Abstract
We describe a simple, robust, and relatively inexpensive non-radioactive in vitro assay for measuring histone acetyl-transferase activity. The assay takes advantage of easy to purify recombinant E. coli-derived fusion proteins containing the NH2-terminal tails of histones H3 and H4 linked to epitope-tagged maltose binding protein (MBP), and immunoblotting with antibodies specific to acetylated H3 and H4. Here we show the specificity and dynamic range of this assay for the histone acetyl-transferases, p300 and PCAF. This assay may be adapted readily for other substrates by simply generating new fusion proteins and for other acetyl-transferases by modifying reaction conditions.
Keywords: non-isotopic, acetyltransferase assay, p300, PCAF, histone H3, histone H4
Introduction
Post-translational modifications of histone proteins are recognized to play a central role in the regulation of gene expression (Li et al., 2007). Specific amino acids within the NH2-terminal tails of core histones, particularly H3 and H4, are susceptible to reversible enzymatic modifications, including acetylation, methylation, and ubiquitination of lysine residues, methylation of arginines, and phosphorylation of serines (Li et al., 2007). Numerous studies have confirmed the idea that different combinations of histone modifications correlate well with dynamic changes in chromatin organization that occur during gene activation or repression (reviewed in (Barrera and Ren, 2006; Shilatifard, 2006)). The complete characterization of the enzymes that mediate these reactions, as well as the identification of potential facilitating or inhibitory molecules, should provide both fundamental mechanistic insights into the biochemistry of gene regulation and be of diagnostic and therapeutic value understanding human biology and disease.
The extent of acetylation of lysine residues within the NH2 tails of H3 and H4 has been linked to the transcriptional potential of a given chromosomal locus (Yang, 2004). Lysine acetylation is generally associated with an open chromatin environment and with a permissive transcriptional template (Yang, 2004). Consistent with this idea, numerous transcriptional co-activator molecules, including CBP, p300, and PCAF, contain intrinsic (histone) acetyl-transferase activity (Kalkhoven, 2004). By contrast, histone de-acetylation has been linked to a closed chromatin architecture and to transcriptional inhibition (Ng and Bird, 2000), and several classes of transcriptional co-repressors have been identified with histone de-acetylase activity (Verdin et al., 2003).
Several methods have been described for evaluating histone acetyl-transferase (HAT) activity. In cells, lysine acetylation of histones H3 and H4 may be assessed within a given genomic region by chromatin immunoprecipitation assay, which takes advantage of antibodies that specifically recognize these modifications (Wilson and Rotwein, 2006). There are also in vitro HAT assays that measure enzymatic activity more directly (Berndsen and Denu, 2005). Most use as substrates either purified core histones or synthetic NH2-terminal histone tails, and typically as the acetyl donor either [³H] or [14C]-labeled acetyl-Co-enzyme A (CoA) (Ait-Si-Ali et al., 1998; Mizzen et al., 1999; Berndsen and Denu, 2005). Alternative assays have been developed in which the appearance of free CoA correlated with acetyltransferase activity. In one version, an enzyme-coupled assay links release of CoA to reduction of NAD+ to NADH by pyruvate dehydrogenase in the presence of pyruvate. NADH is then measured directly at 340 nm or at 440 nm following reduction of a tetrazolium dye as described by Kim et al (Kim et al., 2000) or available commercially (www.biocatbiosciences.com and www.oxfordbiomed.com/). A second assay utilizes the sulfhydryl-sensitive dye, 7-diethylamino-3-(4’-maleimidylphenyl)-4-methylcoumarin, which forms an adduct with CoA and fluoresces at 469 nM (Trievel et al., 2000). These latter assays have the advantage of avoiding the costs and hazards of radioactivity, but are significantly less sensitive than those employing isotopically-labeled acetyl-CoA (Berndsen and Denu, 2005).
Here we describe a simple, inexpensive and sensitive non-radioactive HAT assay for both p300 and PCAF that takes advantage of easy to purify recombinant E. coli-derived fusion proteins containing the NH2-terminal tails of histones H3 and H4 linked to flag epitope-tagged maltose binding protein (MBP). Following incubation with immunoprecipitated or recombinant p300 or PCAF the MBP fusion proteins were subjected to SDS-PAGE and immunoblotting with antibodies specific to acetylated H3 or H4 to assess enzymatic activity and flag antibodies for substrate normalization. This assay is scalable for high-throughput analysis and may be adapted readily for other substrates by simply generating new fusion proteins and for other acetyltransferases by modifying reaction conditions. In addition, after their acetylation, both H3-MBP and H4-MBP potentially may be used as substrates for de-acetylases.
Materials and Methods
Generation of plasmids encoding MBP, H3-MBP and H4-MBP
A modified, Flag-tagged maltose binding protein (MBP) expression cassette was generated by PCR amplification of the 1.2 kb E. coli malE open reading frame from the pMAL-c2 expression plasmid (New England BioLabs). The forward primer incorporated restriction endonuclease sites for NdeI, EcoRI, and XmaI. The NdeI site contains a methionine initiation codon. The reverse primer also included sites for BamHI, SalI, and HindIII, which were followed sequentially by an in-frame flag epitope (codons for amino acids DYKDDDDK), tandem stop codons, and an XhoI site. The modified MBP DNA cassette was sequence verified, subcloned into pET23d (Novagen) and subsequently termed pMBS. H4-MBP was prepared by subcloning a synthetic double-stranded oligonucleotide encoding amino acids 1 – 19 of Tetrahymena histone H4 (codons optimized for expression in E. coli) into the NdeI and XmaI sites of pMBS. H3-MBP was constructed similarly, using a synthetic double-stranded oligonucleotide encoding residues 1 – 22 of Tetrahymena histone H3, also optimized for expression in E. coli.
