Abstract
Background
The silver homologue(SILV) gene plays a major role in melanosome development. SILV is a target for studies concerning melanoma diagnostics and therapy in humans as well as on skin and coat color pigmentation in many species ranging from zebra fish to mammals. However, the precise functional cellular mechanisms, in which SILV is involved, are still not completely understood. While there are many studies addressing SILV function upon a eumelaneic pigment background, there is a substantial lack of information regarding the further relevance of SILV, e.g. for phaeomelanosome development.
Results
In contrast to previous results in other species reporting SILV expression exclusively in pigmented tissues, our experiments provide evidence that the bovine SILV gene is expressed in a variety of tissues independent of pigmentation. Our data show that the bovine SILV gene generates an unexpectedly large number of different transcripts occurring in skin as well as in non-pigmented tissues, e.g. liver or mammary gland. The alternative splice sites are generated by internal splicing and primarily remove complete exons. Alternative splicing predominantly affects the repeat domain of the protein, which has a functional key role in fibril formation during eumelanosome development.
Conclusion
The expression of the bovine SILV gene independent of pigmentation suggests SILV functions exceeding melanosome development in cattle. This hypothesis is further supported by transcript variants lacking functional key elements of the SILV protein relevant for eumelanosome development. Thus, the bovine SILV gene can serve as a model for the investigation of the putative additional functions of SILV. Furthermore, the splice variants of the bovine SILV gene represent a comprehensive natural model to refine the knowledge about functional domains in the SILV protein. Our study exemplifies that the extent of alternative splicing is presumably much higher than previously estimated and that alternatively spliced transcripts presumably can generate molecules of deviating function compared to their constitutive counterpart.
Background
The silver homologue(SILV) gene has been a target for many investigations concerning development of melanosomes, which are the specific pigment carrying compartments within melanophores, e.g. in melanocytes. In humans, SILV plays a major role in studies regarding melanoma diagnosis and therapy, because SILV is a sensitive melanoma marker on transcript and protein level [1,2] and represents a melanoma specific antigen recognized by tumor infiltrating cytotoxic T lymphocytes [3]. The SILV protein also known as PMEL17, GP100 or ME20 [4] is crucial for proper formation and maturation of melanosomes. In stage II melanosomes, processed SILV protein aggregates to form fibrils, to which presumably the eumelanin pigment is attached [5]. Due to its role in melanosome development, SILV has also been subject to several studies investigating the genetic background of coat color.
Coat color phenotype in mammals is dependent on a series of genes determining distribution of melanocytes (e.g. KIT, [6]), synthesis of the two essential pigments, eumelanin (black) and phaeomelanin (red) (e.g. MC1R, [7]), and intra- and intercellular transport mechanisms of proteins relevant for coat color expression (e.g. MATP, [8]). In cattle, variants in the MC1R gene result in exclusively eumelaneic (black) or phaeomelaneic (red) skin [9]. Spotted individuals exhibit delimited, white skin areas lacking melanocytes similar to piebald mice [10]. Similar to other species, dilution loci resulting in a diluted type of the original coat color are known in cattle, e.g. the Dilution locus in the Charolais breed (Dc). The putative role of SILV for various coat color dilution loci was described in a number of species, including mouse, dog, chicken, horse, and cattle [11-16]. There is concurring indication that concerning coat color the effect of a mutation in the SILV gene in mouse, horses and dogs seems to be restricted to the dilution of eumelaneic pigment. This is underlined by experiments in mice indicating that SILV expression seems to be restricted to melanocytes expressing eumelanin [17]. In cattle, however, there is still some controversy regarding the potential role of SILV in phaeomelanosome development, because up to now there is no unequivocal experimental evidence rejecting or propagating SILV mutations as causal background for phaeomelanin dilution [14,18]. First reports on splice variants for the human SILV gene [19] and a retroposon insertion in intron 10 of the canine SILV gene that seems to affect the correct splicing of the gene in merle dogs [12] raise the question whether putative splice variants of the SILV gene may be specifically involved in melanosome development. Comprehensive studies in humans revealed that alternative splicing is a frequent mechanism altering spatial expression pattern and function of proteins [20,21].
In the present study, we present a comprehensive description of the complex expression pattern of the bovine SILV gene in pigmented and non-pigmented tissues. Multiple transcript variants affecting functional key domains of the SILV protein indicate that the bovine SILV gene may serve as a model for investigations about alternative splicing to generate molecules of obviously deviating function compared to constitutive transcripts.
Results
Identification of the SILV transcription start site
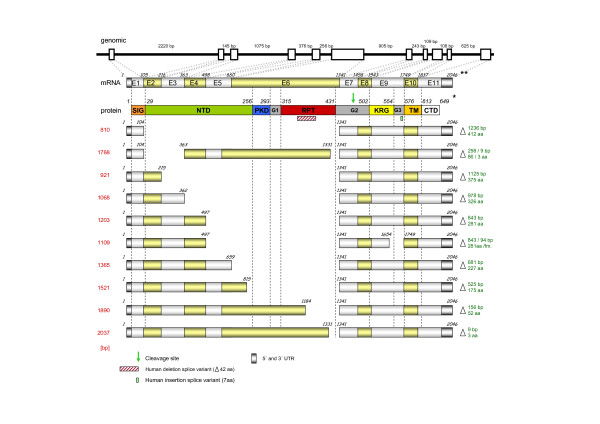
Analysis of several 5'RACE clones from total RNA of eumelaneic, non-dilute (black) skin indicated a sharp peak of transcription start sites (TSS) at position -28 bp to the A of the translation start ATG of the SILV gene (GenBank: EF065525). No further promoters were detected. The SILV transcription start obtained in this study adds an additional 7 bp to the previously deposited bovine SILV cDNA sequence [14]. The cDNA sequence generated by this experiment based on CAP carrying full length mRNA confirmed previous findings about the structure of the bovine SILV gene comprising 2046 bp organized in 11 exons (Figure 1) and encoding 649 amino acids. Aligning the obtained SILV cDNA sequence with the bovine genomic contig NCBI: NW_001495046 [22]) indicated that the bovine SILV gene spans a total of 8107 bp with introns sizes ranging from 108 to 2220 bp. A TTATA motif representing the putative TATA box of the SILV promoter is located 30 bp upstream of the transcription initiation site.
Figure 1.
Structure of the bovine SILV mRNA including alternative transcripts. Position of exons (open boxes) and introns (solid line) in the SILV genomic sequence are indicated. Protein domains according to [4,31] are shown: SIG: signal peptide, NTD: N-terminal domain, PKD: polycystic kidney domain, RPT: repeat domain, KRG: Kringle-like domain, TM: transmembrane domain, CTD: C-terminal domain, G1, G2, G3: undefined domains. E: SILV exon.* Numbers in the SILV protein diagram represent the first amino acid of the respective domain. ** Numbers for the SILV transcripts represent the first nucleotide for the respective exon or the last constitutive nucleotide of an alternative transcript. fm: frame shift mutation. Length [in bp] of the alternative c SILV transcripts is indicated in red (left), the missing part relative to the constitutive SILV transcript (nucleotides and amino acids) is indicated in green (right).
