Abstract
Recent studies have revealed that Escherichia coli possesses an essential targeting system for integral membrane proteins, similar to the mammalian signal recognition particle (SRP) machinery. One essential protein in this system is FtsY, a homologue of the α-subunit of the mammalian SRP-receptor (SR-α). However, E. coli does not possess a close homologue of the integral membrane protein SR-β, which anchors SR-α to the membrane. Moreover, although FtsY can be found as a peripheral membrane protein, the majority is found soluble in the cytoplasm. In this study, we obtained genetic and biochemical evidence that FtsY must be targeted to the membrane for proper function. We demonstrate that the essential membrane targeting activity of FtsY is mediated by a 198-residue-long acidic N-terminal domain. This domain can be functionally replaced by unrelated integral membrane polypeptides, thus avoiding the need for specific FtsY membrane targeting factors. Therefore, the N terminus of FtsY constitutes an independent domain, which is required only for the targeting of the C-terminal NG domain of FtsY to the membrane.
Targeting of newly synthesized membrane and secretory proteins to the endoplasmic reticulum in mammalian cells is mediated cotranslationally by the signal recognition particle (SRP)-machinery (1, 2). In Escherichia coli, the targeting of secretory proteins to the inner membrane can be accomplished posttranslationally (3). Despite this difference, E. coli contains essential genes encoding Ffh and FtsY with a significant similarity to proteins of the eukaryotic SRP machinery (4, 5) and 4.5S RNA which resembles part of the mammalian 7SL RNA (6). Recent studies have elucidated the need for such a system in prokaryotes, demonstrating that two components of the E. coli SRP pathway, FtsY (7) and Ffh (8–10), are utilized preferentially for biogenesis of polytopic membrane proteins. One essential protein in this machinery is FtsY, the E. coli SRP-receptor (SR) which contains a large domain (the NG domain, see Ref. 11 for details) that is homologous to the NG domain of the mammalian SR-α protein (see Fig. 1B). By analogy to SR-α, the α-subunit of the mammalian SRP-receptor, it has been suggested that FtsY acts as a peripheral membrane protein (12, 13), although a large fraction of FtsY is found soluble in the cytoplasm (13). In mammalian cells, SR-α is targeted cotranslationally to the membrane (14) and interacts via a 140-residue-long N-terminal domain with its integral membrane β-subunit (refs. 15 and 16; Fig. 1B). In E. coli, no SR-β homologue has been found and the mechanisms by which FtsY reaches its destination and binds to the membrane are not known. The N-terminal 198-amino acid sequence of FtsY shares no homology with the mammalian receptor and it is highly charged and enriched with acidic residues (Fig. 1A). It has been hypothesized that FtsY might interact directly with membrane phospholipids. However, the observation that a large fraction of FtsY is soluble (13) supports the idea that there may be a limited number of membrane targeting factors or specific binding sites for FtsY.
Figure 1.
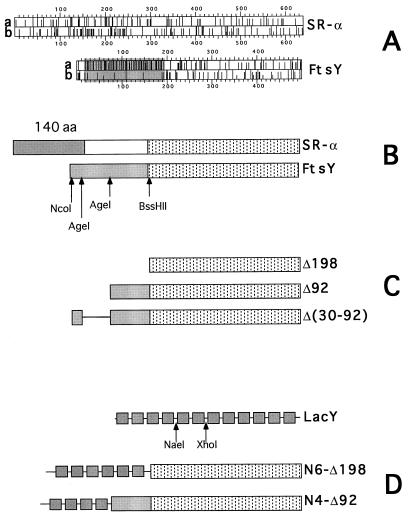
Schematic representation of FtsY and SR-α, FtsY mutants, LacY, and LacY–FtsY hybrids. (A) The distribution of charged residues (line a for Glu or Asp, and line b for Arg or Lys) along the primary sequence of SR-α and FtsY. The figure was adopted from the output of the program dna strider. The shaded region highlights the acidic N-terminal domain of FtsY. (B) Schematic alignment of SR-α and FtsY. Stippled boxes represent the homologous C-terminal NG domains. The shaded (box) N-terminal 140-residue-long domain of SR-α is implicated in membrane attachment (16). The shaded box in FtsY represent the highly acidic N-terminal domain. Restriction enzymes used for mutagenesis are shown under the FtsY box. (C) Schematic presentation of the N-terminal FtsY truncated mutants (Δ198 and Δ92) and the deletion mutant Δ(30–92). The deleted regions are not shown (Δ198 and Δ92) or shown as a straight line [Δ(30–92)]. (D) Schematic picture of lactose permease (LacY) containing 12 transmembrane helices (shown as shaded boxes) and hydrophilic loops (shown as straight lines between the boxes). The enzymes used for the construction of the LacY–FtsY hybrids are shown under the LacY diagram.
