Abstract
Miscoordination of growth and proliferation with the cellular stress response can lead to tumorigenesis. Mammalian target of rapamycin (mTOR), a central cell growth controller, is highly activated in some malignant neoplasms, and its clinical implications are under extensive investigation. We show that constitutive mTOR activity amplifies p53 activation, in vitro and in vivo, by stimulating p53 translation. Thus, loss of TSC1 or TSC2, the negative regulators of mTOR, results in dramatic accumulation of p53 and apoptosis in response to stress conditions. In other words, the inactivation of mTOR prevents cell death by nutrient stress and genomic damage via p53. Consistently, we also show that p53 is elevated in TSC tumors, which rarely become malignant. The coordinated relationship between mTOR and p53 during cellular stress provides a possible explanation for the benign nature of hamartoma syndromes, including TSC. Clinically, this also suggests that the efficacy of mTOR inhibitors in anti-neoplastic therapy may also depend on p53 status, and mTOR inhibitors may antagonize the effects of genotoxic chemotherapeutics.
Keywords: apoptosis, mTOR, p53, rapamycin
Introduction
Tumorigenesis is a complicated process that involves the loss of many mechanisms for regulating cell growth and proliferation in response to stress and growth factor signaling. The mammalian target of rapamycin (mTOR) is a highly conserved serine/threonine kinase, which is central for a diverse set of cellular processes important for growth and proliferation. Furthermore, mTOR is also found to be activated in a variety of malignancies and thus is currently being targeted for anti-neoplastic therapy. mTOR exists in one of two distinct functional complexes, TOR complex 1 (TORC1) and TORC2. TORC1 is potently and specifically inhibited by rapamycin, and it regulates cell size, autophagy, ribosome biogenesis, protein translation, transcription, and cellular viability (Lee et al, 2007). TORC2, which is much less sensitive to rapamycin, is important for cytoskeletal regulation (Loewith et al, 2002; Jacinto et al, 2004; Sarbassov et al, 2004) and AKT activation (Ali and Sabatini, 2005; Hresko and Mueckler, 2005; Sarbassov et al, 2005). Inhibition of TORC2 by rapamycin occurs in a cell type-specific manner and requires far greater concentration and duration than TORC1; thus, inhibition of TORC2 by rapamycin is likely due to an indirect effect on TORC1 (Sarbassov et al, 2006). For the experiments in this paper, references to mTOR activation or inhibition correspond to activation or inhibition of TORC1.
It has been established both genetically and biochemically that the tumor suppressors TSC1 and TSC2 negatively regulate mTOR (Inoki et al, 2002). Loss of either TSC1 or TSC2 results in the autosomal dominant hamartoma syndrome tuberous sclerosis complex (TSC), which is characterized by benign tumor formation in multiple organs, including the kidney, liver, lung, spleen, heart, and brain (Young and Povey, 1998). Loss of either tumor suppressor is sufficient to induce TSC because TSC1 and TSC2 exist as both a physical and functional complex (van Slegtenhorst et al, 1998). The TSC1/TSC2 complex inhibits mTOR by acting as the GAP for the small GTPase Ras-homology enriched in brain (Rheb), and thus decreases Rheb-GTP and prevents stimulation of mTOR activity (Castro et al, 2003; Garami et al, 2003; Inoki et al, 2003a; Tee et al, 2003; Zhang et al, 2003b; Li et al, 2004).
In contrast to other tumor suppressors, loss of either TSC1 or TSC2 leads to tumor formation without malignancy. TSC tumors, which have high mTOR activity, are benign and highly apoptotic (Wataya-Kaneda et al, 2001), suggesting that mTOR activation can sensitize cells to death and mTOR activation alone is insufficient for progression to malignancy. However, the mechanism by which this occurs is not well understood.
Through phosphorylation of TSC2, the low energy response mediator AMPK inactivates mTOR-dependent growth and proliferation. This phosphorylation of TSC2 has a protective role against energy starvation-mediate apoptosis (Inoki et al, 2003b). In addition to inhibiting mTOR, the low energy response coordinated by AMPK also leads to phosphorylation of numerous substrates to inhibit anabolic processes and activate catabolic processes (Hardie et al, 1998; Kahn et al, 2005). To protect the cells from stress, AMPK arrests the cell cycle through phosphorylation of p53 Ser15 (Jones et al, 2005) and arrests protein synthesis through activation of TSC2. In the presence of TSC1/2, cells undergo cell cycle arrest. However, we and others have observed that energy starvation induces cell death in TSC1−/− and TSC2−/− cells in a rapamycin-reversible manner (Inoki et al, 2003b; Shaw et al, 2004), suggesting a role for mTOR inhibition in low energy survival. However, the exact molecular mechanism for this response is not yet known. Here, we also show that activation of mTOR can sensitize cells to the DNA-alkylating agent methyl methanesulfonate (MMS), which causes both base mispairing and replication blocks (Beranek, 1990). Both energy starvation and DNA damage can lead to p53 activation (Duckett et al, 1999; Jones et al, 2005); therefore, we sought to elucidate the mechanism by which mTOR activation sensitizes cells to death, thus, providing greater insight into the relationship between mTOR and viability.