Detection of splice variants
Multiple RT-PCR fragments were detected when amplifying SILV cDNA from RNA of eumelaneic (black), non-diluted skin with primers in the 5' and 3' UTR of the SILV gene (5UTRF1 – E11R2; Table 1, 2). This diversity of transcripts was also obtained with a primer combination spanning exon 1 to exon 7 (E1F3 – E7R1) of the bovine SILV gene (Figure 2). While the constitutive transcripts (2039 bp or 1430 bp, respectively, confirmed by sequencing) were dominating, further fragments up to 900 bp smaller than the constitutive transcript were identified with both primer pairs. In contrast, cloned SILV constitutive cDNA yielded only the expected 2039 or 1430 bp PCR fragment, respectively.
Table 1.
Primers for reverse transcription and amplification in the bovine SILV gene
| Name | Positiona | Exon | Sequence (5' – 3') |
| E1F3 | 69 | 1 | TGATGGGTGTTCTTCTGGCTG |
| E3F1 | 280 | 3 | CTCTATTGCCTTGCACTTTCC |
| E3R1 | 362 | 3 | CATTGATGATGGTGTTGTTGG |
| E5R1 | 656 | 5 | TAATGGTGAAGGCTGAACTGG |
| E5F1 | 575 | 5 | AACATGGAAGTGACTGTCTACC |
| E7R1 | 1430 | 7 | AGCCATAGCGATACAGAACAC |
| E7F1 | 1360 | 7 | GGATGACACTGCCACCTTAG |
| E11R2 | 2046 | 11 | AGGGAAGACCAGAGAAAAGAC |
| E6F2 | 952 | 6 | CACTACAGATAGGCATGTGAC |
| E6R2 | 1030 | 6 | GCCCATGACTTCTGTAGTAGG |
| E10F1 | 1773 | 10 | CTCCTCTGTTCGTGGGCATC |
| E9F1 | 1643 | 9 | GTTTTGCACCAGGTACTGAAG |
| RACE_E7N | 1441 | 7 | CAGGGTGAGGGAAAAGGAGCCATAG |
| 5UTRF1 | 8 | 1 | GTTGCTGGAAGGAAGAACAGG |
| SPex23F1 | 88 | 1/4 | TGTAGGGACCACAGAAGGGAG |
| E5R1 | 656 | 5 | TAATGGTGAAGGCTGAACTGG |
| SPex56F1 | 481 | 4/7 | TGTCTGGAAGACCTGGGGCT |
| E9R2 | 1587 | 9 | TGACACCCTGGCGATGAGATG |
| E5F2 | 565 | 5 | GGGCACATATAACATGGAAGTG |
| SPex6R1 | 1358 | 7/5 | GCAGGGGACTCAGGGAGCCAG |
| SPex6AF1 | 798 | 6/7 | CCTACACCTGGGACTTTGGCT |
| SPex6BF1 | 1167 | 6/7 | CAACTGCAAAAGCTACAGGCT |
| E7F2 | 1409 | 7 | TGTGTTCTGTATCGCTATGGCTC |
| SPex9R1 | 1767 | 9/10 | CTGAGGCCTGCTTCTTGCCCTG |
a Position within the bovine SILV mRNA (according to GenBank: EF065525). Underlined, bold sequence: nucleotides specific for alternative splice site.
Table 2.
Regions of the bovine SILV mRNA tested for expression by RT-PCR in skin and non-pigmented body tissues
| Primer combination | Region | Expected fragment [bp] | Observed fragment [bp] | ||
| Eumelaneic skin | Phaeomelaneic skin | Non-pigmented tissues | |||
| 5UTRF1 – E11R2 | exon 1 – exon 11 | 2039 | 2039, 1102 and multiple further fragments | n.a. | n.a. |
| E1F3 – E7R1 | exon 1 – exon 7 | 1430 | 1430 and further multiple bands | n.a. | n.a. |
| E1F3 – E3R1 | exon 1 – exon 3 | 294 | 294 | 294 | n.a. |
| E3F1 – E5R1 | exon 3 – exon 5 | 377 | 377 | 377 | n.a. |
| E5F1 – E7R1 | exon 5 – exon 7 | 856 | 856, 700, 331, 175 | 856, 700, 331, 175 | 856, 700, 331, 175 (pg, tg, ki, ag, li, lu, he, br, ru, if, sf, sf, mg, fu, co, je, mu) |
| E7F1 – E11R2 | exon 7 – exon 11 | 687 | 687; 593 | 687; 593 | n.a. |
| E6F2 – E7R1 | exon 6 – exon 7 | 479 | 479, 324 | 479, 324 | n.a. |
| E5F1 – E6R2 | exon 5 – exon 6 | 456 | 456 | 456 | n.a. |
| E9F1 – E11R2 | exon 9 – exon 11 | 404 | 404 | 404 | 404 (pg, tg, ki, ag, li, lu, he, br, ru, if, sf, sf, mg, fu, co, je, mu) |
| E10F1 – E11R2 | exon 10 – exon 11 | 274 | 274 | 274 | n.a. |
| E1F3 – E5R1 | exon 1 – exon 5 | 588 | n.a. | n.a. | 588 (pg, tg, ki, ag, li, lu, he, br, ru, if, sf, sf, mg, fu, co, je, mu) |
pg: pituitary gland, tg: thyroid gland, ki: kidney, ag: adrenal gland, li: liver, lu: lung, he: heart, br: brain, ru: rumen, if: intestinal fat, sf: subcutaneous fat, pf: perirenal fat, mg: mammary gland, du: duodenum, co: colon, je: jejunum, mu: skeletal muscle. n.a.: not analyzed
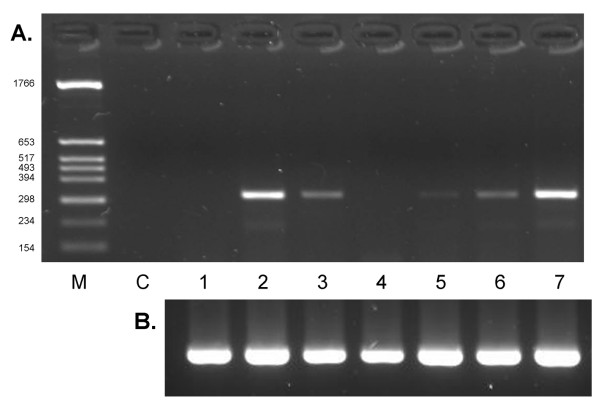
Figure 2.