Here we show that FtsY must be targeted to the membrane for proper function. The membrane localization function is mediated by a 198-residue-long N-terminal domain of the protein. However, the essential targeting and attachment of FtsY to the membrane can be mediated also by unrelated integral membrane polypeptides fused to the N terminus of the NG domain of FtsY.
EXPERIMENTAL PROCEDURES
Materials.
Spectinomycin, ampicillin, arabinose, 3-β-indoleacrylic acid, and phenylmethylsulfonyl fluoride were purchased from Sigma. Restriction enzymes were obtained from New England Biolabs, and modifying enzymes were from Boehringer Mannheim. Antibodies to FtsY were a gift from J. Luirink (Biocentrum Amsterdam). Goat anti-rabbit antibodies conjugated to horseradish peroxidase were obtained from Jackson ImmunoResearch. Antibodies to the C-terminal tail of LacY were kindly provided by H. R. Kaback (University of California, Los Angeles). Prestained protein molecular weight markers were purchased from New England Biolabs, and DNA molecular weight markers were from GIBCO/BRL. GeneClean and Mermaid glassmilk DNA purification kits were obtained from Bio 101, and Wizard Mini Prep kits were from Promega. All other materials were reagent grade and obtained from commercial sources.
Bacterial Strains and Plasmids.
E. coli UT5600[ompT−] obtained from the E. coli Genetic Stock Center at Yale University (strain 7092) was used for mutagenesis and expression studies. E. coli N4156::pAra14-FtsY′ (13) was obtained from J. Luirink. Plasmid pT7-5(lacY) encoding lactose permease under the lac promoter was described elsewhere (17) and served for cloning of ftsY and DNA manipulations. Plasmid pCL1921 (18) (alone or containing various ftsY mutants) was used to transform E. coli N4156::pAra14-FtsY′ (polA−) for growth and expression experiments. Plasmid pATH2 (19) was used as a source for the gene encoding the N-terminal 326-amino acid residues of TrpE which was fused to the 5′ end of the gene encoding a deletion mutant of FtsY (see below).
Construction of ftsY Mutants and lacY–ftsY Gene Fusions.
The ftsY gene was amplified by PCR from the E. coli chromosome using the following sense and antisense deoxyoligonucleotides containing sites for enzymes BamHI and HindIII (underlined), respectively: 5′-TATATGGATCCATGGCGAAAGAAAAAAAACGTGGC and 5′-TATATAAGCTTAATCCTCTCGGGC.
