Abstract
Vanillin (VAN) and cinnamaldehyde (CIN) are dietary antimutagens that effectively inhibit both induced and spontaneous mutations. We have shown previously that VAN and CIN reduced the spontaneous mutant frequency in Salmonella TA104 (hisG428, rfa, ΔuvrB, pKM101) by approximately 50% and that both compounds significantly reduced mutations at GC sites but not at AT sites. Previous studies have suggested that VAN and CIN may reduce mutations in bacterial model systems by modulating DNA repair pathways, particularly by enhancing recombinational repair. To further explore the basis for inhibition of spontaneous mutation by VAN and CIN, we have determined the effects of these compounds on survival and mutant frequency in five Escherichia coli strains derived from the wild-type strain NR9102 with different DNA repair backgrounds. At nontoxic doses, both VAN and CIN significantly reduced mutant frequency in the wild-type strain NR9102, in the nucleotide excision repair-deficient strain NR11634 (uvrB), and in the recombination-proficient but SOS-deficient strain NR11475 (recA430). In contrast, in the recombination-deficient and SOS-deficient strain NR11317 (recA56), both VAN and CIN not only failed to inhibit the spontaneous mutant frequency but actually increased the mutant frequency. In the mismatch repair-defective strain NR9319 (mutL), only CIN was antimutagenic. Our results show that the antimutagenicity of VAN and CIN against spontaneous mutation required the RecA recombination function but was independent of the SOS and nucleotide excision repair pathways. Thus, we propose the counterintuitive notion that these antimutagens actually produce a type of DNA damage that elicits recombinational repair (but not mismatch, SOS, or nucleotide excision repair), which then repairs not only the damage induced by VAN and CIN but also other DNA damage—resulting in an antimutagenic effect on spontaneous mutation.
Keywords: Antimutagens, DNA repair, Recombination, Vanillin, Cinnamaldehyde
1. Introduction
Vanillin (VAN) and cinnamaldehyde (CIN) are natural food flavorings that inhibit mutations induced by chemical and physical mutagens in bacterial test systems [1–5] and mammalian cell culture [6,7]. Both VAN and CIN inhibited micronuclei formation in V79 cells treated with heterocyclic amines [8] and reduced chromosome aberrations in CHO cells exposed to UV-light and X-rays [9]. In addition, VAN and CIN reduced the frequency of spontaneous mutations in Salmonella TA98 and TA104 [10–12].
VAN and CIN have been described as bioantimutagens, a term coined by Kada to describe agents that inhibit mutagenesis by modulating cellular processes of DNA replication and repair [13]. In addition to antimutagenic effects observed in WP2s strains of E. coli, CIN also increased survival of cells exposed to 4-nitroquinoline-1-oxide. This survival-enhancing effect was dependent on recA gene function [1]. Likewise, co-treatment with VAN increased survival in UV-exposed cells except in recA strains [4]. In addition, recombination between plasmids was enhanced with VAN treatment [4]. In Drosophila, VAN modestly inhibited mutations induced by mitomycin C but dramatically increased recombination in mitomycin C-treated flies [14]. Co-treatment with VAN and either ethyl methanesulfonate (EMS) or bleomycin also produced a synergistic effect on recombination in Drosophila [15]. Observations of the recombinogenic effects of VAN, together with the requirement of recA gene function for the survival-enhancing effects of VAN and CIN, have lead to the hypothesis that VAN and CIN may inhibit mutagenesis by increasing repair of DNA damage through the recombinational repair pathway [4].
In previous work examining the effects of VAN and CIN on spontaneous mutations in Salmonella TA104, we showed that both compounds significantly reduced mutations at GC sites but not AT sites and that the antimutagenic effect was dependent on the presence of the pKM101 plasmid in homologues of TA104 [12]. To further elucidate the role of DNA repair pathways on antimutagenesis by VAN and CIN, we have investigated the effects of these compounds on spontaneous mutation in five strains of E. coli that have defects in different DNA repair pathways, including nucleotide excision repair (NR11634), mismatch repair (NR9139), and recombinational repair (NR11317). To investigate whether VAN or CIN reduces spontaneous mutations by inhibiting the error-prone SOS pathway, we tested these compounds in strains that were deficient in SOS response but retained recombination function (NR11475) or were deficient in both SOS response and recombination (NR11317).
2. Materials and methods
2.1. Chemicals and media
Vanillin (VAN) and cinnamaldehyde (CIN) (Sigma–Aldrich, Milwaukee, WI) were dissolved in dimethyl sulfoxide (DMSO, Burdick and Jackson, Muskegon, MI) and evaluated for survival and antimutagenesis using a modified version of the standard plate incorporation assay [16] in the absence of S-9 mix.
Media used were Vogel–Bonner Medium E (VBME) agar plates containing 0.2% glucose and phenyl-β-D-galactopyranoside (P-gal) agar plates (1 × VB salts, 750 μg/ml P-gal, and 1 μg/ml thiamine). Overnight cultures of E. coli strains were grown in LB medium.
2.2. Bacterial strains
The E. coli strains used in this work, listed in Table 1, are derived from parental strain NR9102 [ara, thi, Δpro-lac)X111, F’prolac (F′128-27)], which contains an F′prolac episome carrying two silent base substitution mutations in the lacI gene [17]. Mutations in the lacI gene resulting in constitutive expression of the lac genes can be readily detected on media containing phenyl-β-D-galactopyranoside (P-gal) as a sole source of carbon [18]. NR9139 (mutL::Tn5) lacks mismatch repair (MMR) function due to a mutation in the mutL gene [19].
Table 1.
E. coli strains used in this study
| Strain | Relevant genotypea | Phenotypeb | Reference |
|---|---|---|---|
| NR 9102 | wt | Wild type | [17] |
| NR11634 | uvrB5 | NER− | This work |
| NR11475 | recA430 | SOS−, recombination+ | This work |
| NR11317 | recA56, srl::Tn10 | SOS−, recombination− | This work |
| NR9319 | mutL::Tn5 | MMR− | [19] |
All strains are also ara, thi, Δ(pro-lac), F′prolac (F′128-27); see Section 2.
NER: nucleotide excision repair; MMR: mismatch repair.
The nucleotide repair-deficient (NER−) strain NR11634 harbors a point mutation in the uvrB gene (uvrB5) rather than the large deletion in the region of the uvrB and bio genes found in Salmonella TA98, TA100, and TA104 [20]. NR11634 is a gal+ revertant of NR10234 (galK2), which has been described [21], and was tested for UV sensitivity.