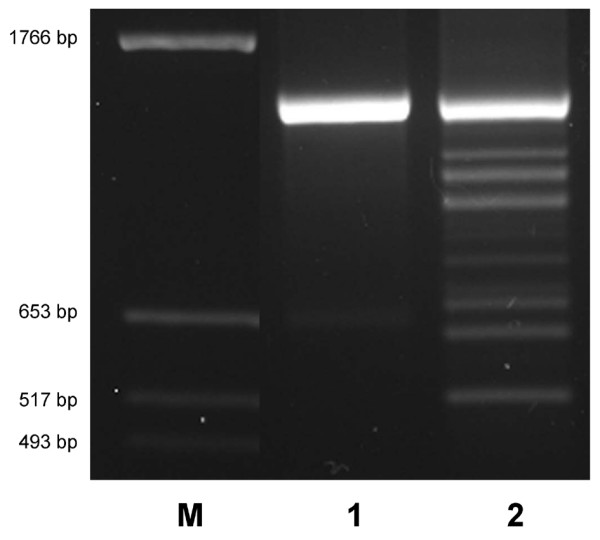
RT-PCR products in the SILV gene in eumelaneic black bovine skin. RT-PCR products were generated from pigmented, eumelaneic (black) bovine skin with primers spanning SILV exon 1 to exon 7 (E1F3/E7R1). M: DNA marker, 1: cloned constitutive SILV cDNA, 2: cDNA from eumelaneic (black), non-dilute (Dcd/Dcd) skin
A series of PCR amplifications in SILV cDNA samples with primers dissecting the transcribed SILV sequence into smaller segments enabled a better discrimination of the generated fragments. Exons 6, 8 and 9 were identified as putative regions for alternative splicing events, because at least two RT-PCR fragments could be unambiguously discriminated with primer combinations spanning exon 5 – 7 (E5F1 – E7R1) and exon 7 – 11 (E7F1 – E11R2). In contrast, only single RT-PCR fragments were observed for the primer combinations covering exon 1 – 3, exon 3 – 5, exon 5 – 6, exon 9 – 11 and exon 10 – 11. Notably, all identified fragments were present in all differentially pigmented skin samples investigated (Table 2, Figure 3). Our data show that the expression of the bovine SILV gene is not restricted to eumelaneic (black), non-dilute (Dcd/Dcd) skin, but also occurs in skin expressing exclusively phaeomelanin. Furthermore, also white skin sections of spotted individuals with eumelaneic or phaeomelaneic background showed SILV gene expression as well as the crème white skin of a eumelaneic homozygous dilute DcD/DcD individual.
Figure 3.
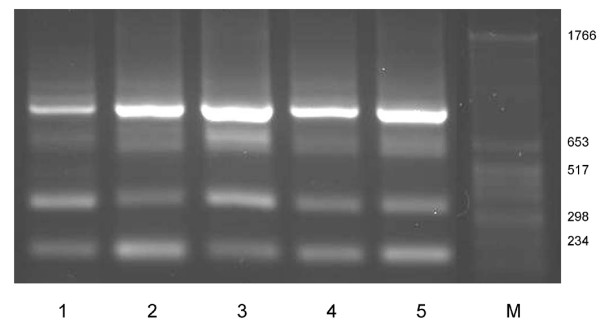
Expression of SILV in eumelaneic and phaeomelaneic bovine skin. RT-PCR products for the bovine SILV region spanning exon 5 to exon 7 were generated with primer E5F1 – E7R1 in differentially pigmented bovine skin. 1: non-pigmented skin from a heterozygous dilute (DcD/Dcd), eumelaneic (ED/ED) spotted individual; 2: pigmented skin from a heterozygous dilute (DcD/Dcd), eumelaneic (ED/ED) spotted individual; 3: crème white skin from a homozygous dilute (DcD/DcD), eumelaneic (ED/ED) non-spotted individual; 4: non-pigmented skin from a heterozygous dilute (DcD/Dcd), phaeomelaneic (Ee/Ee) spotted individual; 5: pigmented skin from a homozygous non-dilute (Dcd/Dcd), eumelaneic (ED/Ee) spotted individual; M: marker
The SILV transcripts additional to the constitutive fragment that had been identified by RT-PCR were isolated from the agarose gel and sequenced to reveal the specific DNA sequence of the different fragments. Alignment of the obtained sequences to the reference constitutive cDNA (GenBank: EF065525) showed that the additional SILV transcripts lacked different parts of the SILV mRNA (Figure 1). With primers spanning exon 5 to exon 7 (E5F1 – E7R1), three alternative transcripts (according to GenBank: EF065525) were characterized: Δ660–1340 lacking 681 bp corresponding to the entire exon 6, Δ816–1340 lacking 525 bp from the 3' end of exon 6 and Δ1185–1340 lacking 156 bp from the 3' end of exon 6. With primers spanning exon 7 to exon 11 (E7R1 – E11R2), we obtained an alternative transcript Δ1655–1748 lacking 94 bp from the 3' end of exon 9. Furthermore, sequencing of an additional RT-PCR transcript amplified with primers from exon 1 and exon 11 (5UTRF1 – E11R2) detected a SILV cDNA Δ498–1340/Δ1655–1748 lacking exon 5, exon 6 and 94 bp from the 3' end of exon 9 (Figure 1).
Diversity in SILV transcripts was also confirmed in the course of our 5'RACE experiments. Several clones containing inserts smaller than the expected size (1441 bp) were detected. Sequencing of the respective clones revealed SILV cDNAs starting from the identified transcription start, but devoid of distinct other regions in the remaining SILV mRNA. On the one hand, we received transcripts confirming already detected alternative splice variants e.g., Δ498–1340, Δ660–1340, Δ816–1340, and Δ1185–1340. Additionally, we identified two further internal splice sites generating a transcript Δ105–362 (skipping exons 2 and 3)/Δ1332–1340 (lacking 9 bp at the 3' end of exon 6 (Figure 1).
In order to obtain indication whether combinations of alternative splicing events occurred, we sequenced a collection of clones from a plasmid library containing SILV RT-PCR products of variable size generated by cDNA amplification of eumelaneic (black), non-dilute (Dcd/Dcd) skin with primers spanning exon 1 to exon 7 (E1F3 – E7R1). Alignment of the generated sequences to the SILV constitutive transcript confirmed previously detected transcripts and yielded an additional new series of transcripts (Figure 1) with deleted exons: Δ363 – 1340 (lacking exon 4 – 6), Δ216 – 1340 (lacking exon 3 – 6) and Δ105 – 1340 (lacking exon 2 – 6).
Only some alternative internal splice sites completely conformed to the GT-AG rule for splice donor and splice acceptor sites. Notably, the alternative splice sites did not affect the reading frame of the transcript except for Δ498–1340/Δ1655–1748. In the transcript Δ498–1340/Δ1655–1748, a premature stop codon is generated at position c.1805.