The amplified DNA was cloned instead of the lacY gene into pT7-5(lacY) under the lac promoter/operator producing plasmid pTftsY. The ftsY mutants and the lacY–ftsY hybrids (see Fig. 1 C and D) were constructed as follows: Δ198 plasmid pTftsY was digested by NcoI and BssHII, treated by Klenow fragment, and the large DNA fragment was ligated on itself. The resulting plasmid was linearized by BssHII, treated by Klenow fragment, and ligated on itself. Δ92 plasmid pTftsY was digested by NcoI and AgeI, treated by Klenow fragment, and the large DNA fragment was ligated on itself. Δ(30–92) plasmid pTftsY was digested by AgeI and the large fragment was ligated on itself. N6-Δ198 plasmid pTftsY was digested by BssHII, treated with Klenow fragment, and then digested by PvuI to create a 1.5-kb DNA fragment. This fragment was ligated to the 2.5-kb DNA fragment released from pT7-5lacY by XhoI, Klenow treatment, and then PvuI. N4-Δ92 plasmid pTftsY was digested by AgeI, treated by Klenow fragment, and then digested by PvuI to create a 1.8-kb DNA fragment. This fragment was ligated to the 2.3-kb DNA fragment released from pT7-5lacY by XhoI, Klenow treatment, and then PvuI. Plasmid pCLtrpE-Δ198 was made by ligating the 5.5-kb DNA fragment of pCLftsY (treated with EcoRI, BssHII, and Klenow fragment) with the 1.4 kb of pATH2 (treated with BamHI, PvuII, and Klenow fragment). All the gene constructs were analyzed by restriction enzymes and DNA sequencing.
Growth of Cells, Expression, and Immunoblotting.
E. coli UT5600 harboring pT7-5 or pT7-5 containing various ftsY gene constructs was grown overnight at 37°C in Luria–Bertani medium (LB) supplemented with ampicillin (100 μg/ml). Cultures were then diluted 1:100, grown to 0.6 OD600 unit, and induced by 0.5 mM isopropyl β-d-thiogalactoside (IPTG) for 2 hr. Cells were harvested, washed in buffer A (7% sucrose/100 mM Tris⋅HCl, pH 8/50 mM NaCl/1 mM EDTA/1 mM phenylmethylsulfonyl fluoride) and resuspended in the same buffer. Cell suspensions (500 μl of 10 OD420 units) were sonicated, and cell debris were removed by centrifugation (2 min at 13,000 rpm). E. coli (N4156::pAra14-FtsY′) cells harboring plasmid pCL1921 or its derivatives as indicated were grown in LB broth with ampicillin (100 μg/ml), spectinomycin (100 μg/ml) and arabinose (0.2%). The overnight cultures were washed once in LB broth and diluted (1:2000) in LB containing the same antibiotics with or without arabinose as indicated. Cell density (OD600) was measured every hour for the construction of growth curves. For expression studies, cells were grown without arabinose to 0.15 OD600 unit and induced by 0.5 mM IPTG for 2 hr. Cells were then harvested and treated as described above. Membranes were collected by ultracentrifugation (30 min at 150,000 × g). Protein concentrations in the extracts and in the membrane fractions were measured according to a modified Lowry procedure in the presence of 2.5% SDS using bovine serum albumin as a standard. An aliquot (10 μg of proteins or as specified) from each sample was then mixed with 2× sample buffer, incubated at 37°C for 15 min, and subjected to SDS/PAGE (7.5–10% as indicated). After electroblotting, the nitrocellulose was incubated with anti-FtsY or anti-LacY serum as indicated, washed twice, and incubated with goat anti-rabbit antibodies conjugated to horseradish peroxidase. The nitrocellulose paper was briefly soaked in the fluorescent substrate solution and exposed to film for ≈10 sec.