Purification of MBP, H3-MBP and H4-MBP
For protein expression, pMBS plasmids were transformed into the E. coli BL21 strain and grown in LB medium. Gene expression was induced by addition of IPTG (300 mM final concentration) to log-phase cultures for 3 hr at 37°C. Recombinant proteins were purified by affinity chromatography with amylose agarose, based on a protocol from the supplier (New England BioLabs) with the following modifications: following binding to amylose resin, MBP proteins were washed 3 times with cold acetyltransferase assay buffer (50 mM Tris Cl pH 8.0, 10% glycerol, 10 mM butyric acid, 0.1 mM EDTA) and eluted by incubation with assay buffer containing 10 mM maltose for 15 min at 22°C, and the supernatants collected after centrifugation for 5 min at 3,000 × g at 4°C in a micro-centrifuge to pellet the resin. The relative purity of each MBP was assessed after SDS-PAGE by staining with Gelcode Blue (Pierce), and the concentration determined by protein assay (BCA, Pierce). A typical yield of purified fusion protein is 10 – 20 mg/L of culture medium. Proteins were stable more than 6 months at 4°C and stored long term in elution buffer supplemented with 10% glycerol at −80°C.
Mammalian cell culture, adenoviral infection, and nuclear protein extract preparation
Murine C3H10T1/2 embryonic fibroblasts (ATCC catalog #CCL-226) were grown at 37°C in humidified air with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, Mediatech-Cellgrow) with 10% fetal calf serum (FCS, Hyclone, Inc). Recombinant adenoviruses (Ad) encoding the following proteins have been described previously: human wild type and acetyltransferase-deficient p300 with COOH-terminal flag epitope tags, human wild type and acetyltransferase-deficient PCAF, each containing a NH2-terminal influenza hemagglutinin (HA) tag (Kuninger et al., 2006), tetracycline-inhibited transcriptional activator (tTA) (Wilson et al., 2003). Expression of p300 and PCAF from these adenoviruses is dependent upon tTA (Kuninger et al., 2006). The p300ATmut contains substitutions at the following amino acids within the catalytic domain: His1415, Glu1423, Tyr1423, Tyr1430, and His1434 changed to Ala, and Leu1428 to Ser (Kraus et al., 1999). The PCAFATmut contains substitutions of amino acids Phe568, Thr569, and Glu570 within the catalytic domain to Ala (Kuninger et al., 2006). Infection conditions have been described elsewhere (Kuninger et al., 2006), with Ad-tTA at a multiplicity of infection (MOI) of 300 and all others at MOIs of 1000. Nuclear protein extracts (NE) were prepared from Ad-infected C3H10T1/2 cells based on a published procedure (Schreiber et al., 1989). The concentration of NE was determined using the BCA assay (Pierce) and proteins were stored in aliquots at −80°C.
In vitro acetylation assays
Recombinant p300 or PCAF proteins were immunoprecipitated from 100 µg of NE using either anti-flag or anti-HA antibodies plus Protein A-agarose (Sigma). Immunoprecipitates were washed in twice in phosphate buffered saline containing 0.1% Tween 20 (PBS-T), and once in acetyl-transferase assay buffer (50 mM Tris-Cl pH 8, 10% glycerol, 10 mM butyric acid, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF). Unless otherwise specified, individual reactions contained immunoprecipitated proteins from 50 µg of NE in 50 µl of assay buffer with 10 µM acetyl CoA (Sigma), and different quantities of purified substrates as stated in individual Figure legends. GST-PCAF (UBI) was used at 500 ng/reaction. Acetylation reactions were incubated for 45 min (unless otherwise specified) at 30°C on a rotating platform, followed by addition of SDS-PAGE sample buffer, electrophoresis through 10% SDS-PAGE gels, and transfer to PVDF membranes. Proteins were detected by immunoblotting followed by image acquisition and quantification using a LiCor Odyssey infrared imaging system. Acetylated histone H3 or H4 were recognized with rabbit anti-acH3 (#06–599) or anti-acH4 (#06–866, UBI, each at 1:1000 dilution). respectively, and Alexa 680-conjugated anti-rabbit IgG (Molecular Probes, 1:4000). Total MBP, H3-MBP, and H4-MBP were detected using anti-flag monoclonal antibodies (Sigma, 1:3000 dilution), as was p300, followed by anti-mouse IRD 800 (Rockland, 1:4000 dilution). PCAF was detected using primary monoclonal antibodies to HA (HA.11, Covance, 1:1000 dilution) followed by anti-mouse IRD 800 (1:4000). Isotopic acetylation assays were performed similarly, except that 10 µg of acid-extracted calf thymus core histones (Sigma) were used as substrates and 10 µM [Acetyl-1-14C] CoA (New England Nuclear, NEC-313; 51 mCi/mmole, 0.4 mM) was added as the acetyl donor. Following incubation at 30°C for 45 min, core histones were precipitated with trichloroacetic acid (TCA) onto GF/C filters (Whatman), washed three times with 10% TCA and once with 100% ethanol, and the amount of 14C incorporated into protein was determined by liquid scintillation counting.