Here we show that increased mTOR activity dramatically enhances p53 activation in response to both glucose starvation and DNA damage. This occurs through stabilizing phosphorylations of p53 by energy starvation or DNA damage and unabated p53 synthesis by constitutive mTOR activation; therefore, rapamycin prevents overactivation of mTOR to protect against both glucose starvation and DNA damage. Furthermore, immunohistochemical staining of angiomyolipomas illustrate in vivo that when mTOR activity is elevated by TSC2 loss, tumors have increased levels of p53. These results may explain why the TSC tumors are highly apoptotic and benign.
Results
Dysregulation of mTOR activation sensitizes cells to p53-dependent insults
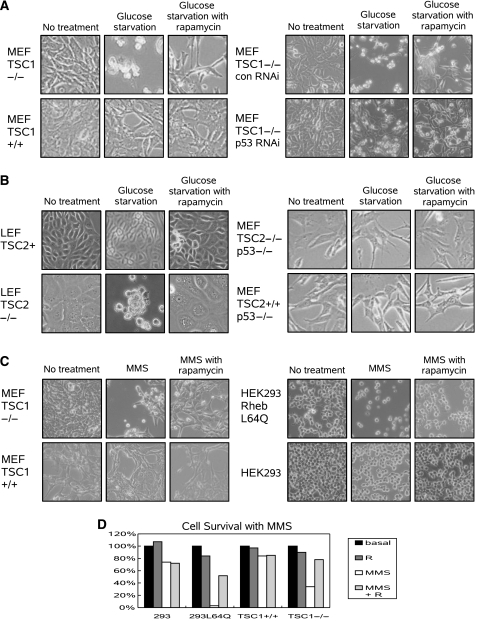
p53 is known to be activated by both energy starvation and DNA damage. To determine the effect of mTOR activation on p53 activation, TSC1−/− and TSC1+/+ MEFs were challenged with glucose starvation. TSC1−/− MEFs, which are unable to downregulate mTOR in response to low starvation, underwent massive amounts of gross cell death, as seen by the appearance of rounded floating cells. Furthermore, inhibition of mTOR by rapamycin protected the TSC1−/− MEFs against glucose starvation-induced cell death. In comparison, TSC1+/+ MEFs, which properly downregulate mTOR in response to low energy, show no evidence of cell death (Figure 1A left).
Figure 1.
Dysregulation of mTOR activation sensitizes cells to p53-dependent insults. (A) TSC1−/− MEFs challenged with glucose starvation (15 h) were more prone to death, which was protected against by rapamycin treatment. RNAi knockdown of p53 decreases sensitivity to glucose starvation (15 h) in TSC1−/− MEFs. (B) LEF TSC2−/− cells were sensitized to glucose starvation (36 h), and both rapamycin and adding back of TSC2 eliminated sensitivity. TSC2−/− p53−/− and TSC2+/+ p53−/− MEFs are resistant to glucose starvation (15 h). (C) TSC1−/− MEFs are more sensitive to MMS treatment (50 μg/ml, 8 h), and rapamycin pretreatment is protective (24 h). Infection of HEK293 cells with Rheb L64Q increases sensitivity to MMS treatment (25 μg/ml, 8 h). Pretreatment of HEK293 Rheb L64Q cells with rapamycin (24 h) protects cells against cell death. (D) Living cells were counted after treatment with MMS (25 μg/ml in 293 Rheb L64Q and 293 cells, 50 μg/ml in TSC1−/− and TSC1+/+ MEFs, 8 h) or rapamycin (R).
To demonstrate that p53 is important for energy starvation-induced cell death during mTOR activation, p53 was knocked down in TSC1−/− MEFs by RNAi (TSC1−/− p53 RNAi MEFs). When TSC1−/− p53 RNAi MEFs are challenged with glucose starvation, they are more resistant to cell death than their control RNAi counterparts. Furthermore, rapamycin treatment of TSC1−/− p53 RNAi MEFs showed no further protection against cell death. In contrast, TSC1−/− control RNAi MEFs were acutely sensitive to glucose starvation, and mTOR inhibition was protective against glucose starvation (Figure 1A right). Together, this suggests that p53 is important for mediating cell death seen by energy starvation in TSC1−/− MEFs.
Consistent with the fact that loss of either TSC1 or TSC2 is sufficient to induce mTOR dysregulation, TSC2−/− LEFs derived from Eker rat kidney tumors also show increased sensitivity to glucose starvation, which can be rescued by rapamycin treatment. Viral infection of TSC2 to restore control of mTOR also protects the LEFs from glucose starvation (Figure 1B left). These results demonstrate that downregulation of mTOR during energy starvation is necessary to prevent cell death.