Confirmation of SILV splice variants in differentially pigmented skin
In order to confirm alternative splice sites in the bovine SILV gene, we developed splice variant specific RT-PCR tests for an exemplary subset of the detected splice variants (Table 3, Figure 4). Splice variant profiling revealed the alternative splice variants Δ498–1340, Δ816–1340 and Δ1185–1340 in all differentially pigmented skins (Table 4). In contrast, we obtained a specific pattern for Δ105–362, which was not detected in non-pigmented, white skin of spotted individuals heterozygous for the dilute locus (DcD/Dcd) regardless whether on a eumelaneic or a phaeomelaneic background. Notably, non-pigmented white skin of a spotted, homozygous non-dilute (Dcd/Dcd) individual exhibited this splice site albeit at a lower level than the pigmented counterpart. Skin with a crème white coat color characteristic for individuals homozygous at the dilution locus DcD/DcD also displayed the alternative splicing Δ105-362. It has to be noted that crème white skin has an essentially pigmented background with melanocytes [23], however, pigmentation is diluted to almost invisibility. In contrast, non-pigmented skin of spotted or piebald individuals is devoid of melanocytes resulting in a white coat color [10].
Table 3.
Splice site specific RT-PCR primer combinations applied for the detection of alternative bovine SILV transcripts
| Alternative splice site | Upstream primer | Downstream primer | Expected alternative transcript [bp] |
| Δ105–362 | SPex23F1 TGTAGGGACCACAGAAGGGAG | E5R1 TAATGGTGAAGGCTGAACTGG | 311 |
| Δ498–1340 | SPex56F1 TGTCTGGAAGACCTGGGGCT | E9R2 TGACACCCTGGCGATGAGATG | 264 |
| Δ660–1340 | E5F2 GGGCACATATAACATGGAAGTG | SPex6R1 GCAGGGGACTCAGGGAGCCAG | 113 |
| Δ816–1340 | SPex6AF1 CCTACACCTGGGACTTTGGCT | E9R2 TGACACCCTGGCGATGAGATG | 265 |
| Δ1185–1340 | SPex6BF1 CAACTGCAAAAGCTACAGGCT | E9R2 TGACACCCTGGCGATGAGATG | 265 |
| Δ1655–1748 | E7F2 TGTGTTCTGTATCGCTATGGCTC | SPex9R1 CTGAGGCCTGCTTCTTGCCCTG | 265 |
Underlined, bold sequence: nucleotides specific for alternative splice site.
Figure 4.
Splice site specific SILV RT-PCR in differentially pigmented bovine skin. A. Detection of bovine SILV splice variant Δ105–362 in differentially pigmented skin. RT-PCR products generated by splice site specific RT-PCR with primers SPex23F1 – E5R1. M: marker; C: negative control, 1: non-pigmented skin from a heterozygous dilute (DcD/Dcd), eumelaneic (ED/ED) spotted individual; 2: pigmented skin from a heterozygous dilute (DcD/Dcd), eumelaneic (ED/ED) spotted individual; 3: crème white skin from a homozygous dilute (DcD/DcD), eumelaneic (ED/ED) non-spotted individual ; 4: non-pigmented skin from a heterozygous dilute (DcD/Dcd), phaeomelaneic (Ee/Ee) spotted individual; 5: pigmented skin from a heterozygous dilute (DcD/Dcd), phaeomelaneic (Ee/Ee) spotted individual; 6: non-pigmented skin from a homozygous non-dilute (Dcd/Dcd), eumelaneic (ED/Ee) spotted individual; 7: pigmented skin from a homozygous non-dilute (Dcd/Dcd), eumelaneic (ED/Ee) spotted individual. B. RT-PCR products generated with GAPDH primers.
Table 4.
Expression of bovine SILV splice variants by splice site specific RT-PCR in differentially pigmented skin and non-pigmented tissues
| Tissues | Δ105–362 | Δ498–1340 | Δ660–1340 | Δ816–1340 | Δ1185–1340 | Δ1655–1748 |
| Non-pigmented, heterozygous dilute, eumelaneic | - | + | + | + | ++ | + |
| pigmented, heterozygous dilute, eumelaneic | ++ | ++ | + | ++ | ++ | ++ |
| pigmented, homozygous dilute, eumelaneic | + | ++ | + | ++ | ++ | ++ |
| Non-pigmented, heterozygous dilute, phaeomelaneic | - | + | + | + | ++ | + |
| Pigmented, heterozygous dilute, phaeomelaneic | + | ++ | + | ++ | ++ | ++ |
| Non-pigmented, homozygous non-dilute, eumelaneic | + | ++ | + | ++ | ++ | ++ |
| Pigmented, homozygous non-dilute, eumelaneic | ++ | ++ | + | ++ | ++ | ++ |
| liver | - | - | - | + | - | - |
| duodenum | - | - | - | + | - | - |
| pituitary gland | - | - | - | + | - | - |
| lung | - | + | + | + | - | - |
| heart | - | - | - | + | - | - |
| kidney | - | + | - | + | - | + |
| mammary gland | - | + | - | + | + | + |
-: no expression detected, + expression detected, ++ enhanced expression detected
Analysis of tissue specific SILV gene expression
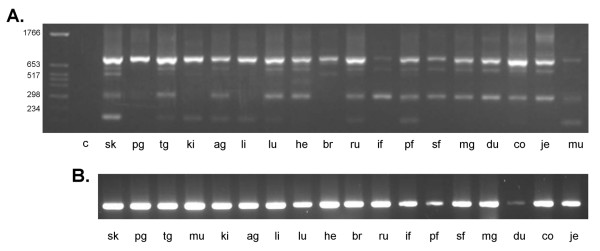
RT-PCR revealed a specific expression of the bovine SILV gene in all 17 investigated tissues from an adult individual: pituitary gland, thyroid gland, kidney, adrenal gland, liver, lung, heart, brain, rumen, intestinal fat, subcutaneous fat, perirenal fat, mammary gland, duodenum, colon, jejunum, skeletal muscle. All three SILV primer combinations tested spanning exon 1 to exon 5 (E1F3 – E5R1), exon 5 to exon 7 (E5F1 – E7R1) and exon 9 to exon 11 (E9F1 – E11R2) confirmed this observation (Table 2). The relative amount as indicated by semi-quantitative RT-PCR differed between tissues: a strong SILV expression equivalent to skin was seen in a number of tissues with very divergent functions, e.g. thyroid gland and colon, whereas SILV was only weakly expressed in brain, muscle and fat tissues (Figure 5). Analysis with primers spanning exon 5 to exon 7 (E5F1 – E7R1) revealed at least two PCR fragments for all tissues with a pattern similar to that obtained for differentially pigmented skin tissues.
Figure 5.
Expression of SILV in a collection of non-pigmented bovine tissues. A: SILV RT-PCR products generated with primers spanning exon 5 to exon 7 (E5F1 – E7R1) in RNA from non-pigmented tissues; sk: skin, pg: pituitary gland, tg: thyroid gland, ki: kidney, ag: adrenal gland, li: liver, lu: lung, he: heart, br: brain, ru: rumen, if: intestinal fat, pf: perirenal fat, sf: subcutaneous fat, mg: mammary gland, du: duodenum, co: colon, je: jejunum, mu: skeletal muscle. B. RT-PCR products generated with GAPDH primers in different non-pigmented tissues.