NR11475, which carries the recA430 allele, is defective in SOS activity due to an inability to cleave the UmuD protein, but retains homologous recombination functions [22]. NR11475 (recA430) was created in a three-step procedure. First, strain KA796 [ara, thi, Δ(pro-lac)] [23] was transduced to become ΔrecA, srl::Tn10 using strain PF1711 as a P1 donor, selecting for tetracycline resistance and testing for UV-sensitivity, yielding NR11476. This strain was then transduced to become recA430, srl+ using strain PF1712 as a P1 donor, selecting for growth on MM-sorbitol medium and scoring for increased UV resistance. PF1711 and PF1712 were kind gifts from P. Foster (Indiana University). Finally, F′128-27 [17] was introduced by conjugation, selecting for proline-proficiency, yielding NF11475.
NR11317 (recA56) lacks both SOS and recombination functions. NR11317 (recA56) was, like NR11475, constructed from KA796. The recA56, srl::Tn10 combination was introduced by P1 transduction using strain UTH2 as P1 donor [24], followed by introduction of F′128-27 as above.
2.3. Antimutagenesis and survival assays
For evaluation of survival, overnight cultures grown in LB medium, diluted 106-fold to yield approximately 200 cells/100 μl, were added to 2 ml of top agar along with varying concentrations of VAN or CIN and poured onto VBME agar plates. For evaluation of antimutagenesis, 100 μl of undiluted overnight cultures (~108 cells) were added to 2 ml of molten top agar along with appropriate concentrations of VAN or CIN, vortexed, and poured onto P-gal agar plates. Both VBME and P-gal plates were incubated for 72 h at 37 °C. Survival and antimutagenesis with VAN and CIN were tested in two independent cultures for each strain, with each dose plated in triplicate. Thus, for each strain, effects on antimutagene-sis and survival were monitored using a total of six plates at each concentration of VAN and CIN. Mutant and surviving colonies were counted using an automatic plate counter (AccuCount 1000, Biologics, Manassas, VA). Effects of VAN and CIN on mutant colonies were reported as mutant frequencies (MF), i.e., the number of mutant colonies/108 surviving colonies.
2.4. Statistical analyses
We conducted a separate analysis for each strain of E. coli with each compound, 10 analyses in all. Each analysis used data from the duplicate experimental runs involving the two independent overnight cultures, and each run included a survival arm and a mutant-yield arm. Counts of mutants/plate or survivors/plate for each analysis were modeled using generalized linear mixed models with Poisson error distribution and its canonical link function (logarithm). Thus, our analysis was a mixed-effects analysis of variance for Poisson-distributed data. The mean response for each plate was modeled as depending on the dose of the compound and the arm. We needed a mixed-effects analysis of variance, rather than a simpler fixed-effects one, to accommodate multiple sources of extra-Poisson variation, such as the run-to-run variation in MF that several lacI strains derived from KA796 had exhibited previously [25]. Our mixed-effects analysis allowed for extra-Poisson variation due to differing response levels between independent cultures within each arm (random effect for run × arm), between the precise shape of the dose trajectory in a given arm between independent cultures (random effect for run × arm × dose), and between replicate plates (residual). Thus, our analysis explicitly accounted for relevant sources of variation when assessing MF associated with increasing doses of these compounds.
The statistical model is illustrated in Fig. 1A for VAN treatment in NR9102 (wild type). In this case, survival remains unaffected for all doses. On the other hand, the mutant frequency clearly decreases at higher doses of VAN. As a general rule, if a decrease in mutants/108 survivors (mutant-yield arm) begins at doses lower than those at which there is any decrease in the survivors/plate (survival arm), then we conclude that the compound is antimutagenic. In contrast, if the trajectories in both arms simply parallel each other, we conclude that the compound is not antimutagenic in that strain. Thus, a statistical test for an antimutagenic effect in the models is a test for an arm × dose interaction.
Fig. 1.
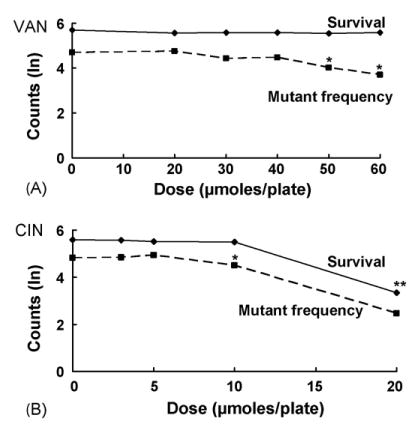
Effects on survival and lacI mutant frequency of VAN (A) and CIN (B) in E. coli NR9102 (wild type). Single asterisks indicate VAN or CIN dose levels at which significant antimutagenic effects were observed (p < 0.001 in all indicated cases). Double asterisks indicate a toxic dose. Plotted are the natural log(ln) of the geometric means of surviving colonies per plate (n = 6) and the ln of the geometric means of the mutant frequency.
In addition to a single overall test for arm × dose interaction in each analysis, the overall statistic was partitioned into a series of one-degree-of-freedom interaction statistics to find the lowest dose at which the arm × dose interaction was evident. The first one-degree-of-freedom statistic tested whether the change in response between the second dose level and the first differed across arms; the second statistic tested whether the change in response between the third dose level and the average of the lowest two dose levels differed across arms, and so on. The final test in the series examined whether the change in response between the highest dose level and the average of all the lower dose levels differed between arms.
The statistical contrasts involved in constructing this series of tests are orthogonal so that the one-degree-of-freedom tests partition the overall test of interaction into independent aspects. Likewise, contrasts were constructed within either the survival arm or the antimutagenesis arm to infer the lowest dose at which the mean response dropped off significantly and to estimate the magnitude of the decrease as a proportion of the baseline response (average response among the lower doses). All statistical analyses were carried out using SAS software (SAS Institute, Cary, NC). In particular, we fitted the generalized linear mixed models with the GLIMMIX macro <http://support.sas.com/ctx/samples/index.jsp?sid=536>.