Since RNAi of p53 in the TSC1−/− MEFs incompletely knocked down p53, we wanted to test the effects of mTOR activation in a p53-null background. To test whether p53 is important for regulating cellular viability in the presence of constitutive mTOR activation, TSC2−/− p53−/− and TSC2+/+ p53−/− MEFs were challenged with energy starvation. Loss of either TSC1 or TSC2 impairs the ability to downregulate mTOR in response to glucose starvation. However, when p53 is also missing, TSC2−/− p53−/− MEFs and TSC2+/+ p53−/− MEFs showed equal sensitivity to glucose starvation (Figure 1B right). Furthermore, rapamycin had no effect on either cell type. Therefore, loss of p53 eliminated the increased sensitivity to energy starvation induced by aberrant mTOR activation, which implies that p53 is important for mediating cell death, and mTOR may be an upstream regulator of p53.
To test whether mTOR activation also sensitized cells to other activators of p53, various cell types were used to examine the sensitivity to DNA damage. p53 is potently activated by DNA damage induced by alkylating agents such as MMS and topoisomerase inhibitors such as etoposide. Like energy starvation, MMS treatment also induced cell death in TSC1−/− MEFs but not in WT counterparts. Furthermore, inhibition of mTOR by rapamycin pretreatment also protected against MMS-induced DNA damage (Figure 1C left, D). Interestingly, MMS treatment inhibits mTOR activation, as determined by S6K phosphorylation in TSC1+/+ and TSC2+/+ p53−/− MEFs, but not in TSC1−/− and TSC2+/+ p53−/−MEFs (Supplementary Figure 1). The role of the TSC complex in mediating mTOR inhibition by MMS was further established by TSC2 RNAi in HEK293 cells. Knockdown of TSC2 significantly compromised MMS-induced mTOR inactivation (Supplementary Figure 2). Consistently, in HEK293 cells, the activation of mTOR by infection of an active mutant of Rheb (Rheb L64Q) also sensitized the cells to MMS, which was also inhibited by pretreatment with rapamycin (Figure 1C right, D). Thus, aberrant mTOR activation sensitizes cells to DNA damage.
mTOR activation enhances p53 phosphorylation and accumulation
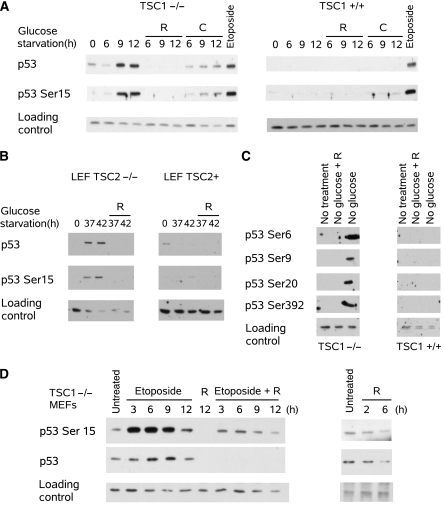
The observations that loss of p53-reduced sensitivity to energy starvation in TSC cells (Figure 1A versus B) and activation of mTOR increased sensitivity to DNA damage (Figure 1C) suggested that p53 activation could be important for mediating mTOR's proapoptotic role. To test this possibility, p53 activation was determined by phosphorylation and accumulation. In the TSC1−/− MEFs, glucose starvation induced phosphorylation of p53 on Ser15 (mouse p53 Ser18) and accumulation of p53 protein (Figure 2A). However, the phosphorylation and accumulation of p53 could be prevented by inhibition of mTOR or AMPK with rapamycin or compound C, respectively. Rapamycin lowered p53 levels below basal levels, while compound C maintained p53 levels similar to basal levels. In comparison, p53 showed little activation by glucose starvation in the TSC1+/+ MEFs. However, DNA damage by etoposide activated p53 in both TSC1+/+ and TSC1−/− MEFs; therefore, it is unlikely that the TSC1+/+ MEFs are defective in p53 activation (Figure 2A). Inhibition of p53 accumulation by rapamycin suggests that constitutive activation of mTOR was responsible for the difference in p53 response between the TSC1−/− and TSC1+/+ MEFs.