Analysis of MBP and H3-MBP by isoelectric focusing, SDS-PAGE, and immunoblotting
MBP or H3-MBP (0.5 µg) were each incubated with or without GST-PCAF for 45 min under conditions described above. After completion of the reaction, 20% (10 µl) of each sample was diluted in a total volume of 125 µl of rehydration buffer (6 M urea, 2 M thiourea, 4% CHAPS, 60 mM DTT, 1X ready strip buffer pH 3 – 10 (BioRad) and 0.1% bromophenol blue), and incubated at 23°C for 30 min. Protein samples were applied to a 7 cm, linear pH 4.7 – 5.9 IPG strip (ReadyStrip, BioRad), hydrated for 12 hr at 20°C and focused at 4000 V for a total of 10,000 V-hr using an Amersham Ettan IPGphor isoelectric focusing system. The IPG strips were then equilibrated in 5 ml of buffer (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS, 0.002% bromophenol blue) with 20 mg/ml of DTT for 15 min at 23°C. Buffer was replaced with 5 ml of new rehydration buffer containing 50 mg/ml of iodoacetamide, and incubated for 15 min at 23°C. The equilibrated IPG strips were then subjected to SDS-PAGE (10% separating gel), followed by immunoblotting and detection, as above.
Results
Generation and purification of fusion proteins encoding the NH2-terminal tails of core histones H3 and H4
Fig. 1A shows the sequences of the NH2-terminal 22 amino acids of Tetrahymena histone H3 and the 19 NH2-terminal residues of histone H4, and the strategy for fusing each histone tail to flag epitope-tagged maltose binding protein (MBP). As the MBP cassette was constructed with multiple cloning sites at both 5’ and 3’ ends, new fusion proteins may be engineered readily. As seen in Fig. 1B, each chimeric protein was readily induced in the E. coli BL21 strain by addition of IPTG to bacteria transformed with a pET23d expression plasmid encoding the appropriate fusion gene, and was purified to near homogeneity from the soluble bacterial lysate after affinity chromatography with amylose agarose.
Figure 1. Purification of recombinant chimeric proteins encoding the NH2-terminus of histones H3 or H4.
A. Diagram illustrating the linear structure of fusion proteins containing the NH2-terminal part of Tetrahymena histone H3 (amino acids 1 – 22) or H4 (residues 1 – 19) joined to maltose binding protein (MBP), followed by a flag epitope. Lysine resides in each histone segment are indicated with bold type. Positions of 5’ and 3’ multiple cloning sites (mcs) also are indicated. B. Purification of E. coli-derived H3- and H4-MBP. Pictured is an image of a stained SDS-PAGE gel of soluble E. coli protein lysates before and after induction of the fusion gene with IPTG (U - un-induced, I - induced), and following purification of the chimeric proteins using amylose resin (P). For details see “Materials and Methods”.
In vitro acetylation of H4-MBP by p300
Fig. 2 depicts results of in vitro assays using purified H4-MBP or MBP as substrates for the histone acetyl-transferase, p300, which was enriched by immunoprecipitation from soluble nuclear protein extracts of mouse 10T1/2 mesenchymal stem cells infected with a recombinant adenovirus encoding flag-epitope tagged human p300. Acetylated H4-MBP was detected by immunoblotting using a commercial polyclonal antibody that raised against a synthetic peptide (coupled to KLH) containing acetyl-lysine residues at positions 5, 8, 12, and 16 of histone H4. As seen in the dose-response experiments in Fig. 2A, acetylated H4-MBP could be detected by this antibody after incubation of as little as 50 ng of the purified protein with p300 for 45 min. In contrast, exposure of as much as 5 µg of MBP to p300 for 45 min led to a much weaker signal, while both MBP and H4-MBP were detected with identical sensitivity by immunoblotting with an antibody to the flag epitope.
Figure 2. Acetylation of H4-MBP by p300.
A. Dose-dependent acetylation of purified H4-MBP (upper panels) after incubation for 45 min with p300 immunoprecipitated from 50 µg of nuclear protein extracts from C3H10T1/2 cells infected with Ad-p300 (lower panels). Equivalent amounts of MBP or H4-MBP proteins were used (middle panels). Results from 2 independent experiments are displayed to the right in graphical form. B. Kinetics of H4-MBP acetylation by p300. Time course studies show increases in H4 acetylation (upper panel) after incubation of 0.5 µg of purified H4-MBP fusion protein (middle panel) for 0 – 240 min with Flag-tagged p300 immunoprecipitated from 50 µg of C3H10T1/2 cell nuclear protein extracts (lower panels). Results of 3 independent experiments (mean ± SEM) are graphed to the right. C. Saturable acetylation of H4-MBP by p300. Flag-tagged p300 was isolated by immunoprecipitation from different amounts of nuclear protein extracts (NE) obtained from C3H10T1/2 cells infected with Ad-p300 (upper panel), and was incubated with 0.5 µg of H4-MBP (middle panel) for 45 min, followed by analysis of acetylated H4 by immunoblotting (lower panel). Results from 2 independent experiments are graphed to the right.