RESULTS AND DISCUSSION
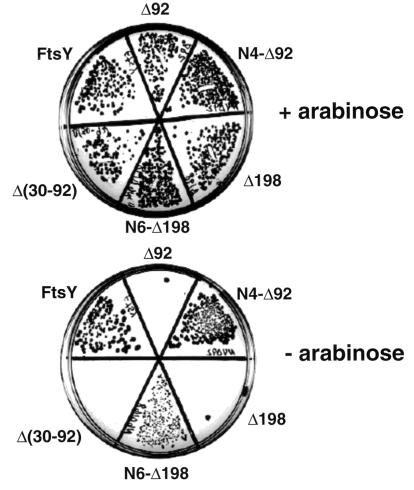
We have speculated that like in SR-α (16), the membrane targeting and attachment domain of FtsY is located within the N terminus. To identify this domain, full ftsY DNA was amplified from the chromosome and inserted into pT7-5(lacY) (17), instead of the lacY coding region, under the control of the lac promoter/operator (plasmid pTftsY). This plasmid was used to construct N-terminally manipulated FtsY mutants, leaving the conservative C-terminal NG domain intact (Fig. 1B, stippled boxes). One mutant contains an internal deletion of codons 30–92 of ftsY [Fig. 1C, Δ(30–92)] and the other two are N-terminally truncated versions of FtsY (Fig. 1C, Δ92 and Δ198). To test the function of the mutants we used the E. coli strain N4156::pAra14-FtsY′ (13) which harbors a chromosomal copy of the essential ftsY gene under the control of the tight araB promoter (instead of the native ftsY) and therefore requires arabinose for growth. The deleterious effects of arabinose depletion on cell growth and on the expression of FtsY have been analyzed previously (7, 13). One technical point worth mentioning is that the ftsY gene constructs were transferred with their lac promoters to the low copy-number plasmid, pCL1921 (18), which is compatible with the polA− phenotype of strain N4156::pAra14-FtsY′ [thereby generating plasmids pCLftsY, pCLΔ(30–92), pCLΔ92 and pCLΔ198]. When competent N4156::pAra14-FtsY′ cells are transformed with pCLftsY, they form wild-type-like single colonies on LB plates without arabinose and with 0.1 mM IPTG (Fig. 2). However, when transformed with plasmids encoding the deletion mutants, single colonies are formed only on arabinose supplemented LB plates (Fig. 2).
Figure 2.
Formation of single colonies on agar plates with or without arabinose. Competent E. coli N4156::pAra14-FtsY′ (13) was transformed with pCL1291 derivatives harboring the various ftsY gene constructs as indicated. The transformants were incubated in LB supplemented with 0.2% arabinose at 37°C. One hour later, the recovered transformants were centrifuged, washed once in LB without arabinose, and spread on LB plates containing ampicillin (100 μg/ml), spectinomycin (100 μg/ml), IPTG (0.1 mM), and with or without arabinose. Photographs were taken after incubation for 36 hr at 37°C.
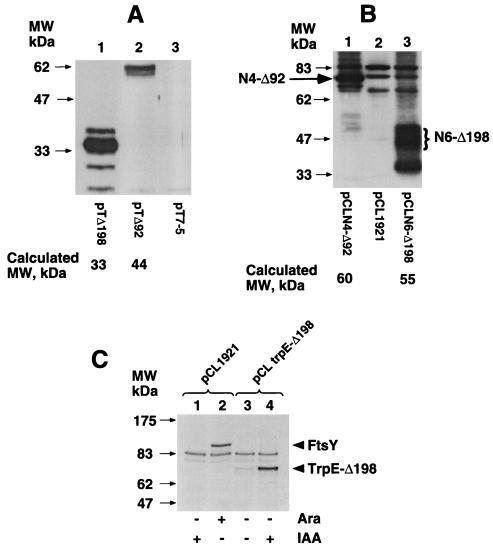
Clearly, one explanation for the observation that the mutants do not support growth of FtsY-depleted cells is that the deleted sequences are essential for enzymatic activity, or alternatively are required for targeting and attachment of FtsY to the membrane. To exclude a more trivial possibility that the mutants are simply not expressed from the lac promoter, immunoblots were carried out with extracts prepared from E. coli cells (ftsY+) transformed with pT7-5 (control), pTΔ92, or pTΔ198, using anti-FtsY antibodies. The results demonstrate that mutants Δ198 and Δ92 are expressed from the lac promoter (Fig. 3A, lanes 1 and 2, respectively). Interestingly, unlike wild-type FtsY which migrates extremely slowly (apparent molecular weight 97 kDa compared with a calculated molecular weight of 54 kDa; Fig. 3C, lane 2; see also refs. 13 and 20), the migration of mutant Δ92 is only moderately slower (≈58 kDa) compared with a calculated molecular weight of 44 kDa and Δ198 migrates according to its calculated molecular weight (33 kDa). Therefore, the aberrant migration of FtsY in SDS/PAGE is determined by the N-terminal sequence.