Figure 2.
mTOR activation enhances p53 phosphorylation and accumulation. (A) Glucose starvation time course showed both p53 Ser15 phosphorylation and p53 accumulation in TSC1−/− MEFs, which was reversed by rapamycin (R) and compound C (C, 10 μM); however, this was not seen in TSC1+/+ MEFs. Etoposide treatment (6 μg/ml, 6 h) induced p53 in both TSC1−/− and TSC1+/+ MEFs. Tubulin was used as loading control. TSC1+/+ immunoblots were exposed longer than TSC1−/− immunoblots to help normalize the signals because p53 levels are low in TSC1+/+ MEFs. (B) Glucose starvation induced p53 Ser15 phosphorylation and p53 accumulation in TSC2−/− LEFs, which was reversed by rapamycin treatment; however, TSC2+ LEFs did not show induction of p53. Actin was used as loading control. Exposure of TSC2+ LEF p53 immunoblot was increased due to low basal p53 levels. (C) Glucose starvation of TSC1−/− MEFs also increased phosphorylation on p53 Ser6, Ser9, Ser20, and Ser392, which was reversed by rapamycin. However, TSC1+/+ MEFs did not show p53 phosphorylation during glucose starvation. Nonspecific band was used as a loading control (D) and p53 phosphorylation and accumulation was stimulated by etoposide. Concurrent rapamycin treatment decreased p53 protein levels and detected p53 Ser15 phosphorylation. Rapamycin treatment alone also decreased basal p53 levels. Tubulin was used as a loading control.
To verify that the p53 response was not limited to just the TSC1−/− MEFs, the TSC2−/− LEFs were also tested. Similarly, in the TSC2−/− LEFs, glucose starvation induced activation of p53 as seen by phosphorylation and accumulation of p53, which was eliminated by mTOR inhibition. Consistently, the add back of TSC2 also prevented p53 activation by glucose starvation and restores repression of total p53 protein level by glucose starvation (Figure 2B).
To determine whether p53 was fully activated, other p53 phosphorylation sites were also analyzed by immunoblot. Ser15 phosphorylation induces dissociation between p53 and its ubiquitin E3 ligase Mdm2; however, it is insufficient to induce p53 DNA binding, which is induced by phosphorylation on p53 Ser392 (Kapoor et al, 2000). Therefore, multiple phosphorylations are necessary to fully activate p53. We show that glucose starvation induced phosphorylation on several sites, including Ser6, Ser9, Ser20, and Ser392 (Figure 2C). Furthermore, phosphorylation on those sites was eliminated by the addition of rapamycin. However, the p53 protein level was also inhibited by rapamycin, therefore, it is possible that p53 phosphorylation decreased indirectly by decreasing total p53 protein.
To determine whether DNA damage also induced rapamycin reversible p53 accumulation, TSC1−/− MEFs were treated with etoposide. Etoposide induced phosphorylation and accumulation of p53, and similar to glucose starvation-dependent p53 activation, rapamycin decreases the p53 protein level and the detected phosphorylation on Ser15. Moreover, the treatment of TSC1−/− MEFs with rapamycin alone also decreases p53 protein levels (Figure 2D). Taken together, this suggests that mTOR is a positive regulator of p53, and inhibition of mTOR attenuates p53 accumulation irrespective of the stimulus used to stabilize p53.
p53 is stabilized by energy starvation
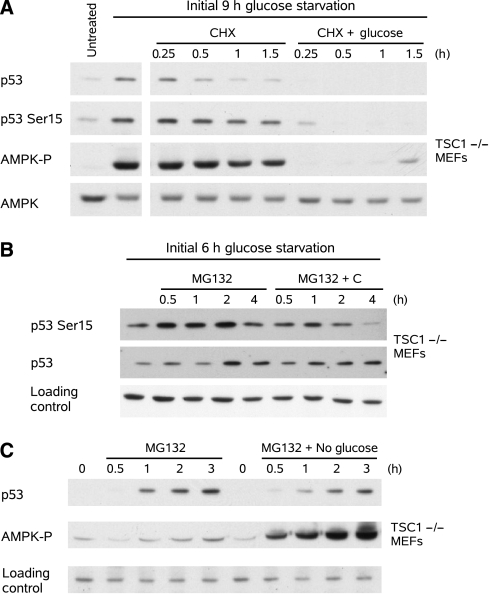
As seen earlier, the activation of p53 by energy starvation requires both AMPK activation and dysregulation of mTOR (Figure 2A). The accumulation of p53 by energy starvation could be due to either stabilization or increased synthesis of p53. To better understand the role of energy starvation on p53 activation, both p53 synthesis and degradation were examined in the TSC1−/− MEFs. To test the effect of energy starvation on p53 stability, p53 was accumulated by glucose starvation in TSC1−/− MEFs, and translation was then blocked by cycloheximide treatment, after which the cells were maintained in either glucose-free media or switched to glucose-containing media. Restoration of glucose decreased p53 stability. This decrease in p53 stability was correlated with AMPK inactivation (Figure 3A). Similar results were also seen in the absence of cycloheximide; however, the change in p53 stability was partially masked by continued p53 synthesis (Supplementary Figure 3). Furthermore, glucose starvation-induced phosphorylation of p53 Ser15 was also eliminated by the restoration of glucose (Figure 3A). The concurrent change in p53 protein level confounds conclusions about AMPK and p53 Ser15 phosphorylation with this experiment; therefore, p53 Ser15 phosphorylation was also subsequently determined in the absence of degradation.
Figure 3.