3. Results
To investigate the antimutagenic effects of VAN and CIN in E. coli, we used the lacI forward-mutation system, in which the mutational target is the lacI gene, which encodes the repressor of the lac operon. lacI mutants, lacking a functional repressor, can be detected based on their constitutive expression of the lac operon. Because the sugar analog P-gal is a substrate for the lac enzymes but not an inducer of the operon, such lacI mutants can be isolated and scored by their ability to form colonies on plates containing P-gal as the sole carbon source [18]. Because the entire lacI gene is the target, a large variety of mutations at many sites throughout the gene can be detected, making it a suitable target for investigation of mutational processes in E. coli, including spontaneous mutagenesis [17,25,26].
In the present case, we examined the effect of VAN and CIN on the frequency of spontaneous lacI mutants using a plate-incorporation assay. We plated aliquots of overnight cultures of E. coli on P-gal plates using soft top agar containing various concentrations of the two chemicals and determined the effect of VAN or CIN on the number of lacI mutants. This method is similar to the plate-incorporation assays used in the Salmonella (Ames) assay, which we used previously to detect antimutagenic effects of VAN and CIN [12]. In parallel, we also determined the effect of the chemicals on the survival of the strains (see Section 2). We showed in our study in Salmonella [12] that VAN and CIN did not cause growth inhibition. Although we did not reassess this issue in E. coli, we have assumed that reductions in survival by VAN and CIN observed in the present study were not due to growth inhibition.
The results are presented in Figs. 1–5, and Tables 2 and 3 provide summaries of all the data, including the highest non-toxic dose for each of the strains. We note that the data presented in Figs. 1–5 are plotted on a natural log scale. In Tables 2 and 3, the colony counts for survival, mutant yield, and mutant frequency for the highest nontoxic doses are compared to averages of all lower doses in each strain to reflect the statistical analysis. Confidence intervals for mutant frequency values are included. The numbers of colonies on selective and nonselective plates are presented in Appendices A and B.
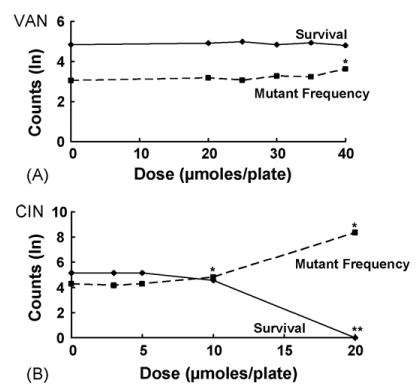
Fig. 5.
Effects on survival and mutant frequency of VAN (A) and CIN (B) in MMR-deficient E. coli NR9319 (mutL); see legend to Fig. 1 for details. Asterisk indicates a significant mutagenic effect for VAN (p < 0.001 at 40 and 60 μmol/plate) and a significant antimutagenic effect for CIN (p < 0.05 for CIN at 10 μmol/plate).
Table 2.
Survival and mutant frequency of E. coli strains plated in the presence of VAN
| Survival
|
Mutant yield
|
Mutant frequencyd |
||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Dosea (μmol/plate) | Colony countb | %c | Colony countb | %c | MF | %c (95% CI)e | % Reduction (〈% increase〉) |
| NR 9102 (wt) | 0–50 | 271.86 | 100 | 239.94 | 100 | 88.25 | 100 | |
| 60 | 266.66 | 98 | 114.18 | 48 | 42.81 | 49 (37, 64) | 51 | |
| NR11634 (uvrB) | 0–35 | 227.35 | 100 | 443.77 | 100 | 195.19 | 100 | |
| 40 | 237.41 | 104 | 354.53 | 80 | 149.47 | 77 (66, 89) | 23 | |
| NR11475 (recA430) | 0–35 | 323.24 | 100 | 582.77 | 100 | 180.29 | 100 | |
| 40 | 332.42 | 103 | 390.25 | 67 | 117.40 | 65 (49, 86) | 35 | |
| NR11317 (recA56) | 0–35 | 136.29 | 100 | 32.43 | 100 | 23.79 | 100 | |
| 40 | 121.35 | 89 | 46.42 | 143 | 38.25 | 161 (124, 209) | 〈61〉 | |
| NR9319 (mutL) | 0–30 | 573.07 | 100 | 4855.17 | 100 | 847.22 | 100 | |
| 40 | 565.72 | 99 | 5161.07 | 106 | 912.30 | 108 (84, 138) | 〈8〉 | |
Control range (average of all lower doses) and highest nontoxic dose.
Geometric mean colony count estimated from fitted Poisson mixed model.
Percent of control value.
Mutants/108 survivors.
95% confidence intervals for mutant frequency values.
Table 3.
Survival and mutant frequency of E. coli strains plated in the presence of CIN
| Survival
|
Mutant yield
|
Mutant frequencyd |
||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Dosea (μmol/plate) | Colony countb | %c | Colony countb | %c | MF | %c (95% CI)e | % Reduction (〈% increase〉) |
| NR 9102 (wt) | 0–5 | 258.73 | 100 | 337.61 | 100 | 130.49 | 100 | |
| 10 | 241.29 | 93 | 219.55 | 65 | 90.99 | 70 (59, 82) | 30 | |
| NR11634 (uvrB) | 0–10 | 226.56 | 100 | 486.00 | 100 | 214.51 | 100 | |
| 20 | 178.54 | 79 | 15.69 | 3 | 8.79 | 4 (2, 6) | 96 | |
| NR11475 (recA430) | 0–5 | 305.24 | 100 | 668.28 | 100 | 218.93 | 100 | |
| 10 | 292.94 | 96 | 372.19 | 56 | 127.05 | 58 (41, 82) | 42 | |
| NR11317 (recA56) | 0–5 | 170.00 | 100 | 117.54 | 100 | 69.14 | 100 | |
| 10 | 105.28 | 62f | 117.64 | 100 | 111.75 | 162 (117, 223) | 〈62〉 | |
| NR9319 (mutL) | 0–5 | 580.51 | 100 | 5451.25 | 100 | 939.87 | 100 | |
| 10 | 551.86 | 95 | 3319.60 | 61 | 601.53 | 64 (41, 100) | 36 | |
Control range (average of all lower doses) and highest nontoxic dose.
Geometric mean colony count estimated from fitted Poisson mixed model.
Percent of control value.
Mutants/108 survivors.
95% confidence intervals for mutant frequency values.
At 10 μmol/plate, CIN was toxic in this strain.