Results of time course experiments illustrated in Fig. 2B demonstrate that acetylated H4-MBP can be detected within 5 min after incubation with p300. Maximal acetylation was seen at 120 min, and half-maximal between 15 and 30 min. Results of dose-response experiments for p300 are shown in Fig. 2C. Acetylation of H4-MBP was seen with p300 immunoprecipitated from as little as 5 µg of soluble nuclear proteins from adenovirus infected 10T1/2 cells, and was maximal with 25 µg. Overall, these studies demonstrate that the in vitro acetylation of H4-MBP by human p300 is rapid, specific, and saturable.
In vitro acetylation of H3-MBP by PCAF
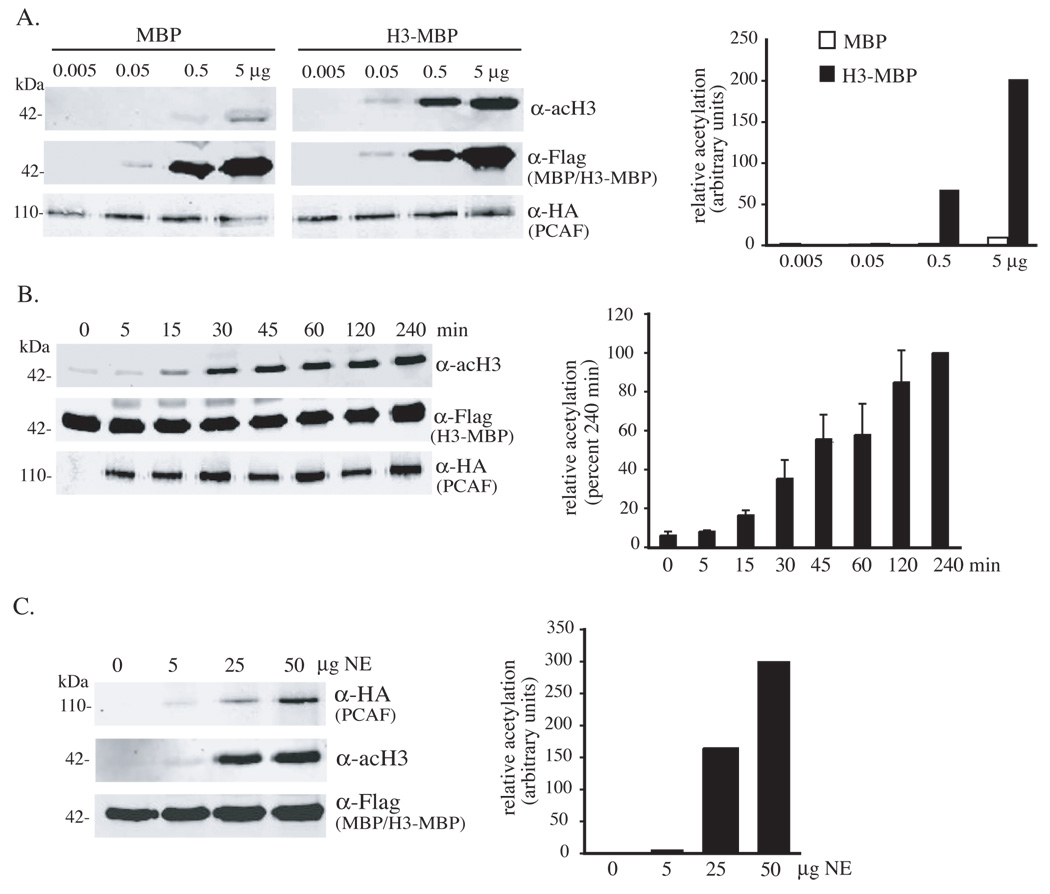
The acetyl-transferase PCAF has been shown to acetylate histone H1 and H3 in vitro (Herrera et al., 1997). As seen in Fig. 3A, after incubation of as little as 50 ng of purified H3-MBP with immunoprecipitated HA-tagged human PCAF the acetylated protein could be detected by immunoblotting with a commercial polyclonal antibody raised against a synthetic histone H3 peptide containing acetyl-lysine residues at positions 10 and 15. When 0.5 or 5 µg of purified substrate was used, the extent of its acetylation was ∼100 times greater than was seen with equivalent amounts of MBP alone, although both proteins could be detected to an equal extent after immunoblotting with an antibody to the flag epitope (Fig. 3A). Time course experiments using PCAF immunoprecipitated from 50 µg of 10T1/2 cell nuclear protein extracts showed that acetylation of H3-MBP was detectable within 15 min, was maximal at 120 min, and reached half-maximal levels within 30 – 45 min (Fig. 3B). Dose-response studies in which the amount of PCAF was varied indicated that the extent of acetylation of H3-MBP increased between 5 and 50 µg of added enzyme (Fig. 3C). Taken together, the results in Fig. 3 show that acetylation of H3-MBP by PCAF is rapid and specific.
Figure 3. Acetylation of H3-MBP by PCAF.