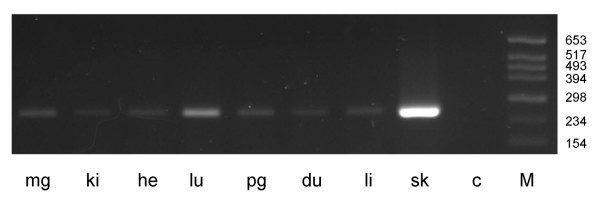
While alternative splice variants were rather uniformly distributed across the differentially pigmented skins, substantial variation was seen for the other tissues (Table 4). Only transcript variant Δ105–362 was tissue-specific for skin. Whereas splice variant Δ816–1340 could be detected in all seven tissues tested (Figure 6), Δ1185–1340 was only found in mammary gland at a low level.
Figure 6.
Splice site specific SILV RT-PCR in non-pigmented bovine tissues. Detection of SILV splice variant Δ816–1340 by splice site specific RT-PCR with primers SPex6AF1 – E9R2 in RNA from non-pigmented tissues. mg: mammary gland, ki: kidney, he: heart, lu: lung, pg: pituitary gland, du: duodenum, li: liver, sk: skin. M: marker; C: negative control
Discussion
Structure of the bovine SILV gene
The bovine SILV gene shows a strong structural homology with the respective gene in human and mouse. The bovine mRNA (GenBank: EF065525) is separated into 11 exons and revealed a homology of 87% to the human (NM_006928.3) and 83% to mouse mRNA (NM_021882.4). At protein level, the similarities were 78 or 73%, respectively. The sharp peak of transcription start sites detected in CAP finding RACE experiments and the putative TATA box of the SILV promoter located 30 bp upstream correspond to the classical pattern of TATA box promoter architecture characteristic for genes highly conserved in evolution [24]. Our RACE experiments in the eumelaneic (black) non-dilute (Dcd/Dcd) skin yielded essentially an identical transcription start compared to a SILV mRNA obtained by 5'RACE from a homozygous dilute (DcD/DcD), crème-white skin [18]. Thus, the use of alternative transcription starts can be formally excluded as the background for differences in coat color between dilute and non-dilute individuals.
SILV transcription in phaeomelaneic and non-pigmented tissues
Taking advantage of our animal model with individuals expressing exclusively eumelanin or phaeomelanin, our RT-PCR results prove that in cattle SILV expression in skin is not restricted to areas with eumelanocytes, because we also found SILV transcripts in phaeomelaneic and non-melaneic skin. This result is in contrast to previous reports in mice [17] indicating that SILV expression could only be detected in cells synthesizing eumelanin. Whereas there are many studies about the melanophore-specific expression of SILV in the literature (reviewed by [4]), only one report in humans also describes SILV to be ubiquitously expressed [25]. This result is supported by 112 ESTs isolated from non-pigmented tissues in a total of 785 human SILV ESTs listed in the NCBI Unigene database [26]). Although no conclusion regarding SILV transcription can be drawn from bovine Unigene data base entries, our own experiments clearly reject the hypothesis that SILV expression is specific to eumelanocytes in cattle. Expression outside of pigmented tissues indicates that products of the bovine SILV gene seem to have a new, up to now unknown function additional to fibril formation in the course of melanosome development. Chakraborty et al. postulate a catalytic role of SILV in melanin synthesis [27]. But as melanin synthesis is restricted to melanophores in skin, uvea and other pigmented tissues [28], involvement of SILV in melanin synthesis would not explain SILV expression in tissues without melanophores. Consequently, the additional role of SILV expression postulated from our study has to exceed melanophore specific functions. Up to now, it is completely unclear, which role SILV transcription may play in e.g. thyroid gland or colon, two tissues with extremely different cell type composition, for which a high SILV expression was detected in our experiments. If indeed human SILV expression is also not restricted to melanophores as indicated by [25], cattle may serve as an appropriate model to investigate its potential function in non-melanophores.
Alternative splicing in SILV affecting functionally relevant domains
RT-PCR with an overlapping panel of primer pairs indicated nine alterative splice sites in the bovine SILV gene resulting in splicing of cryptic introns predominantly affecting exon 6, but also exons 2, 3, 4, 5 and exon 9. Thus, the splicing pattern comprises the use of single or combined cassette exons and alternative 5' splice sites resulting predominantly in deletions of single or multiple exons, which is in line with other comprehensive studies investigating alternative splicing [21]. Sequence alignment of the bovine SILV cDNA with the nine cattle ESTs homologous to SILV, which are deposited in the Unigene database [26], did not indicate alternative splicing. However, the confirmed alternative SILV transcripts in cattle from our study are in line with previous experiments in humans describing two alternative splice sites in exon 6 and exon 9 of the SILV gene [19]. The extent of alternative splicing seen in cattle exceeds the number of published alternative transcripts in humans substantially, and we even cannot exclude that further splice variation is present in cattle. The Alternative Splicing Database [29,30] collecting electronic data on constitutive and alternative transcripts lists entries exclusively for human SILV and indicates that the number of alternative transcripts in humans is also presumably higher than presently considered. Interestingly, humans and cattle are the only species for which alternative transcripts have been experimentally established [19,29]. No additional transcripts are published e.g. in mice.
Comparative analysis of the bovine SILV protein with the respective counterpart in human according [4] suggested that several functional domains of the SILV protein as defined by [31] should be affected by alternative splicing in cattle. Splice variant Δ105–362 characterized by the complete skipping of exons 2 and 3 results in loss of the proximal part of the N terminal domain (NTD) of the bovine SILV protein. Thus, skipping of exons 2 and 3 would affect the posttranslational modification of the protein substantially, because the NTD carries the majority of SILVs' glycosylation sites as reviewed by [4]. The alternative transcripts Δ363–1340, Δ498–1340 and Δ660–1340 lack 326, 281 or 227 amino acids of the constitutive transcript, respectively, representing the C terminal part of the N terminal domain and the entire Polycystic kidney disease (PKD) and Repeat (RPT) domains. While the PKD has an immunoglobulin folding structure and is thought to mediate protein-protein interactions, the RPT domains seems to be necessary for fibril formation in melanosomes [31]. The entire RPT domain is also missing in the Δ816–1340 transcript in addition to the distal PKD domain. Transcript Δ1185–1340 lacks the distal part of the RPT domain, whereas Δ1332–1340 is devoid of 3 amino acids of the Gap2 domain. Splice variant Δ1655–1748 is the only transcript with a disrupted reading frame and a premature stop codon generating a truncated protein without transmembrane and C terminal domain or possibly inducing a nonsense-mediated mRNA decay [32]. While the Kringle-like domain and the transmembrane domain seem to be rather invariable, the RPT domain is most frequently affected by the different splice variants in our data set. Experiments with human SILV deletion constructs showed that a protein lacking the Repeat domain (ΔRPT SILV) showed an appropriate intracellular trafficking [31]. However, HELA cells overexpressing ΔRPT SILV formed only abnormal fibrils, indicating that the RPT domain plays a crucial role in the development of the striated fibrillar structure in melanosomes. Hoashi et al. [31] used HMB45, an antibody frequently employed for melanoma diagnostic as a probe specific for melanosomal fibrils and showed that its epitope is located in the second and third amino acid repeat of the RPT domain. Thus, bovine transcripts lacking the respective part of the RPT domain (like 498–1340, Δ660–1340 and Δ816–1340 in our study) should not be able to form intact fibrils, a process central to appropriate melanosome development as determined for human SILV. Interestingly, the two of the four isoforms of SILV in humans due to alternative splicing also affect the RPT domain albeit not the epitope for HMB45 [31]. The alternative splice variants lacking entire functional domains as confirmed for the bovine SILV gene represent ideal candidates for further study into the potential effects of alternative splicing on function [21].