Both VAN and CIN were antimutagenic against spontaneous mutations in the wild-type strain NR9102, significantly reducing the calculated MF by 51% and 30%, respectively, at the highest nontoxic concentrations tested (Fig. 1A, Table 2, Fig. 1B, Table 3, for p-values in all strains, see Figs. 1–4). Likewise, in the nucleotide excision repair-deficient (NER−) strain NR11634, both VAN and CIN effectively inhibited spontaneous mutations. At 40 μmol/plate, VAN reduced the MF by 23% without any cytotoxicity (Fig. 2A, Table 2), whereas CIN at 20 μmol/plate dramatically reduced the MF by 96% with 79% survival. Although this strain was somewhat sensitive to cell killing at 20 μmol/plate, survival in NR11634 was still considerably higher at this dose than in any of the other strains tested.
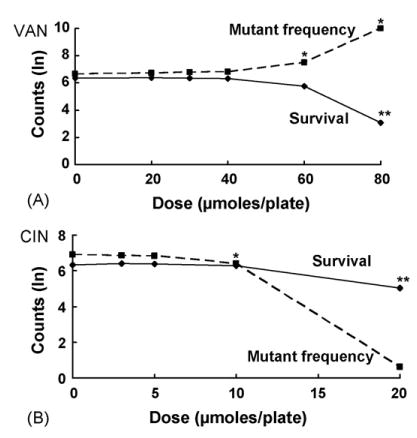
Fig. 4.
Effects on survival and mutant frequency of VAN (A) and CIN (B) in SOS-deficient and recombination-deficient E. coli NR11317 (recA56); see legend to Fig. 1 for details. Asterisk indicates a statistically significant mutagenic effect (p < 0.05 for CIN at 10 μmol/plate; p < 0.001 for CIN at 20 μmol/plate and VAN at 40 μmol/plate).
Fig. 2.
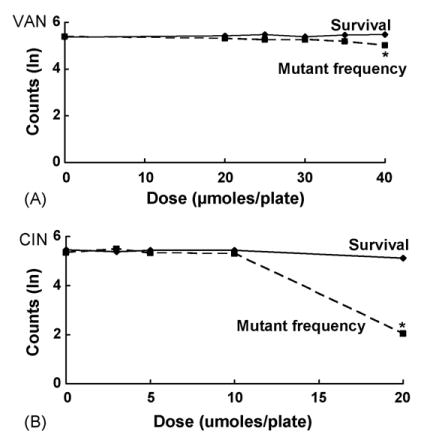
Effects on survival and mutant frequency of VAN (A) and CIN (B) in NER-deficient E. coli NR11634 (uvrB); see legend to Fig. 1 for details.
To examine the role of the SOS repair pathway in antimutagenesis by VAN and CIN, we first tested these chemicals in NR11475 (recA430), which is deficient in SOS activity but is recombination proficient [22]. Both VAN and CIN were antimutagenic in this strain, reducing the MF by 35 and 42%, respectively (Fig. 3, Tables 2 and 3). Interestingly CIN was a more effective antimutagen at the same doses in this strain than in the wild-type strain NR9102. At 10 μmol/plate, CIN decreased the MF by 30% in NR9102 compared to a 42% decrease in NR11475. As in the NER− strain NR11634, CIN at 20 μmol/plate had a dramatic effect on the MF in NR11475, reducing mutations by nearly 99%. However, survival was reduced by 40% at this dose, which we considered a toxic.
Fig. 3.
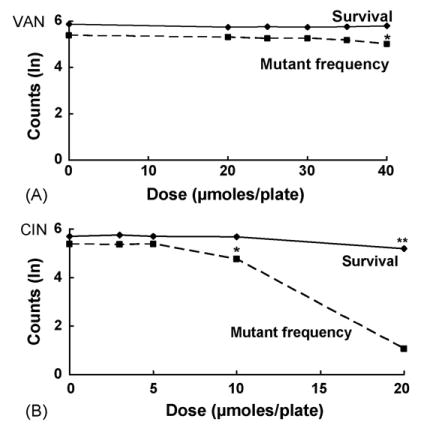
Effects on survival and mutant frequency of VAN (A) and CIN (B) in SOS-deficient E. coli NR11475 (recA430); see legend to Fig. 1 for details. Asterisk indicates a statistically significant antimutagenic effect (p < 0.001 for VAN at 40 μmol/plate; p < 0.05 for CIN at 10 μmol/plate).
In contrast, no antimutagenic effects were observed in the recA56 strain NR11317, which is defective in both SOS and recombination functions [24]. In fact, both chemicals were mutagenic in this strain, producing increases in the MF. At 40 μmol/plate, VAN increased the MF by 61%, and CIN at 10 μmol/plate increased the MF by 62% (Fig. 4, Tables 2 and 3). In addition, CIN was cytotoxic at this dose, reducing survival by 38%. Thus, when comparing the results in the recA430 and recA56 strains, it appears that the antimutagenic effects of VAN and CIN were dependent on the recA recombination function but independent of the recA SOS function.
We tested VAN and CIN in the mismatch-repair-(MMR-) defective strain NR9319 to examine whether MMR plays a role in the antimutagenic effects of these chemicals. Only CIN proved antimutagenic in this strain, reducing the MF by 36% at 10 μmol/plate (Fig. 5B, Table 3). In contrast, VAN at 40 μmol/plate was not significantly antimutagenic or mutagenic, increasing the MF by only 8% (Fig. 5A, Table 2). At higher VAN doses, however, MF in NR9139 increased dramatically, with a two-fold increase at 60 μmol/plate, and a 28-fold increase at 80 μmol/plate (Appendix A).
To examine whether the apparent antimutagenic effects of VAN and CIN might be due to differences in cytotoxicity on VBME versus P-gal plates, a reconstruction experiment was conducted with the wild-type strain NR9102. Briefly, several independently isolated lacI− mutants of NR9102 were purified on P-gal plates, grown in liquid culture for 16 h, and diluted 106-fold to yield approximately 200 cells/100 μl. Then 100 μl of this dilution were plated with varying concentrations of VAN or CIN on either P-gal or VBME plates, and the plates were incubated for 72 h at 37 °C. In parallel, cultures of NR9102 were plated with VAN or CIN on P-gal plates to reproduce the antimutagenic effects described above. Although there was a slightly reduced survival of pre-established lacI− mutants on P-gal versus VBME plates at 60 μmol of VAN/plate and 15 μmol of CIN/plate, this effect did not account for the clear reduction in the MF observed for NR9102 cultures (results not shown). Thus, the antimutagenic effect observed in NR9102, NR11634, and NR11475 was not likely due to differential cytotox-icity of VAN or CIN on selective P-gal plates versus the nonselective VBME plates.