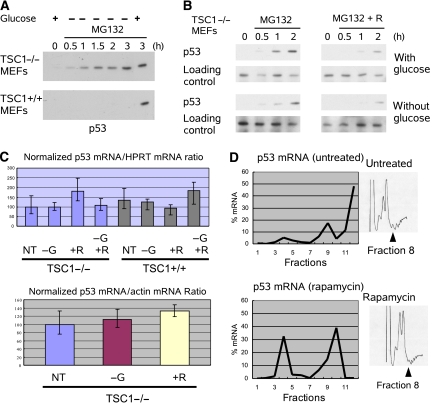
Stabilization of p53 during glucose starvation is due to AMPK. (A) p53 was stabilized by glucose starvation in TSC1−/− MEFs, and further synthesis was blocked by cycloheximide (50 ng/ml). Reintroduction of glucose 30 min before cycloheximide treatment decreased p53 stability. (B) p53 was accumulated with glucose starvation in TSC1−/− MEFs, and degradation was blocked by MG132 (20 μM). Compound C decreased phosphorylation on p53 Ser15 when degradation of p53 was blocked. Tubulin was used as a loading control. (C) Degradation of p53 was blocked by MG132 in TSC1−/− MEFs. Glucose starvation did not increase p53 synthesis. Tubulin was used as a loading control.
To test the effect of AMPK activity on p53 phosphorylation during glucose starvation, p53 was accumulated by glucose starvation in TSC1−/− MEFs, and MG132, a proteosome inhibitor, was used to prevent p53 degradation. Block of AMPK by Compound C reduced p53 Ser15 phosphorylation even in the absence of p53 degradation. This suggested that activation of AMPK was responsible for p53 Ser15 phosphorylation during energy starvation (Figure 3B).
To rule out the possibility that accumulation of p53 protein during energy starvation was due to changes in p53 protein synthesis, we treated TSC1−/− MEFs with MG132 in the presence or absence of glucose. p53 synthesis was measured indirectly by observing its rate of accumulation in the absence of degradation. Glucose starvation slightly decreased the rate of p53 accumulation (Figure 3C). Taken together, this suggests that energy starvation induces p53 accumulation in TSC1−/− MEFs by increasing p53 stability but not synthesis.
Inhibition of mTOR decreases p53 synthesis without increasing degradation
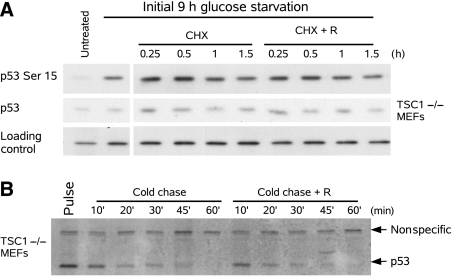
Rapamycin treatment of TSC1−/− MEFs decreases p53 levels, which can be due to decreased synthesis, increased degradation, or a combination of both. To distinguish between these possibilities, we first tested the effect of mTOR activity on p53 phosphorylation during glucose starvation. p53 was accumulated by glucose starvation in TSC1−/− MEFs and further synthesis was blocked by cycloheximide. Not only did rapamycin have no effect on p53 Ser15 phosphorylation, but rapamycin also did not have a significant effect on p53 half-life (Figure 4A). In comparison, 30 min of rapamycin treatment is sufficient to completely eliminate mTOR-dependent phosphorylation of ribosomal S6 kinase 1 (S6K), a direct downstream target (Supplementary Figure 4). This suggested that mTOR is not responsible for p53 Ser15 phosphorylation and indicates that rapamycin does not destabilize p53 after prolonged glucose starvation.
Figure 4.
Rapamycin does not affect p53 stability or phosphorylation. (A) p53 was stabilized by glucose starvation in TSC1−/− MEFs, and further synthesis was blocked by cycloheximide (50 ng/ml). Addition of rapamycin 30 min before cycloheximide treatment did not affect p53 stability or phosphorylation on Ser15. Tubulin was used as a loading control. (B) 35S Pulse-chase both in the presence and absence of rapamycin in glucose-rich media of TSC1−/− MEFs. Rapamycin did not enhance the degradation of p53.
To more directly test the effect of rapamycin on p53 stability, an 35S-pulse-chase was used to determine p53 stability. In glucose-containing media, TSC1−/− MEFs were labeled with 35S-methionine, and then it was chased with cold methionine. Consistent with the results observed by cycloheximide treatment in the absence of glucose, rapamycin also did not significantly reduce the half-life of p53 in the presence of glucose (Figure 4B). The addition of excess methionine during the cold chase also had no effect on mTOR activity, as assayed by S6K1 phosphorylation (Supplementary Figure 4). Together, this suggests that mTOR inhibition does not stimulate p53 degradation; therefore, the protective role of rapamycin during glucose starvation is not due to destabilization of p53.