Variation in alternative splicing of SILV with respect to coat color phenotype
In cattle, we did not find any mutation in the coding SILV sequence that could convincingly be associated with phaeomelanin coat color dilution in a F2 resource population based on the Charolais and the Holstein breed [14]. In dogs, Clark et al., [12] described a mutation in intron 10 of the canine SILV gene that was associated with coat color dilution and concluded that the mutation might impair correct splicing of the canine SILV transcript. Thus, another as yet undetected mutation in the genomic sequence of the bovine SILV gene could possibly affect the structure of the SILV mRNA, representing the genetic background for phaeomelanin dilution in cattle. Because we could confirm SILV expression in eumelaneic and also phaeomelaneic skin, any genetic variant affecting regulation or coding structure of the gene might theoretically also affect phaeomelanin dilution in cattle, although presently there is no indication, which function SILV may exert on maturation of phaeomelanosomes. Thus, given our expression data the SILV gene could not formally be rejected as the background for phaeomelanin dilution in cattle. However, neither the transcription start, which was conserved between homozygous dilute (DcD/DcD) and non-dilute (Dcd/Dcd) individuals, nor the distribution of splice variants in the differentially pigmented skins, convincingly explained the differences in dilution phenotype. Hence, there is no indication that variation in the primary sequence of the SILV protein either due to variation in the coding sequence or due to alternative splicing is responsible for dilution of phaeomelanin in cattle.
Whereas the pattern of transcript variants is rather similar across the panel of differentially pigmented skin, the variability of splice variants detected in other tissues points towards a tissue specific splicing mechanism. This may represent a tool for adapting SILV expression to the requirements of the respective cells [20].
Conclusion
Although the structure of the SILV gene is conserved across a variety of species including cattle, its pattern of transcription shows substantial differences regarding alternative splicing between cattle and human on the one side and mice on the other. Our experiments provide evidence for a ubiquitous transcription of the bovine SILV gene not restricted to pigmented cells and show a striking variety of alternative splice sites. These results indicate that potentially SILV may have functions exceeding melanosome development. This would have to be considered in future investigations concerning melanoma diagnostic and therapy and also in studies taking SILV as a model for amyloid formation [33] or for intracellular transport mechanisms. The similarity in expression pattern compared to human and the variety of alternative transcripts predestine bovine SILV as an adequate animal model.
Whereas alternative splicing is a well established regulatory mechanism for gene expression [20], the confirmed alternative SILV transcripts are in line with the hypothesis of the new postulated additional SILV functions exceeding synthesis and deposition of melanin. This hypothesis is supported by the detection of SILV transcripts in non-melanophores in our study. However, it has to be considered that our analyses are restricted to the transcription level. Thus, subsequent steps modulating SILV expression due to e.g. nonsense mediated decay [32] and translational and posttranslational modification require further investigation to confirm the postulated additional functions of the SILV gene. Due to its extensive, up to now unique pattern of alternative splicing, bovine splice variants represent a naturally occurring model for the function of the domains located in the spliced regions of the SILV and for the alternative functions postulated for SILV. Furthermore, factors regulating the processes of alternative splicing in the different tissues can be investigated exemplarily.
Methods
Tissue samples
For our study, we included individuals from a Charolais × German Holstein F2 cattle resource population [34]. The resource population segregates for the coat color trait loci (i) Dilution (Dc) responsible for coat color dilution, (ii) Extension (E) responsible for an eumelaneic (black) or a phaeomelaneic (red) phenotype, and (iii) Spotted (S) resulting in a pigmented-white spotting pattern similar to piebald in mice. We collected differentially pigmented neck skin after slaughter: (1) black and white sections of a homozygous non-dilute (Dcd/Dcd), spotted (s/s) eumelaneic (ED/Ee) animal, (2) diluted colored and white sections of an eumelaneic (ED/ED), heterozygous DcD/Dcd, spotted (s/s) individual, (3) diluted colored and white sections of a phaeomelaneic (Ee/Ee), heterozygous DcD/Dcd, spotted (s/s) individual and (4) the crème white skin of a homozygous non-dilute (DcD/DcD), non-spotted, eumelaneic (ED/ED) individual. Hair and subcutaneous tissue was removed and the remaining cutis was snap frozen. Genotypes of the individuals for the SILV c64G>A mutation, presumably responsible for dilution of eumelanin, were determined as described by [14]. Genotypes at the dominant-recessive Extension locus were determined by sequencing of the MC1R gene. Primers MC1RF1 (5'-TACTACTTTATCTGCTGCCTG-3') and MC1RR1 (5'-GCGTAGAAGATGGAGATGTAG-3') flanking the causal mutations for the alleles ED (dominant black, eumelanin) and Ee (recessive red, phaeomelanin) were used for amplification in genomic DNA of individuals tested and consecutive sequencing of the resulting PCR products. In addition to the differentially pigmented skin samples, 17 tissues (pituitary gland, thyroid gland, kidney, adrenal gland, liver, lung, heart, brain, rumen, intestinal fat, subcutaneous fat, perirenal fat, mammary gland, duodenum, colon, jejunum, skeletal muscle) from an adult female Charolais × German Holstein F2 individual were collected at slaughter and immediately snap frozen. Total RNA was isolated with the NucleoSpin® RNAII kit (Macherey and Nagel) except all fat tissues, which were isolated with the RNeasy Lipid tissues kit (Qiagen), essentially as described by the manufacturers.