4. Discussion
Both VAN and CIN were antimutagenic for spontaneous mutation in the wild-type strain NR9102. At the highest nontoxic doses tested, VAN produced a more dramatic effect than CIN, reducing the MF by 51%, compared to a 30% reduction by CIN (Tables 2 and 3). In the NER− strain NR11634, both VAN and CIN were antimutagenic. However, in this strain, the inhibitory effect of CIN was much more dramatic, reducing the MF by nearly 96% at 20 μmol/plate, a dose that was cytotoxic in all other strains tested. The basis for enhanced survival with CIN at this dose in the absence of NER is not known.
VAN and CIN had similar antimutagenic effects in NR11475 (reca430), which lacks SOS activity but retains recombination function. Both chemicals reduced the MF by approximately 40%. In the absence of recombination function in NR11317 (recA56), not only did VAN and CIN fail to inhibit spontaneous mutagenesis, both compounds were in fact mutagenic, increasing the MF by approximately 61 and 62%, respectively.
In the MMR− strain NR9319 (mutL), VAN and CIN had opposite effects. VAN was slightly mutagenic at 40 μmol/plate, and this effect was more pronounced at higher doses with a 28-fold increase in MF at 80 μmol/plate, due mainly to cytotoxicity (Appendix A). In contrast, CIN was antimutagenic in NR9319, reducing the MF even more effectively than in the wild-type strain. This suggests that the mechanism by which VAN and CIN reduce the spontaneous MF in wild-type strains differs with respect to the requirement of these compounds for mismatch repair.
Spontaneous mutations in E. coli occur at a low rate, ~10−10 mutations per base pair per generation [27]. Although a lower rate (~10−11 mutations per base pair per generation) is estimated for humans [27], spontaneous mutations may play a causal role in human diseases, e.g., cancer and birth defects [28]. Among the sources of spontaneous mutation are replication errors, which occur occasionally despite normal proofreading functions and mismatch correction, and various other types of unrepaired DNA damage, including depurination, deamination of cytosine and 5-methyl cytosine, and exposure to endogenous alkylating agents and reactive oxygen species [17]. Unrepaired single-strand lesions, including apurinic/apyrimidinic (AP) sites, 8-oxoguanine, thymine glycol, and 3-methyl adenine, can lead to collapse or stalling of a replication fork. The resulting nicks and gaps in DNA can lead to the formation of double strand breaks (DSBs), which are estimated to occur spontaneously in mammalian cells at a rate of 50 DSBs/cell/cell cycle or 1 DSB/108 base pairs/cell cycle [29]. In bacteria, repair of DNA damage, including DSBs, which results in stalled or collapsed replication forks, is mediated by the RecA protein in conjunction with either the RecBCD or RecFOR pathways [30].
Our results demonstrate that VAN and CIN are antimutagenic, even in the absence of NER or SOS activity. Interestingly, both compounds were mutagenic in the recombination-deficient strain NR11317, suggesting that VAN and CIN may induce some type of DNA damage that is repaired primarily by recombinational repair. Although the nature of the DNA damage induced by VAN and CIN in bacteria is unknown, studies in mammalian cells suggest several types of possible damage.
CIN may damage DNA indirectly by binding to intracellular thiol groups in proteins and to glutathione. Weibel et al. [31] showed that CIN forms protein conjugates by binding almost exclusively to thiol groups within cysteine residues. Ka et al. [32] demonstrated that treatment with CIN rapidly depleted intracellular glutathione and protein thiol levels in human HL-60 leukemia cells. At the same concentration, CIN also increased reactive oxygen species (ROS) levels and induced apoptosis in this cell line. The authors hypothesized that depletion of intracellular glutathione levels results in increased ROS levels that, in turn, alter the cellular redox status and mitochondrial membrane potential. The subsequent release of cytochrome c activates the pro-apoptotic proteins caspase-9 and caspase-3 and thereby induces apoptosis. This result is supported by similar work in V79 cells demonstrating glutathione depletion and oxidative damage after CIN exposure [33]. In addition to oxidant effects, CIN induced DNA damage in mammalian cells as measured by the comet assay [33,34]. We also found that CIN increased DNA migration in the comet assay in human HepG2 hepatocellular carcinoma and HCT116 colon cancer cell lines (unpublished data). The nature of this CIN-induced DNA damage is unknown.
In contrast to the possible oxidant damage induced by CIN, there is evidence that VAN has antioxidant activity. VAN effectively inhibited lipid peroxidation [35–37], scavenged free radicals [38,39], and reduced the number of DNA single-strand breaks induced by singlet oxygen [40]. In addition to these antioxidant effects, VAN has been shown to inhibit mutagenicity induced by a variety of chemical and physical agents [1–3,8,9]. However, VAN has been reported [41] to increase the Hprt mutant frequency in V79 cells exposed to EMS. Co-treatment with VAN also had a synergistic effect on recombination on MMC- and EMS-exposed Drosophila in the SMART assay [14,15]. O-vanillin, a compound related structurally to VAN, induced DNA damage as detected by the comet assay [42]. As with CIN, we found increases in DNA migration in HCT116 cells after 4-h exposures to nontoxic concentrations of VAN (unpublished data). Collectively, these findings suggest that, in addition to antioxidant activity, VAN also may induce some type of DNA damage that enhances recombinational repair.
In summary, we found that VAN and CIN were effective antimutagens against spontaneous mutation in the wild-type strain NR9102 of E. coli and that this antimutagenic effect was independent of both the SOS and NER pathways. Given the DNA damage induced in mammalian cells by CIN and VAN as detected by the comet assay and the mutagenic effect of these compounds in the recombination-defective E. coli strain NR11317, we propose the following model to explain the antimutagenic effects of VAN and CIN. VAN and CIN induce some type(s) of DNA damage, either by direct interaction with DNA or indirectly through modulating intracellular protein or glutathione levels, which elicits recombinational repair. This activation of recombinational repair permits repair not only of VAN- or CIN-induced damage but also of other DNA lesions, thus reducing the frequency of spontaneous mutation.