Figure 3.
Expression of FtsY mutants, LacY–FtsY hybrids, and TrpE-Δ198 hybrid. (A) E. coli UT5600 cells transformed with the indicated plasmid were grown overnight, diluted, and induced by IPTG at the exponential growth phase. Cell extracts were subjected to immunoblot analysis with anti FtsY antibodies. (B) E. coli N4156::pAra14-FtsY′ transformed with the indicated plasmids were grown overnight, diluted into LB without arabinose, and induced by IPTG at OD600 of ≈0.15. Membrane fractions were separated by SDS/PAGE and subjected to immunoblot analysis with anti-FtsY antibodies. (C) E. coli N4156::pAra14-FtsY′ transformed with the indicated plasmids were grown overnight, diluted into LB without arabinose, and 3 hr later induced for 2 hr by 0.2% arabinose (Ara, for expression of FtsY, lane 2) or by 100 μg per ml of indoleacrylic acid (IAA, for expression of hybrid TrpE-Δ198, lane 4) (19).
The observations that we have described lead to the conclusion that the N-terminal domain of FtsY carries an essential function. One possibility referred to above is that this domain mediates membrane targeting and attachment of FtsY. To test this hypothesis we constructed hybrids between the N-terminally truncated FtsY mutants and unrelated integral membrane proteins which have the ability to guide and anchor the mutant proteins to the membrane. We used integral membrane polypeptides from the LacY of E. coli because it is well characterized structurally (21) and contains 12 hydrophobic membrane spanning segments (Fig. 1D). The fusion joints, connecting N-terminal fragments of LacY with the FtsY mutants, were placed at known cytoplasmic locations according to the secondary structure of the permease which has been recently updated (22). Specifically, mutants Δ92 and Δ198 were fused to the cytoplasmic loops between transmembrane segments 4 and 5 (to form N4-Δ92), and 6 and 7 (to form N6-Δ198), respectively (Fig. 1D). Immunoblot analysis with anti-FtsY antibodies demonstrates, despite of the occasional, nonspecific cross reactivity of the polyclonal anti FtsY antibodies with some unrelated proteins (Fig. 3B, lane 2), that the two fusion proteins are expressed and present in the membrane fraction (Fig. 3B, lanes 1 and 3). As expected, hybrid N4-Δ92 contains part of the acidic N-terminal domain of FtsY and therefore migrates slower (as a 74-kDa band, Fig. 3B, lane 1) than its theoretical molecular weight (60 kDa). Hybrid N6-Δ198 which does not contain the N-terminal acidic residues of FtsY, migrates as expected (55 kDa, Fig. 3B, lane 3), with a characteristic broad band, as do many other integral membrane proteins. A putative proteolytic product which migrates similarly to mutant Δ198 is also apparent (Fig. 3B, lane 3).