A. Dose-dependent acetylation of purified H3-MBP (upper panels) after incubation for 45 min with HA-tagged PCAF immunoprecipitated from 50 µg of nuclear protein extracts from C3H10T1/2 cells infected with Ad-PCAF (lower panels). Equivalent amounts of MBP and H3-MBP proteins were used (middle panels). Results from 2 independent experiments are displayed to the right. B. Kinetics of H3-MBP acetylation by PCAF. Time course studies demonstrate increases in H4 acetylation (upper panel) after incubation of 0.5 µg of purified H3-MBP fusion protein (middle panel) with HA-tagged PCAF immunoprecipitated from 50 µg of C3H10T1/2 cell nuclear protein extracts (lower panels). Results of 3 independent experiments (mean ± SEM) are graphed to the right. C. Saturable acetylation of H3-MBP by PCAF. HA-tagged PCAF was isolated by immunoprecipitation from different amounts of NE from C3H10T1/2 cells infected with Ad-PCAF (upper panel), and was incubated with 0.5 µg of H3-MBP (middle panel) for 45 min. Acetylated H3 was assessed by immunoblotting (lower panel). Results from 2 independent experiments are displayed to the right.
Assessing the specificity and sensitivity of in vitro acetylation by p300 and PCAF of H3-MBP and H4-MBP
We next examined the specificity of each acetyl-transferase for each histone substrate. As seen in Fig. 4A, p300 was able to acetylate both H3-MBP and H4-MBP to a comparable extent, while PCAF, or a bacterial fusion protein containing its catalytic domain, could only acetylate H3-MBP. These results are consistent with what has been published about each enzyme (Schiltz et al., 1999). As shown in Fig. 4B, mutations in the catalytic domains of p300 or PCAF rendered both enzymes non-functional toward their substrates. Thus, the non-radioactive in vitro acetylation assay may be used to assess the enzymatic activity of modified versions of these acetyl-transferases.
Figure 4. Specificity of PCAF for H3-MBP, and p300 for both H3- and H4-MBP.
A. Comparison of acetylation of MBP, H3-MBP, and H4-MBP (upper panels) by p300 and PCAF, and by an E. coli-derived GST-PCAF fusion protein. Equivalent amounts of each fusion protein (0.5 µg) were used in each assay (lower panels). B. Recombinant p300 with mutations in the acetyltransferase domain (ATmut) cannot acetylate H4-MBP (upper panel). Identical amounts of H4-MBP (0.5 µg) were used in each experiment (middle panels), and concentrations of wild type and mutant p300 were equivalent (lower panels). C. Recombinant PCAF with mutations in the acetyltransferase domain (ATmut) cannot acetylate H3-MBP (upper panel). Identical amounts of H3-MBP (0.5 µg) were used in each experiment (middle panels), and concentrations of wild type (wt) and mutant PCAF were comparable (lower panels).
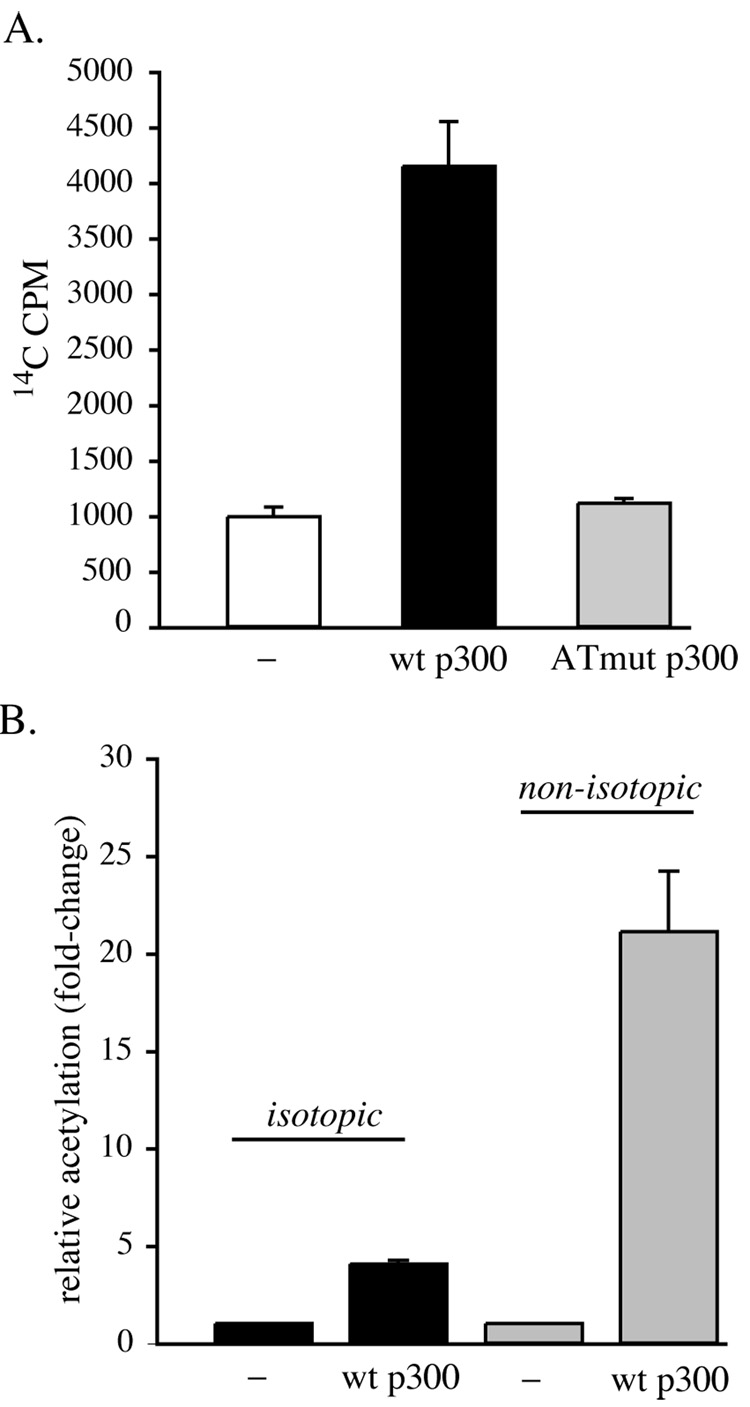