Acknowledgments
This research was supported in part by the NIH Intramural Research Program of the National Institute of Environmental Health Sciences. In particular, D.T. Shaughnessy acknowledges support from an Intramural Research Training Award. We thank Peggy Mathews (US EPA) and Sarah Warren (US EPA) for their assistance and laboratory support in this work. We thank D.A. Bell (NIEHS), K. Witt (NIEHS), L. Recio (ILS), and W. Ward (US EPA) for their helpful comments on the manuscript. This manuscript was reviewed by the National Health and Environmental Effects Research Laboratory, US Environmental Protection Agency, and approved for publication. Approval does not signify that the contents necessarily reflect the views and policies of the agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
Appendix A
Mutant yield and survival in E. coli strains plated in the presence of VAN.
| Strain | Dose (μmol/plate) | Mutants, Aa | Mutants, Ba | Mutant yield (mean)b | Survival, Aa | Survival, Ba | Survival (mean)b |
|---|---|---|---|---|---|---|---|
| NR9102 (wt) | 0 | 458.19 | 237.69 | 330.01 | 295.07 | 308.76 | 301.84 |
| 20 | 457.73 | 198.16 | 301.17 | 268.22 | 256.23 | 262.15 | |
| 30 | 413.10 | 123.92 | 226.26 | 265.69 | 271.29 | 268.48 | |
| 40 | 365.02 | 150.93 | 234.72 | 271.42 | 264.19 | 267.78 | |
| 50 | 240.35 | 83.78 | 141.90 | 257.01 | 256.51 | 256.76 | |
| 60 | 196.59 | 58.45 | 107.20 | 264.76 | 267.24 | 266.00 | |
| NR11634 (uvrB) | 0 | 630.13 | 356.72 | 474.11 | 383.09 | 121.11 | 215.40 |
| 20 | 625.85 | 331.65 | 455.59 | 378.19 | 135.62 | 226.48 | |
| 25 | 634.38 | 330.62 | 457.97 | 367.75 | 156.05 | 239.56 | |
| 30 | 529.73 | 323.79 | 414.15 | 391.33 | 119.33 | 216.10 | |
| 35 | 541.28 | 315.89 | 413.50 | 389.15 | 140.17 | 233.55 | |
| 40 | 454.94 | 278.24 | 355.79 | 396.07 | 142.84 | 237.86 | |
| NR11475 (recA430) | 0 | 943.72 | 488.59 | 679.04 | 358.98 | 347.27 | 353.07 |
| 20 | 890.46 | 482.73 | 655.63 | 321.23 | 303.53 | 312.25 | |
| 25 | 765.56 | 430.25 | 573.92 | 330.31 | 306.56 | 318.21 | |
| 30 | 826.32 | 383.09 | 562.63 | 320.81 | 304.45 | 312.52 | |
| 35 | 772.67 | 250.64 | 440.07 | 331.58 | 306.75 | 318.92 | |
| 40 | 520.95 | 236.70 | 351.16 | 333.02 | 330.66 | 331.84 | |
| NR11317 (recA56) | 0 | 18.17 | 39.33 | 26.73 | 50.83 | 314.02 | 126.34 |
| 20 | 27.92 | 39.37 | 33.15 | 52.28 | 361.46 | 137.47 | |
| 25 | 28.48 | 34.59 | 31.39 | 58.59 | 367.24 | 146.69 | |
| 30 | 28.73 | 39.02 | 33.48 | 50.01 | 318.34 | 126.18 | |
| 35 | 31.08 | 39.92 | 35.23 | 50.57 | 381.61 | 138.92 | |
| 40 | 37.23 | 54.93 | 45.22 | 47.45 | 311.14 | 121.51 | |
| NR9319 (mutL) | 0 | 4521.28 | 4513.01 | 4517.15 | 340.98 | 964.26 | 573.41 |
| 20 | 5073.04 | 4773.98 | 4921.24 | 338.66 | 1039.27 | 593.26 | |
| 30 | 4991.86 | 5226.46 | 5107.81 | 319.54 | 1018.56 | 570.50 | |
| 40 | 5210.38 | 5013.11 | 5110.79 | 311.16 | 1010.54 | 560.75 | |
| 60 | 6160.81 | 5561.39 | 5853.43 | 166.97 | 608.95 | 318.86 | |
| 80 | 5155.92 | 4413.12 | 4770.08 | 5.31 | 89.59 | 21.82 |
Unadjusted geometric mean of mutants/plate or survivors/plate from three replicate plates.
Unadjusted geometric mean of mutants/plate or survivors/plate from six plates (two independent cultures).
Appendix B
Mutant yield and survival in E. coli strains plated in the presence of CIN.
| Strain | Dose (μmol/plate) | Mutants, Aa | Mutants, Ba | Mutant yield (mean)b | Survival, Aa | Survival, Ba | Survival (mean)b |
|---|---|---|---|---|---|---|---|
| NR9102 | 0 | 479.11 | 231.60 | 333.11 | 264.41 | 265.51 | 264.96 |
| 3 | 504.19 | 216.01 | 330.01 | 258.71 | 264.90 | 261.79 | |
| 5 | 497.77 | 238.18 | 344.33 | 236.52 | 259.18 | 247.59 | |
| 10 | 306.89 | 155.30 | 218.31 | 243.50 | 238.98 | 241.23 | |
| 20 | 2.41 | 4.64 | 3.34 | 23.26 | 35.27 | 28.64 | |
| NR11634 | 0 | 671.55 | 349.01 | 484.13 | 396.78 | 135.85 | 232.17 |
| 3 | 665.40 | 423.83 | 531.05 | 389.44 | 120.32 | 216.46 | |
| 5 | 616.70 | 371.52 | 478.66 | 398.13 | 133.84 | 230.84 | |
| 10 | 612.92 | 355.67 | 466.90 | 387.34 | 137.45 | 230.74 | |
| 20 | 23.99 | 7.00 | 12.96 | 329.28 | 87.31 | 169.56 | |
| NR11475 | 0 | 871.29 | 492.77 | 655.25 | 307.88 | 290.68 | 299.16 |
| 3 | 953.86 | 473.87 | 672.31 | 319.03 | 310.32 | 314.65 | |
| 5 | 922.21 | 473.84 | 661.04 | 312.92 | 289.65 | 301.06 | |
| 10 | 342.35 | 343.29 | 342.82 | 306.84 | 279.10 | 292.64 | |
| 20 | 7.11 | 3.73 | 5.15 | 184.28 | 173.51 | 178.82 | |
| NR11317 | 0 | 127.32 | 119.18 | 123.18 | 112.03 | 259.72 | 170.58 |
| 3 | 103.24 | 112.27 | 107.66 | 120.49 | 239.93 | 170.03 | |
| 5 | 116.72 | 125.72 | 121.14 | 115.49 | 253.43 | 171.09 | |
| 10 | 121.00 | 113.16 | 117.02 | 49.91 | 183.13 | 95.60 | |
| 20 | 41.85 | 42.17 | 42.01 | 0.67 | 0.67 | 0.67 | |
| NR9319 | 0 | 6121.46 | 4707.27 | 5368.00 | 327.45 | 984.96 | 567.92 |
| 3 | 5536.03 | 5186.12 | 5358.22 | 350.57 | 1031.62 | 601.38 | |
| 5 | 6491.66 | 4709.50 | 5529.23 | 338.16 | 1041.50 | 593.46 | |
| 10 | 3010.35 | 3532.95 | 3261.20 | 289.06 | 1012.23 | 540.92 | |
| 20 | 5.67 | 0.00 | 2.83 | 66.42 | 351.67 | 152.83 |
Geometric mean of mutants/plate or survivors/plate from three replicate plates.