To compare the effects of mTOR activity on p53 synthesis, MG132 was used to block degradation in both TSC1−/− and TSC1+/+ MEFs. p53 accumulation was determined both in the presence and absence of glucose. In the TSC1+/+ MEFs, p53 synthesis was inhibited by glucose starvation. In contrast, in the TSC1−/− MEFs, p53 synthesis continues despite the absence of glucose (Figure 5A). In other words, when the mTOR pathway can be shut down by energy starvation, p53 synthesis is abated; however, when glucose starvation cannot shut down mTOR, p53 synthesis remains unaffected.
Figure 5.
Inhibition of mTOR decreases p53 synthesis. (A) p53 degradation was blocked by MG132. Accumulation of p53 was examined under various conditions in TSC1−/− and TSC1+/+ MEFs. In TSC1−/− MEFs, glucose starvation is unable to shut down p53 synthesis. In TSC1+/+ MEFs, glucose starvation decreases the rate of p53 synthesis. (B) TSC1−/− MEFs were pretreated with rapamycin for 6 h prior to MG132 treatment. Accumulation of p53 was decreased by rapamycin pretreatment regardless of whether glucose was present. Tubulin was used as a loading control. (C) p53 mRNA was normalized to either Actin mRNA or HPRT mRNA in TSC1−/− and TSC1+/+ MEFs. Neither glucose starvation (−G, 6 h) nor rapamycin (+R, 6 h) treatment had significant effects on p53 mRNA level. (D) p53 mRNA was fractionated over a sucrose gradient in WT MEFs to examine the p53 mRNA association with polysomes. Fractions 8–12 represent polysome-associated fractions. Rapamycin decreased polysome association of p53 mRNA. A full colour version of this figure is available at EMBO Journal Online.
To demonstrate that inhibition of mTOR in the TSC1−/− MEFs can indeed reduce the accumulation of p53, rapamycin was used to inhibit mTOR before the addition of MG132. Pretreatment with rapamycin decreased the rate of p53 accumulation, which suggests that rapamycin indeed reduces p53 synthesis. Furthermore, whether glucose was present in the media has no effect on the rapamycin induced reduction in p53 (Figure 5B). Therefore, mTOR activity seems to be critical for regulating p53 synthesis.
Together, inhibition of mTOR decreases p53 synthesis but does not affect p53 stability. This suggested that the robust activation of p53 by glucose starvation in the TSC1−/− MEFs was due to unabated p53 synthesis by constitutive mTOR activation. Conversely, the lack of p53 response in the TSC1+/+ MEFs could be explained by inactivation of mTOR by AMPK-dependent phosphorylation of TSC2, which leads to inhibition of p53 synthesis (Inoki et al, 2003b).
mTOR regulates the association of p53 mRNA with polysomes
mTOR plays a role in the regulation of both transcription and translation; therefore, to clarify the mechanism by which mTOR affects p53 synthesis, both p53 transcription and translation were examined. In order to determine the effects of mTOR inhibition on TP53 transcription, quantitative RT–PCR (qRT–PCR) was used to determine p53 mRNA level. After glucose starvation or rapamycin treatment, the level of p53 mRNA was determined and normalized to either actin mRNA or hypoxanthine-guanine phosphoribosyltransferase (HPRT) mRNA (Figure 5C). Our data indicate that neither rapamycin nor glucose starvation significantly changed p53 mRNA levels.
To examine the effect of mTOR on p53 translation, polysome fractionation was used to determine the fraction of p53 mRNA being actively translated. Lysates were fractionated in a sucrose gradient, and mRNA was collected and analyzed by qRT–PCR to determine the relative distribution of the mRNA. In the untreated TSC1+/+ MEFs, the p53 mRNA was predominately associated with the polysome fractions (Figure 5D). However, rapamycin decreased the percentage of p53 mRNA in the polysome-associated fractions and increased the percentage of p53 mRNA in the non-polysome fractions. This shift in p53 mRNA indicated that rapamycin treatment was able to decrease the fraction of p53 mRNA being actively translated (Figure 5D). Although the decrease in p53 translation by rapamycin was not specific to p53, as seen by a corresponding decrease in polysome-associated actin mRNA (Supplementary Figure 5), the distribution of mRNA across the different fractions was different between p53 and actin. Together, the lack of change in total p53 mRNA and the shift of p53 mRNA away from the polysome by rapamycin suggests that regulation of p53 protein levels by mTOR activation is primarily due to increased translation.