Analysis of skin SILV gene expression
The isolated RNA was reverse transcribed into cDNA according to [35] with a primer mix containing Oligo dT(12–18) and a gene specific primer from the 3'UTR of the SILV mRNA (E11R2, Table 1). The resulting cDNAs were amplified with GO-Taq polymerase (Promega) under standard conditions and primers as indicated in Table 1 and Table 2. Due to confirmed splice variants in the RPT and in the GAP3 domain of the human SILV gene and because we obtained suggestive additional bovine SILV transcripts in initial investigations, the entire bovine SILV cDNA was examined for additional splice sites using overlapping PCR primer combinations (Table 2). These investigations were carried out in skin of different coat color phenotypes. The generated PCR fragments were separated on agarose gels. As a control, expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was determined by RT-PCR as described [35].
Sequencing of SILV transcripts
The analysis of SILV transcripts in differently pigmented skin and non-pigmented body tissues detected more than one RT-PCR fragment for several primer combinations. The respective fragments were excised from the agarose gel, purified using the Nucleospin Extract II kit (Macherey and Nagel) and sequenced on a capillary sequencer (ABI 310, Applied Biosystems; MEGABACE, GE Healthcare) using BigDye® (Applied Biosystems) chemistry. Two prominent RT-PCR fragments > 1000 bp generated with primers spanning exon 1 to exon 11 (5UTRF1 – E11R2, Table 1) were excised from the gel, purified, and cloned into the pCR4 Blunt TOPO vector (Invitrogen) according the manufacturers instructions. Seven size fractionated subsets of RT-PCR fragments generated by primers enclosing exon 1 to exon 7 (E1F3 – E7R1, Table 1) were excised from the agarose gel, purified and cloned into pDrive vector (Qiagen) according to the manufacturers instructions. Clones with different insert sizes according to colony PCR were sequenced on a capillary sequencer (ABI 310, Applied Biosystems; MEGABACE, GE Healthcare) using BigDye® (Applied Biosystems) chemistry.
SILV RNA 5' RACE experiments
To determine the transcription start site for the bovine SILV gene and to test for alternative promoters, 5' RACE experiments with the GeneRacer Kit (Invitrogen) based on RNA ligase mediated and oligo-capping methods were performed. After CAP selection in a preparation of total RNA from black skin of a eumelaneic (ED/ED), non-dilute (Dcd/Dcd) individual, reverse transcription with oligo dT and re-amplification with the RACE 5' oligo and a SILV specific primer from exon 7 (RACE_E7N, Table 1) was carried out according to the manufacturers instructions. The generated fragments were cloned into pDrive vector using the Qiagen PCR cloning kit (Qiagen). Clones with different insert sizes according to colony PCR were sequenced. Sequences obtained were aligned to the deposited SILV mRNA (GenBank: EF065525).
Confirmation of SILV splice variants
To confirm the detected SILV splice variants, SILV cDNA samples were investigated by splice site specific RT-PCR. For this purpose, SILV cDNAs were generated by reverse transcription with the E11R2 primer from total RNA of skins differing in coat color phenotype. Afterwards, the cDNAs were subjected to PCR amplification with one SILV primer corresponding to the sequence of the constitutive SILV mRNA and another primer specific for each splice variant, respectively (Table 1, Table 3). Splice variant specific primers were designed to bridge the splice ends of the addressed alternative splice site. For negative control, DNA of a plasmid with a constitutive SILV cDNA insert was included.
SILV expression in non-pigmented tissues
Finally, SILV expression was screened in a collection of 17 total RNAs from non-pigmented tissues (pituitary gland, thyroid gland, kidney, adrenal gland, liver, lung, heart, brain, rumen, intestinal fat, subcutaneous fat, perirenal fat, mammary gland, duodenum, colon, jejunum, skeletal muscle). Analogous to the RT-PCR in skin tissue, the different RNAs were reversely transcribed and amplified by using primers corresponding to the constitutive SILV cDNA sequence in combinations covering exon 1 to exon 5 (E1F3/E5R1), exon 5 to exon 7 (E5F1/E7R1) and exon 9 to exon 11 (E9F1/E11R2, Table 2).
Because there was evidence on alternative splicing in non-pigmented skin, a collection of eight non-pigmented other tissues (liver, duodenum, pituitary gland, lung, heart, kidney, mammary gland) was further investigated for alterative splicing by splice variant specific RT-PCR analogous to the procedure described for skin. The respective primer combinations are listed in Table 3.
Competing interests
The author(s) declares that there are no competing interests.
Authors' contributions
CK conceived, designed and coordinated the study, carried out PCR analysis of RNAs, participated in sequence alignments and drafted the manuscript. RW participated in coordination of the study, participated in sequence alignments, carried out the RACE and splice variant specific experiments, and helped drafting the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Astrid Kühl, Simone Wöhl, Marlies Deutscher and Oda Haufft for skilful technical assistance. The continuous support of our colleagues at the FBN Dummerstorf involved in generation and care of the SEGFAM F2 resource population is thankfully acknowledged.
Contributor Information
Christa Kuehn, Email: kuehn@fbn-dummerstorf.de.
Rosemarie Weikard, Email: weikard@fbn-dummerstorf.de.