Geometric mean of mutants/plate or survivors/plate from six plates (two independent cultures).
References
- 1.Ohta T, Watanabe K, Moriya M, Shirasu Y, Kada T. Analysis of the antimutagenic effect of cinnamaldehyde on chemically induced mutagenesis in Escherichia coli. Mol Gen Genet. 1983;192:309–315. doi: 10.1007/BF00392167. [DOI] [PubMed] [Google Scholar]
- 2.Ohta T, Watanabe K, Moriya M, Shirasu Y, Kada T. Antimutagenic effects of cinnamaldehyde on chemical mutagenesis in Escherichia coli. Mutat Res. 1983;107:219–227. doi: 10.1016/0027-5107(83)90164-1. [DOI] [PubMed] [Google Scholar]
- 3.Ohta T, Watanabe M, Watanabe K, Shirasu Y, Kada T. Inhibitory effects of flavourings on mutagenesis induced by chemicals in bacteria. Food Chem Toxicol. 1986;24:51–54. doi: 10.1016/0278-6915(86)90264-4. [DOI] [PubMed] [Google Scholar]
- 4.Ohta T, Watanabe M, Shirasu Y, Inoue T. Post-replication repair and recombination in uvrA umuC strains of Escherichia coli are enhanced by vanillin, an antimutagenic compound. Mutat Res. 1988;201:107–112. doi: 10.1016/0027-5107(88)90116-9. [DOI] [PubMed] [Google Scholar]
- 5.Ohta T. Modification of genotoxicity by naturally occurring flavorings and their derivatives. Crit Rev Toxicol. 1993;23:127–146. doi: 10.3109/10408449309117114. [DOI] [PubMed] [Google Scholar]
- 6.Imanishi H, Sasaki YF, Matsumoto K, Watanabe M, Ohta T, Shirasu Y, Tutikawa K. Suppression of 6-TG-resistant mutations in V79 cells and recessive spot formations in mice by vanillin. Mutat Res. 1990;243:151–158. doi: 10.1016/0165-7992(90)90038-l. [DOI] [PubMed] [Google Scholar]
- 7.Fiorio R, Bronzetti G. Effects of cinnamaldehyde on survival and formation of HGPRT-mutants in V79 cells treated with methyl methanesulfonate, N-nitroso-N-methylurea, ethyl methanesulfonate and UV light. Mutat Res. 1994;324:51–57. doi: 10.1016/0165-7992(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 8.Sanyal R, Darroudi F, Parzefall W, Nagao M, Knasmuller S. Inhibition of the genotoxic effects of heterocyclic amines in human derived hepatoma cells by dietary bioantimutagens. Mutagenesis. 1997;12:297–303. doi: 10.1093/mutage/12.4.297. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki YF, Imanishi H, Watanabe M, Ohta T, Shirasu Y. Suppressing effect of antimutagenic flavorings on chromosome aberrations induced by UV-light or X-rays in cultured Chinese hamster cells. Mutat Res. 1990;229:1–10. doi: 10.1016/0027-5107(90)90002-l. [DOI] [PubMed] [Google Scholar]
- 10.de Silva HV, Shankel DM. Effects of the antimutagen cinnamaldehyde on reversion and survival of selected Salmonella tester strains. Mutat Res. 1987;187:11–19. doi: 10.1016/0165-1218(87)90071-1. [DOI] [PubMed] [Google Scholar]
- 11.De Flora S, Bennicelli C, Rovida A, Scatolini L, Camoirano A. Inhibition of the ‘spontaneous’ mutagenicity in Salmonella typhimurium TA102 and TA104. Mutat Res. 1994;307:157–167. doi: 10.1016/0027-5107(94)90288-7. [DOI] [PubMed] [Google Scholar]
- 12.Shaughnessy DT, Setzer RW, DeMarini DM. The antimutagenic effect of vanillin and cinnamaldehyde on spontaneous mutation in Salmonella TA104 is due to a reduction in mutations at GC but not AT sites. Mutat Res. 2001;480–481:55–69. doi: 10.1016/s0027-5107(01)00169-5. [DOI] [PubMed] [Google Scholar]
- 13.Kada T, Inoue T, Ohta T, Shirasu Y. Antimutagens and their modes of action. Basic Life Sci. 1986;39:181–196. doi: 10.1007/978-1-4684-5182-5_15. [DOI] [PubMed] [Google Scholar]
- 14.Santos JH, Graf U, Reguly ML, Rodrigues de Andrade HH. The synergistic effects of vanillin on recombination predominate over its antimutagenic action in relation to MMC-induced lesions in somatic cells of Drosophila melanogaste. Mutat Res. 1999;444:355–365. doi: 10.1016/s1383-5718(99)00101-1. [DOI] [PubMed] [Google Scholar]
- 15.Sinigaglia M, Reguly ML, de Andrade HH. Effect of vanillin on toxicant-induced mutation and mitotic recombination in proliferating somatic cells of Drosophila melanogaster. Environ Mol Mutagen. 2004;44:394–400. doi: 10.1002/em.20067. [DOI] [PubMed] [Google Scholar]
- 16.Maron DM, Ames BN. Revised methods for the Salmonella mutagenicity test. Mutat Res. 1983;113:173–215. doi: 10.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- 17.Schaaper RM, Dunn RL. Spontaneous mutation in the Escherichia coli lacI gene. Genetics. 1991;129:317–326. doi: 10.1093/genetics/129.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Press; 1972. [Google Scholar]
- 19.Schaaper RM. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J Biol Chem. 1993;268:23762–23765. [PubMed] [Google Scholar]
- 20.Porwollik S, Wong RM, Sims SH, Schaaper RM, DeMarini DM, McClelland M. The Delta uvrB mutations in the Ames strains of Salmonella span 15 to 119 genes. Mutat Res. 2001;483:1–11. doi: 10.1016/s0027-5107(01)00239-1. [DOI] [PubMed] [Google Scholar]