We also compared the ability of immunoprecipitated p300 to acetylate core histones using 14C-acetyl CoA as the donor with the sensitivity and dynamic range of our non-radioactive assay. As illustrated in Fig. 5A, wild type p300 was able to acetylate calf thymus core histones, causing a 4-fold rise in 14C incorporation, while p300 with mutations in its catalytic segment stimulated no increase over background values. As graphed in Fig. 5B, the non-isotopic assay using p300 and H4-MBP was 5 times more sensitive than the isotopic assay with core histones as the substrate. Also, the amount of substrate needed was less, as we used 10 µg of calf thymus core histones and 0.5 µg of H4-MBP to generate the data in Fig. 5B.
Figure 5. Comparison of the acetyltransferase activity of p300 assessed using isotopic labeling of core histones versus detection of acetylated H4-MBP by immunoblotting.
A. Core histones (10 µg) were incubated with immunoprecipitated wild-type (wt) p300, the acetyltransferase domain mutant (ATmut), or no p300 (−) for 45 min in the presence of 14C-acetyl CoA, as described in “Materials and Methods”. Results of 3 independent experiments (mean ± SEM) are graphed. B. Comparison of isotopic acetylation of 10 µg of core histones by p300 with non-isotopic labeling of 0.5 µg of H4-MBP. Equivalent amounts of immunoprecipitated p300 were used in each group of experiments. Results of 3 independent assays (mean ± SEM) are depicted.
Evaluating the extent of acetylation of H3-MBP by PCAF
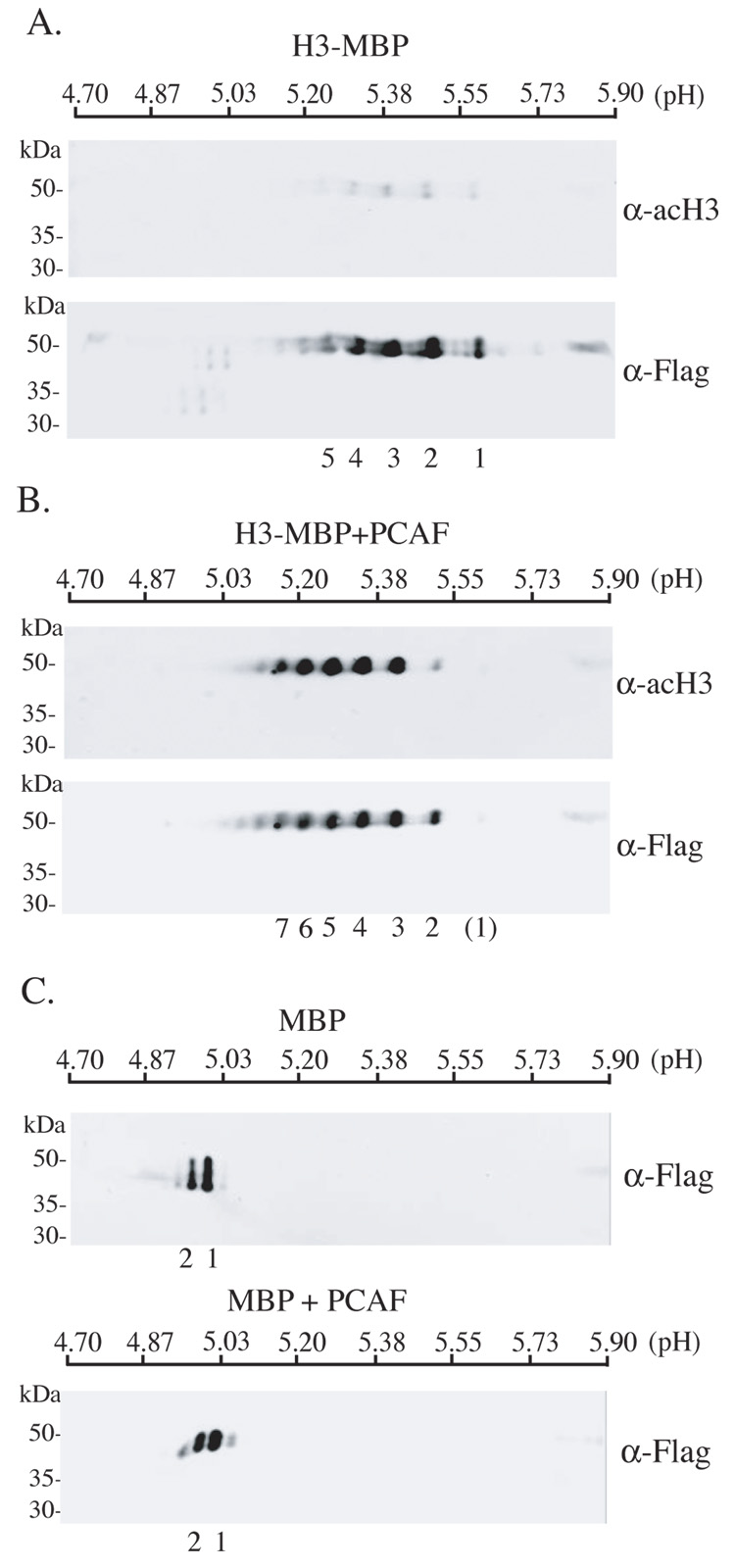
We next employed isoelectric focusing followed by SDS-PAGE and immunoblotting to examine the extent of acetylation of H3-MBP by PCAF. As depicted in Fig. 6A, although at least 4 major isoforms of H3-MBP differing in their isoelectric points (pI) could be recognized by the anti-flag antibody, in the absence of incubation with PCAF none of these protein species were detected by the acetyl-H3 antibody, indicating that they represented the presence of other post-translational modifications, such as phosphorylation, which would alter the charge of H3-MBP. As demonstrated in Fig. 6B, in vitro incubation of H3-MBP with PCAF led to the recognition of at least 5 major isoforms by the acetyl-H3 antibody (spots 3 – 7, Fig. 6B), and these proteins have been shifted to lower isoelectric points compared with those detected by the flag antibody in either Fig. 6B (spots 1 and 2 react minimally with anti-acetyl H3) or in Fig. 6A (spots 5 – 7 barely detected by anti-flag), clearly demonstrating the neutralization of positive charges on multiple lysine residues after their acetylation. Unmodified H3-MBP has a predicted pI of 5.46 (at neutral pH) calculated using Scansite (http://scansite.mit.edu/). In silico estimations of pI shifts, based on individual amino acid substitutions designed to mimic neutralization of side chains on lysine residues, yielded a result of minus 0.08 pH units/Lys substitution, which is very similar in magnitude to the observed pI shifts in Fig 6B. Interestingly, theoretical changes in pI following addition of phosphate groups to Ser/Thr residues in H3-MBP were also predicted to decrease the pI by ~0.08 pH units/phosphorylation, consistent with the range