References
- Spanknebel K, Coit DG, Bieligk SC, Gonen M, Rosai J, Klimstra DS. Characterization of micrometastatic disease in melanoma sentinel lymph nodes by enhanced pathology - Recommendations for standardizing pathologic analysis. Am J Surg Pathol. 2005;29:305–317. doi: 10.1097/01.pas.0000152134.36030.b7. [DOI] [PubMed] [Google Scholar]
- Tsukamoto K, Ueda M, Hirata S, Osada A, Kitamura R, Takahashi T, Ichihashi M, Shimada S. gp100 mRNA is more sensitive than tyrosinase mRNA for RT-PCR amplification to detect circulating melanoma cells in peripheral blood of melanoma patients. J Dermatol Sci. 2000;23:126–131. doi: 10.1016/S0923-1811(99)00098-5. [DOI] [PubMed] [Google Scholar]
- Bakker ABH, Schreurs MWJ, Deboer AJ, Kawakami Y, Rosenberg SA, Adema GJ, Figdor CG. Melanocyte Lineage-Specific Antigen Gp100 Is Recognized by Melanoma-Derived Tumor-Infiltrating Lymphocytes. J Exp Med. 1994;179:1005–1009. doi: 10.1084/jem.179.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos AC, Truschel ST, Raposo G, Marks MS. The Silver locus product Pmel17/gp100/Silv/ME20: controversial in name and in function. Pigment Cell Res. 2005;18:322–336. doi: 10.1111/j.1600-0749.2005.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson JF, Harper DC, Tenza D, Raposo G, Marks MS. Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol Biol Cell. 2001;12:3451–3464. doi: 10.1091/mbc.12.11.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA, Terry RB, Bailey E. A PCR-RFLP for KIT associated with tobiano spotting pattern in horses. Anim Genet. 2002;33:301–303. doi: 10.1046/j.1365-2052.2002.00854.x. [DOI] [PubMed] [Google Scholar]
- Rees JL. Genetics of hair and skin color. Ann Rev Genet. 2003;37:67–90. doi: 10.1146/annurev.genet.37.110801.143233. [DOI] [PubMed] [Google Scholar]
- Du JY, Fisher DE. Identification of aim-1 as the underwhite mouse mutant and its transcriptional regulation by MITF. J Biol Chem. 2002;277:402–406. doi: 10.1074/jbc.M110229200. [DOI] [PubMed] [Google Scholar]
- Olson TA. Genetics of colour variation. In: Fries R and Ruvinsky A, editor. The Genetics of Cattle. Wallingford, UK, CABI; 1999. pp. 33–54. [Google Scholar]
- Schaible RH. Clonal Distribution of Melanocytes in Piebald-Spotted and Variegated Mice. J Exp Zool. 1969;172:181–199. doi: 10.1002/jez.1401720205. [DOI] [PubMed] [Google Scholar]
- Brunberg E, Andersson L, Cothran G, Sandberg K, Mikko S, Lindgren G. A missense mutation in PMEL17 is associated with the Silver coat color in the horse. BMC Genetics. 2006;7:46. doi: 10.1186/1471-2156-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LA, Wahl JM, Rees CA, Murphy KE. Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc Natl Acad Sci U S A. 2006;103:1376–1381. doi: 10.1073/pnas.0506940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerje S, Sharma P, Gunnarsson U, Kim H, Bagchi S, Fredriksson R, Schutz K, Jensen P, von Heijne G, Okimoto R, Andersson L. The Dominant white, Dun and Smoky color variants in chicken are associated with insertion/deletion polymorphisms in the PMEL17 gene. Genetics. 2004;168:1507–1518. doi: 10.1534/genetics.104.027995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn C, Weikard R. An investigation into the genetic background of coat colour dilution in a Charolais · German Holstein F2 resource population. Anim Genet. 2007;38:109–113. doi: 10.1111/j.1365-2052.2007.01569.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Esparza MM, Jimenez-Cervantes C, Bennett DC, Lozano JA, Solano F, Garcia-Borron JC. The mouse silver locus encodes a single transcript truncated by the silver mutation. Mamm Genome. 1999;10:1168–1171. doi: 10.1007/s003359901184. [DOI] [PubMed] [Google Scholar]
- Reissmann M, Bierwolf J, Brockmann GA. Two SNPs in the SILV gene are associated with silver coat colour in ponies. Anim Genet. 2007;38:1–6. doi: 10.1111/j.1365-2052.2006.01553.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Vieira WD, Potterf B, Sakai C, Imokawa G, Hearing VJ. Modulation of melanogenic protein expression during the switch from eumelanogenesis to pheomelanogenesis. J Cell Sci. 1995;108:2301–2309. doi: 10.1242/jcs.108.6.2301. [DOI] [PubMed] [Google Scholar]
- Oulmouden A, Julien R, Laforet JM, Leveziel H. Use of silver gene for authentication of the racial origin of animal populations, and of the derivative products thereof. WO2005/019473. 2005 [Google Scholar]
- Nichols SE, Harper DC, Berson JF, Marks MS. A novel splice variant of Pmel17 expressed by human melanocytes and melanoma cells lacking some of the internal repeats. J Invest Dermatol. 2003;121:821–830. doi: 10.1046/j.1523-1747.2003.12474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang YS, Zhang ZY, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Tress ML, Martelli PL, Frankish A, Reeves GA, Wesseling JJ, Yeats C, Olason PI, Albrecht M, Hegyi H, Raimondo AGD, Lagarde J, Laskowski RA, Lopez G, Sadowski MI, Watson JD, Fariselli P, Rossi I, Nagy A, Kai W, Storling Z, Orsini M, Assenov Y, Blankenburg H, Huthmacher C, Ramirez F, Schlicker A, Denoeud F, Jones P, Kerrien F, Orchard S, Antonarakis FE, Reymond A, Birney E, Brunak S, Casadio R, Guigo R, Harrow J, Hermjakob H, Jones DT, Lengauer T, Orengo CA, Patthy L, Thornton JL, Tramontano A, Valencia A. The implications of alternative splicing in the ENCODE protein complement. Proc Natl Acad Sci U S A. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI map viewer. 2007. http://www.ncbi.nlm.nih.gov/mapview/
- Guibert S, Girardot M, Leveziel H, Julien R, Oulmouden A. Pheomelanin coat colour dilution in french cattle breeds is not correlated with the TYR, TYRP1 and DCT transcription levels. Pigment Cell Res. 2004;17:337–345. doi: 10.1111/j.1600-0749.2004.00152.x. [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engstrom PG, Frith MC, Forrest ARR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y. Genome-wide analysis of mammalian promoter architecture and evolution. Nature Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Brouwenstijn N, Slager EH, Bakker ABH, Schreurs MWJ, Van der Spek CW, Adema GJ, Schrier PI, Figdor CG. Transcription of the gene encoding melanoma-associated antigen gp100 in tissues and cell lines other than those of the melanocytic lineage. Br J Cancer. 1997;76:1562–1566. doi: 10.1038/bjc.1997.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Unigene database. 2007. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene
- Chakraborty AK, Platt JT, Kim KK, Kwon BS, Bennet DC, Pawelek JM. Polymerization of 5,6-dihydroxyindole-2-carboxylic acid to melanin by the pmel 17 silver locus protein. Eur J Biochem. 1996;236:180–188. doi: 10.1111/j.1432-1033.1996.t01-1-00180.x. [DOI] [PubMed] [Google Scholar]
- Hearing VJ, Tsukamoto K. Enzymatic Control of Pigmentation in Mammals. FASEB Journal. 1991;5:2902–2909. [PubMed] [Google Scholar]
- The Alternative Splicing Database. 2007. http://www.ebi.ac.uk/asd/index.html
- Thanaraj TA, Stamm S, Clark F, Riethoven JJ, Le Texier V, Muilu J. ASD: the Alternative Splicing Database. Nucl Acids Res. 2004;32:D64–D69. doi: 10.1093/nar/gkh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoashi T, Muller J, Vieira WD, Rouzaud F, Kikuchi K, Tamaki K, Hearing VJ. The repeat domain of the melanosomal matrix protein PMEL17/GP100 is required for the formation of organellar fibers. J Biol Chem. 2006;281:21198–21208. doi: 10.1074/jbc.M601643200. [DOI] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:100–107. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn C, Bellmann O, Voigt J, Wegner J, Guiard V, Ender K. An experimental approach for studying the genetic and physiological background of nutrient transformation in cattle with respect to nutrient secretion and accretion type. Arch Anim Breed. 2002;45:317–330. [Google Scholar]
- Weikard R, Kuhn C, Goldammer T, Freyer G, Schwerin M. The bovine PPARGC1A gene: molecular characterization and association of an SNP with variation of milk fat synthesis. Physiol Genomics. 2005;21:1–13. doi: 10.1152/physiolgenomics.00103.2004. [DOI] [PubMed] [Google Scholar]