- 21.Pavlov YI, Suslov VV, Shcherbakova PV, Kunkel TA, Ono A, Matsuda A, Schaaper RM. Base analog N6-hydroxylaminopurine mutagenesis in Escherichia coli: genetic control and molecular specificity. Mutat Res. 1996;357:1–15. doi: 10.1016/0027-5107(96)00060-7. [DOI] [PubMed] [Google Scholar]
- 22.Ennis DG, Levine AS, Koch WH, Woodgate R. Analysis of recA mutants with altered SOS functions. Mutat Res. 1995;336:39–48. doi: 10.1016/0921-8777(94)00045-8. [DOI] [PubMed] [Google Scholar]
- 23.Schaaper RM, Danforth BN, Glickman BW. Rapid repeated cloning of mutant lac repressor genes. Gene. 1985;39:181–189. doi: 10.1016/0378-1119(85)90312-9. [DOI] [PubMed] [Google Scholar]
- 24.Welch MM, McHenry CS. Cloning and identification of the product of the dnaE gene of Escherichia coli. J Bacteriol. 1982;152:351–356. doi: 10.1128/jb.152.1.351-356.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oller AR, Schaaper RM. Spontaneous mutation in Escherichia coli containing the dnaE911 DNA polymerase antimutator allele. Genetics. 1994;138:263–270. doi: 10.1093/genetics/138.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schaaper RM, Danforth BN, Glickman BW. Mechanisms of spontaneous mutagenesis: an analysis of the spectrum of spontaneous mutation in the Escherichia coli lacI gene. J Mol Biol. 1986;189:273–284. doi: 10.1016/0022-2836(86)90509-7. [DOI] [PubMed] [Google Scholar]
- 27.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 29.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci USA. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox MM. Recombinational DNA repair in bacteria and the RecA protein. Prog Nucleic Acid Res Mol Biol. 1999;63:311–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 31.Weibel H, Hansen J. Interaction of cinnamaldehyde (a sensitizer in fragrance) with protein. Contact Dermatitis. 1989;20:161–166. doi: 10.1111/j.1600-0536.1989.tb04650.x. [DOI] [PubMed] [Google Scholar]
- 32.Ka H, Park HJ, Jung HJ, Choi JW, Cho KS, Ha J, Lee KT. Cinnamaldehyde induces apoptosis by ROS-mediated mitochondrial permeability transition in human promyelocytic leukemia HL-60 cells. Cancer Lett. 2003;196:143–152. doi: 10.1016/s0304-3835(03)00238-6. [DOI] [PubMed] [Google Scholar]
- 33.Janzowski C, Glaab V, Mueller C, Straesser U, Kamp HG, Eisenbrand G. Alpha,beta-unsaturated carbonyl compounds: induction of oxidative DNA damage in mammalian cells. Mutagenesis. 2003;18:465–470. doi: 10.1093/mutage/geg018. [DOI] [PubMed] [Google Scholar]
- 34.Glaab V, Collins AR, Eisenbrand G, Janzowski C. DNA-damaging potential and glutathione depletion of 2-cyclohexene-1-one in mammalian cells, compared to food relevant 2-alkenals. Mutat Res. 2001;497:185–197. doi: 10.1016/s1383-5718(01)00260-1. [DOI] [PubMed] [Google Scholar]
- 35.Zhou YC, Zheng RL. Phenolic compounds and an analog as superoxide anion scavengers and antioxidants. Biochem Pharmacol. 1991;42:1177–1179. doi: 10.1016/0006-2952(91)90251-y. [DOI] [PubMed] [Google Scholar]
- 36.Murcia MA, Martinez-Tome M. Antioxidant activity of resveratrol compared with common food additives. J Food Prot. 2001;64:379–384. doi: 10.4315/0362-028x-64.3.379. [DOI] [PubMed] [Google Scholar]
- 37.Kamat JP, Ghosh A, Devasagayam TP. Vanillin as an antioxidant in rat liver mitochondria: inhibition of protein oxidation and lipid peroxidation induced by photosensitization. Mol Cell Biochem. 2000;209:47–53. doi: 10.1023/a:1007048313556. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Mori A. Antioxidant and pro-oxidant activities of p-hydroxybenzyl alcohol and vanillin: effects on free radicals, brain peroxidation and degradation of benzoate, deoxyribose, amino acids and DNA. Neuropharmacology. 1993;32:659–669. doi: 10.1016/0028-3908(93)90079-i. [DOI] [PubMed] [Google Scholar]
- 39.Santosh Kumar S, Priyadarsini KI, Sainis KB. Free radical scavenging activity of vanillin and o-vanillin using 1,1-diphenyl-2-picrylhydrazyl (DPPH) radical. Redox Rep. 2002;7:35–40. doi: 10.1179/135100002125000163. [DOI] [PubMed] [Google Scholar]
- 40.Kumar SS, Ghosh A, Devasagayam TP, Chauhan PS. Effect of vanillin on methylene blue plus light-induced single-strand breaks in plasmid pBR322 DNA. Mutat Res. 2000;469:207–214. doi: 10.1016/s1383-5718(00)00074-7. [DOI] [PubMed] [Google Scholar]
- 41.Tamai K, Tezuka H, Kuroda Y. Different modifications by vanillin in cytotoxicity and genetic changes induced by EMS and H2O2 in cultured Chinese hamster cells. Mutat Res. 1992;268:231–237. doi: 10.1016/0027-5107(92)90229-u. [DOI] [PubMed] [Google Scholar]
- 42.Frenzilli G, Bosco E, Barale R. Validation of single cell gel assay in human leukocytes with 18 reference compounds. Mutat Res. 2000;468:93–108. doi: 10.1016/s1383-5718(00)00042-5. [DOI] [PubMed] [Google Scholar]