of H3-MBP species detected in Fig 6A and B. It is also clear from examination of Figs. 6A and 6B that acetylation of H3-MBP is not complete, as otherwise there would be a greater shift toward lower isoelectric points. This may reflect preferential lysine acetylation, which has been described for Lys14 in yeast H3 (corresponds to Lys15 in H3-MBP) (Mizzen et al., 1999; Trievel et al., 2000). We also tested the hypothesis that MBP was a substrate for PCAF by examining its pattern of isoelectric focusing either before or after incubation with the enzyme. As seen in Fig. 6C, the pI of MBP, predicted to be 4.95, was unaltered following exposure to PCAF. The positions of the two isoforms of MBP did not change after incubation with PCAF and were not recognized by the anti-acetyl H3 antibody or by an antibody to acetyl-lysine (data not shown). Along with results depicted in Fig. 3 and Fig. 4, these data indicate the specificity of PCAF toward the H3 peptide sequence within the fusion protein.
Figure 6. Multi-site acetylation of H3-MBP by PCAF.
A. Detection of H3-MBP (0.5 µg) by immunoblotting using anti-acH3 (upper panel) or anti-Flag antibodies (lower panel) after isoelectric focusing using linear pH 4.7 to 5.9 gradient gels and SDS-PAGE. B. Detection of H3-MBP (0.5 µg) by immunoblotting using anti-acH3 (upper panel) or anti-Flag antibodies (lower panel) after incubation with GST-PCAF, followed by isoelectric focusing and SDS-PAGE, as described in A. C. Detection of MBP by immunoblotting using anti-Flag antibodies before (upper panel) or after (lower panel) incubation with GST-PCAF, and followed by isoelectric focusing and SDS-PAGE, as described in A. For A – C, individual spots are numbered. The numbers correspond to one another in A and B, and in both parts of C.
Discussion
When considered together, the results presented in this manuscript demonstrate that we have developed a robust in vitro assay for assessing histone acetylation. Our data show that both p300 and PCAF can readily and quantitatively acetylate core histone substrates in vitro that can be detected by immunoblotting with specific antibodies. Under the conditions used for these assays, histone acetylation is rapid and saturable, being first detectable within minutes of initiation of the reaction and rising to maximal values of at least 100 times greater than baseline levels within 2 hrs. The assay also is specific, as neither p300 nor PCAF are able to measurably acetylate MBP alone. In addition, PCAF can acetylate only H3-MBP, while p300 is able to use both H3-MBP and H4-MBP as substrates. As mutant forms of both proteins with crippled catalytic domains (Kuninger et al., 2006) are ineffective enzymes in our hands (Fig. 4B, C), this assay also offers the potential to be used for rapid screening of modified versions of different histone acetyl-transferases for enzymatic activity and additionally could be employed to evaluate the efficacy of compounds or cofactors that may modulate acetyl transferase activity.