Energy starvation triggers apoptosis by intrinsic pathway in TSC cells
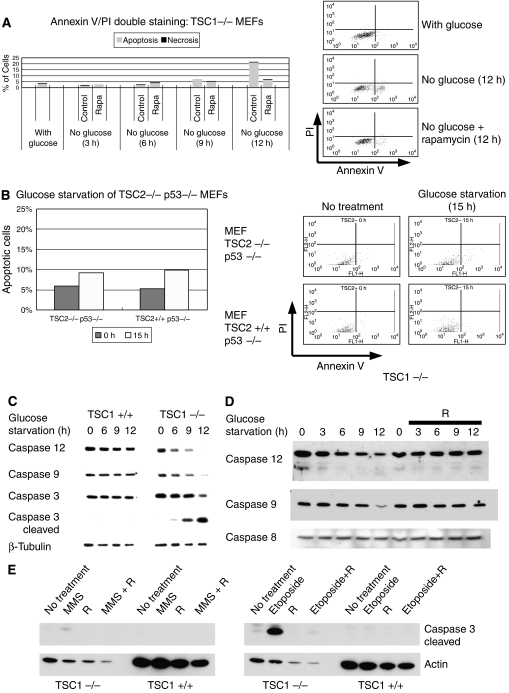
p53 is a potent activator of the intrinsic apoptotic pathway; however, energy starvation can induce cell death via both necrosis or apoptosis. To confirm that induction of p53 was triggering apoptosis in the TSC1−/− MEFs, Annexin V/propidium iodide (PI) double staining was measured by FACS analysis. TSC1−/− MEFs were glucose starved both in the presence and absence of rapamycin. FACS analysis demonstrated that the dying cells induced by glucose starvation were predominately stained by Annexin V and not PI; thus, demonstrating death was predominately apoptotic (Figure 6A). Furthermore, consistent with the protection against gross cell death (Figure 1A left) and inhibition of p53 synthesis (Figure 2A) seen earlier, rapamycin decreased apoptosis in response to glucose starvation.
Figure 6.
Energy starvation triggers apoptosis via intrinsic pathway in TSC cells. (A) Annexin V (X-axis)/propidium iodide (Y-axis) double staining showed that glucose starvation (12 h) induced cell death predominately through apoptosis as opposed to necrosis in TSC1−/− MEFs. Early apoptotic cells can be stained by Annexin V, which binds to phosphotidyl-serines normally found in the inner -aspect of the cell membrane but can be found on the outer-aspect of the cell membrane in apoptotic cells. On the other hand, during early apoptosis, PI stains DNA and is excluded from the nucleus, so staining does not occur. During necrosis and late apoptosis, membrane integrity is compromised, and cells are stained by both Annexin V and PI. (B) FACS analysis showed that TSC2−/− p53−/− and TSC2+/+ p53−/− MEFs were equally resistant to energy starvation (15 h). (C) Glucose starvation induced cleavage of caspases 12, 9, and 3 in TSC1−/− MEFs but not in TSC1+/+ MEFs. (D) Rapamycin treatment during glucose starvation prevented caspase 12 and 9 cleavage. Caspase 8 was not cleaved by glucose starvation. (E) Rapamycin pretreatment (24 h) protects against DNA damage by MMS (50 μM, 8 h) or etoposide (50 μM, 8 h) as seen by caspase 3 cleavage.
To show that p53 was important for inducing apoptosis, FACS analysis was also performed on TSC2−/− p53−/− and TSC2+/+ p53−/− MEFs. Like what was seen by gross visualization (Figure 1B right), both cell types were equally resistant to glucose starvation (Figure 6B). To exclude the possibility that the TSC2−/− MEFs had a delayed apoptotic response, Annexin V/PI doubling staining was also carried out after 24 h of glucose starvation. Again, the TSC2−/− p53−/− cells did not show enhanced sensitivity to glucose starvation (Supplementary Figure 6). In comparison, increased sensitivity to glucose starvation in the TSC1−/− MEFs was readily apparent at 12 h (Figure 6A). Together, these data suggest that p53 triggers apoptosis induced by glucose starvation, when mTOR is misregulated.
To further confirm that the intrinsic pathway was activated by glucose starvation, caspase activation was assayed by immunoblot. In the TSC1−/− MEFs, glucose starvation induced the intrinsic death pathways as seen by cleavage of caspases 9 and 12 (Figure 6C). Furthermore, the executioner caspase 3 was also activated. In contrast, glucose starvation did not activate caspase 3, 9, or 12 in the TSC1+/+ MEFs.
To demonstrate that rapamycin also prevented the induction of caspase cleavage in the TSC1−/− MEFs, mTOR was inhibited by rapamycin during glucose starvation. When TSC1−/− MEFs were rescued with rapamycin, cleavage of caspases 12 and 9 did not occur; thus, the intrinsic apoptotic pathway was not activated. Furthermore, glucose starvation had no effect on caspase 8 (Figure 6D). Taken together, glucose starvation of TSC1−/− MEFs induces apoptosis consistent with the observed changes in p53 activation.
