces the expression of heme oxygenase-2 in various types of human cell lines. A possible strategy for the maintenance of intracellular heme level. FEBS Lett. 273:3136-3147. [DOI] [PubMed] [Google Scholar]