As the generation and purification of MBP fusion proteins in E. coli is simple and straightforward, we envision opportunities to produce a variety of potential substrates for many different protein acetyl-transferases. For example, published studies indicate that the myogenic transcription factor, MyoD, is acetylated by PCAF in conjunction with p300 in vivo, and that in vitro, both enzymes acetylate one or more of three lysine residues within the DNA binding domain of the protein (Puri et al., 1997; Sartorelli et al., 1999; Polesskaya et al., 2001). With our assay and antibodies recognizing either acetyl-lysine or the flag epitope, the pattern and extent of acetylation at each lysine by each enzyme could be investigated after isoelectric focusing and SDS-PAGE. Similarly detailed analyses could be performed with domains of other proteins that are subjected to reversible acetylation as part of their regulation. It also is possible to use H3-MBP, H4-MBP, or other MBP fusion proteins to study different de-acetylases, as in our preliminary investigations, acetyl-H3-MBP and acetyl-H4-MBP are relatively stable modified proteins, yet under appropriate conditions can become de-acetylated (data not shown).
In conclusion, we have developed a simple, robust, and relatively inexpensive non-radioactive in vitro assay for detecting and quantifying histone acetylation. New substrates may be generated readily and the assay may be modified easily for investigation of reversible protein acetylation for a variety of purposes.
Acknowledgements
These studies were supported by NIH Grants RO1 DK42748 and RO1 DK63073 (to P. R.). We thank Ryan Kuzmickas for technical assistance during the early phases of this project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ait-Si-Ali S, Ramirez S, Robin P, Trouche D, Harel-Bellan A. A rapid and sensitive assay for histone acetyl-transferase activity. Nucleic Acids Res. 1998;26:3869–3870. doi: 10.1093/nar/26.16.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera LO, Ren B. The transcriptional regulatory code of eukaryotic cells--insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr. Opin. Cell Biol. 2006;18:291–298. doi: 10.1016/j.ceb.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Berndsen CE, Denu JM. Assays for mechanistic investigations of protein/histone acetyltransferases. Methods. 2005;36:321–331. doi: 10.1016/j.ymeth.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Herrera JE, Bergel M, Yang XJ, Nakatani Y, Bustin M. The histone acetyltransferase activity of human GCN5 and PCAF is stabilized by coenzymes. J. Biol. Chem. 1997;272:27253–27258. doi: 10.1074/jbc.272.43.27253. [DOI] [PubMed] [Google Scholar]
- Kalkhoven E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- Kim Y, Tanner KG, Denu JM. A continuous, nonradioactive assay for histone acetyltransferases. Anal. Biochem. 2000;280:308–314. doi: 10.1006/abio.2000.4546. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Manning ET, Kadonaga JT. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuninger D, Wright A, Rotwein P. Muscle cell survival mediated by the transcriptional coactivators p300 and PCAF displays different requirements for acetyltransferase activity. Am. J. Physiol. Cell Physiol. 2006;291:C699–C709. doi: 10.1152/ajpcell.00056.2006. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The Role of Chromatin during Transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Mizzen CA, Brownell JE, Cook RG, Allis CD. Histone acetyltransferases: preparation of substrates and assay procedures. Methods Enzymol. 1999;304:675–696. doi: 10.1016/s0076-6879(99)04041-0. [DOI] [PubMed] [Google Scholar]
- Ng HH, Bird A. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 2000;25:121–126. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- Polesskaya A, Naguibneva I, Fritsch L, Duquet A, Ait-Si-Ali S, Robin P, Vervisch A, Pritchard LL, Cole P, Harel-Bellan A. CBP/p300 and muscle differentiation: no HAT, no muscle. EMBO J. 2001;20:6816–6825. doi: 10.1093/emboj/20.23.6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, Nakatani Y, Levrero M. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts”, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Trievel RC, Li FY, Marmorstein R. Application of a fluorescent histone acetyltransferase assay to probe the substrate specificity of the human p300/CBP-associated factor. Anal. Biochem. 2000;287:319–328. doi: 10.1006/abio.2000.4855. [DOI] [PubMed] [Google Scholar]
- Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- Wilson EM, Hsieh MM, Rotwein P. Autocrine growth factor signaling by insulin-like growth factor-II mediates MyoD-stimulated myocyte maturation. J. Biol. Chem. 2003;278:41109–41113. doi: 10.1074/jbc.C300299200. [DOI] [PubMed] [Google Scholar]
- Wilson EM, Rotwein P. Control of MyoD function during initiation of muscle differentiation by an autocrine signaling pathway activated by insulin-like growth factor-II. J. Biol. Chem. 2006;281:29962–29971. doi: 10.1074/jbc.M605445200. [DOI] [PubMed] [Google Scholar]
- Yang XJ. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays. 2004;26:1076–1087. doi: 10.1002/bies.20104. [DOI] [PubMed] [Google Scholar]