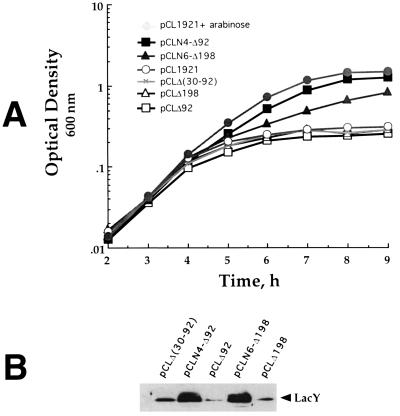
To test the activity of these hybrids, the genes were transferred into pCL1921 (as described above for the deletion mutants) and the resulting plasmids were used to transform N4156::pAra14-FtsY′ cells. The transformants were plated on LB plates with or without arabinose and IPTG. Both LacY–FtsY expressing transformants were able to form colonies on plates without arabinose as well as on plates with arabinose (Fig. 2). Cells expressing N6-Δ198 produce smaller colonies, indicating that although this hybrid complements FtsY-depleted cells, it might not be fully functional or alternatively its expression might be toxic to some extent. The ability of the truncated FtsY mutants and of the LacY–FtsY hybrids to support growth of FtsY-depleted cells was assayed also quantitatively, in liquid media (Fig. 4A). For this purpose, overnight cultures grown in LB containing arabinose were harvested, washed twice, and diluted 1:2,000 into fresh LB without arabinose. The growth of cells harboring only the vector (pCL1921) or mutants Δ92, Δ198, and Δ(30–92) ceases after 4–5 hr at 0.3 OD600 unit. Mutants N4-Δ92 and N6-Δ198 continue to grow up to cell densities of 1.5, 1.4, and 0.8 OD600 unit, respectively, in the absence of arabinose. In fact, the growth of N4-Δ92 in the absence of arabinose is similar to the growth in the presence of arabinose of control cells harboring the vector alone. To exclude the possibility that any protein can “activate” the NG domain of FtsY if fused to its N terminus, an important control experiment was conducted. We constructed a hybrid composed of the N-terminal 326-amino acid residues of the cytoplasmic protein, TrpE, fused to the N terminus of Δ198. Western blot analysis indicates that the hybrid (TrpE-Δ198, Fig. 3C, lane 4) and wild-type FtsY (Fig. 3C, lane 2) are expressed to a comparable level. However, TrpE-Δ198 is unable to support growth of FtsY depleted cells (data not shown). Therefore, the NG domain of FtsY is functional only when attached to a membrane protein but not to a soluble protein.
Figure 4.
Functional complementation of FtsY depleted cells by LacY–FtsY hybrids. E. coli (N4156::pAra14-FtsY′) cells harboring the indicated plasmids were grown overnight in LB with arabinose, washed once in LB, and diluted to OD600 = 0.005 in LB with or without arabinose as indicated. Growth curves (A) were constructed from the average of two independent experiments. After 4 hr, 10 ml samples were transferred from chosen cultures into separate Erlenmeyers and induced by 0.5 mM IPTG for 2 hr. Membranes were prepared from the induced cells and subjected to Western blot analysis using anti-LacY antibodies.
Since it has been shown that FtsY is essential for expression of integral membrane proteins, including LacY (7), we tested the ability of the FtsY mutants and the LacY–FtsY hybrids to support LacY expression from the chromosome, in FtsY-depleted cells. Cultures were treated as in the growth experiments and 4 hr after FtsY depletion the cultures were induced by IPTG, and membrane preparations were analyzed by immunoblotting using anti-LacY antibodies (7). As shown in Fig. 4B, the amount of LacY in membranes prepared from cells harboring pCLN4-Δ92 or pCLN6-Δ198 is significantly higher than in cells harboring pCLΔ(30–92), pCLΔ198, or pCLΔ92 (see also ref. 7). Under the same conditions, the expression of the cytoplasmic protein, β-galactosidase, is not affected by FtsY depletion (data not shown; see also ref. 7). Therefore, the function of the inactive FtsY mutants, Δ92 and Δ198, is restored when they are fused to cytoplasmic loops of an integral membrane protein. We conclude from these experiments that for proper function, FtsY must be targeted to the membrane and that its N-terminal domain is indeed involved in this process.
Despite the functional similarity between the N-terminal domains of SR-α (16) and FtsY (shown here) in mediating peripheral interaction with the membrane, they differ markedly in their primary structures and overall chemical properties: The N-terminal domain of FtsY contains a large number of acidic amino acid residues (mainly glutamic acids) throughout the entire sequence, whereas the membrane binding domain of SR-α contains two hydrophobic stretches and a basic region. These differences support a suggestion that the two proteins use different membrane targeting and attachment mechanisms or recognize and interact with different integral membrane counterparts. In both cases, however, the SRP receptor is probably responsible for an essential step late during the targeting process, by placing the ribosome–nascent chain–SRP complex near the translocation machinery on the target membrane. An important challenge is to clarify the mechanism and to identify the proteins that mediate the following step in the targeting pathway where the translation machinery is transferred from the SRP system to the translocation apparatus.
Acknowledgments
This work was supported by the Leo and Julia Forchheimer Center for Molecular Genetics, the Weizmann Institute of Science, and the Dr. Josef Cohn Minerva Center for Biomembrane Research.
ABBREVIATIONS
- SRP
signal recognition particle
- SR
SRP-receptor
- LB
Luria–Bertani medium
- IPTG
isopropyl β-d-thiogalactoside
References
- 1.Nunnari J, Walter P. Curr Opin Cell Biol. 1992;4:573–580. doi: 10.1016/0955-0674(92)90074-m. [DOI] [PubMed] [Google Scholar]
- 2.Rapoport T A. Science. 1992;258:931–936. doi: 10.1126/science.1332192. [DOI] [PubMed] [Google Scholar]
- 3.Wickner W T, Lodish H F. Science. 1985;230:400–407. doi: 10.1126/science.4048938. [DOI] [PubMed] [Google Scholar]
- 4.Römisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B. Nature (London) 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein H D, Poritz M A, Strub K, Hoben P J, Brenner S, Walter P. Nature (London) 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 6.Ribes V, Römisch K, Giner A, Dobberstein B, Tollervey D. Cell. 1990;63:591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- 7.Seluanov A, Bibi E. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 8.Ulbrandt N D, Newitt J A, Bernstein H D. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 9.de Gier J-W L, Mansournia P, Valent Q A, Phillips G J, Lurink J, von Heijne G. FEBS Lett. 1996;399:307–309. doi: 10.1016/s0014-5793(96)01354-3. [DOI] [PubMed] [Google Scholar]
- 10.Macfarlane J, Muller M. Eur J Biochem. 1995;233:766–771. doi: 10.1111/j.1432-1033.1995.766_3.x. [DOI] [PubMed] [Google Scholar]
- 11.Montoya G, Svensson C, Luirink J, Sinning I. Nature (London) 1997;385:365–368. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- 12.Gill D R, Salmond G P C. Mol Gen Genet. 1987;210:504–508. doi: 10.1007/BF00327204. [DOI] [PubMed] [Google Scholar]
- 13.Luirink J, Hagen-Jongman C M, der Weijden C C, Oudega B, High S, Dobberstein B, Kusters R. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young J C, Andrews D W. EMBO J. 1996;15:172–181. [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J D, Tajima S, Lauffer L, Walter P. J Cell Biol. 1995;128:273–282. doi: 10.1083/jcb.128.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young J C, Ursini J, Legate K R, Miller J D, Walter P, Andrews D W. J Biol Chem. 1995;270:15650–15657. doi: 10.1074/jbc.270.26.15650. [DOI] [PubMed] [Google Scholar]
- 17.Bibi E, Kaback H R. Proc Natl Acad Sci USA. 1990;87:4325–4329. doi: 10.1073/pnas.87.11.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lerner C G, Inouye M. Nucleic Acids Res. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koerner T J, Hill J E, Myers A M, Tzagoloff A. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- 20.Gill D R, Salmond G P C. Mol Microbiol. 1990;4:575–583. doi: 10.1111/j.1365-2958.1990.tb00626.x. [DOI] [PubMed] [Google Scholar]
- 21.Kaback H R, Frillingos S, Jung H, Jung K, Prive G G, Ujwal M L, Weitzman C, Wu J, Zen K. J Exp Biol. 1994;196:183–195. doi: 10.1242/jeb.196.1.183. [DOI] [PubMed] [Google Scholar]
- 22.Ujwal M L, Jung H, Bibi E, Manoil C, Altenbach C, Hubbell W L, Kaback H R. Biochemistry. 1995;34:14909–14917. doi: 10.1021/bi00045a036. [DOI] [PubMed] [Google Scholar]