To further confirm biochemically that rapamycin protects against DNA damage, both TSC1−/− and TSC1+/+ MEFs were treated with either MMS or etoposide. TSC1−/− MEFs were more sensitive to DNA damage by both MMS and etoposide, as seen by the induced caspase 3 cleavage. Consistently, rapamycin treatment protected TSC1−/− MEFs against DNA damage-induced cell death (Figure 6E). Taken together, DNA damage by either MMS or etoposide induces apoptosis of TSC1−/− MEFs consistent with gross observation.
p53 accumulation associated with energy stress in angiomyolipomas
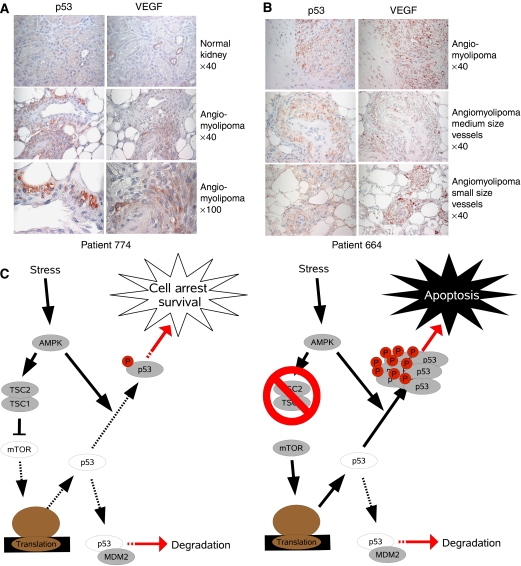
To determine whether our model of regulation of p53 by mTOR was also reflected in vivo, angiomyolipomas were stained by immunohistochemistry. Angiomyolipomas are benign tumors consisting of smooth muscle cells, adipose tissue, and blood vessels of which both the stromal cells and the vasculature demonstrate loss of heterozygosity for either TSC1 or TSC2, and thus mTOR activation (Karbowniczek et al, 2003). Like what was seen in the TSC1−/− MEFs, both sporadic and TSC disease-associated angiomyolipomas showed high levels of p53 and VEGF. It has been shown that VEGF expression can be induced by either hypoxia or loss of TSC (Brugarolas et al, 2003; El-Hashemite et al, 2003); therefore, VEGF staining may indicate areas of energy stress or TSC loss. In patient 774, who has a sporadic angiomyolipoma, both tumor and normal tissue can be compared (Figure 7A). Patients with sporadic angiomyolipomas do not have associated Tuberous Sclerosis disease, but they have Loss of Heterozygosity of TSC2; therefore, they show upregulated mTOR (Henske et al, 1995). In normal kidney cells, both VEGF and p53 staining are very low. It is interesting to note that there are small areas of VEGF upregulation, which may reflect areas of energy stress; however, in the absence of mTOR activation, p53 levels are universally low. In comparison, within the angiomyolipoma, both VEGF expression and p53 levels are correspondingly elevated. Consistently, in patient 663, who has a TSC-associated angiomyolipoma, both VEGF and p53 are elevated (Figure 7B). Furthermore, the distribution of p53 and VEGF upregulation are also strikingly similar. Together, co-elevation of p53 and VEGF in angiomyolipomas, and the lack of elevation of p53 in normal tissue may suggest that loss of TSC1/2 may also contribute to p53 accumulation during energy stress in vivo.
Figure 7.
Energy stress in angiomyolipomas is associated with p53 upregulation and model of p53 activation by energy starvation in TSC−/− cells. (A) Tissues from both normal kidney and sporadically arising angiomyolipomas were stained for p53 and VEGF. Normal tissue showed little upregulation of either p53 or VEGF, while in the angiomyolipoma, both p53 and VEGF staining were dramatically increased. (B) Tissues from TSC patient derived angiomyolipomas were stained for p53 and VEGF. Both p53 and VEGF were correspondingly increased. (C) Model for negative regulation of p53 by mTOR to promote survival during stress. When the mTOR pathway is intact, AMPK activation downregulates p53 synthesis via the mTOR pathway and stabilizes p53 via phosphorylation. However, in the absence of TSC, p53 synthesis cannot be downregulated; therefore, when AMPK stabilizes p53, p53 is greatly elevated and apoptosis is induced.
Discussion
We have shown that mTOR regulates p53 synthesis, and continued synthesis of p53 by mTOR activation sensitizes cells to p53 activators. Aberrant mTOR activation leads to increased sensitivity to both DNA damage and energy starvation. Interestingly, both DNA damage and energy starvation can also lead to AMPK activation. DNA damage by etoposide induces p53-dependent activation of AMPK, which in turn inhibits mTOR though AMPK-TSC2 (Feng et al, 2005). Similarly, we also see inhibition of mTOR via TSC2 in MMS-induced DNA damage. Therefore, our evidence suggests that through the AMPK-TSC-mTOR pathway, p53 forms a negative feedback loop to keep its own synthesis in check. Consequently, cells that cannot inhibit mTOR are faced with runaway p53 activation when stimulated.
Using energy starvation as an example, we propose a model where low energy activates AMPK to phosphorylate and stabilize p53 (Figure 7C). However, p53 induction is controlled because AMPK activation also inhibits mTOR and thereby inhibits p53 translation. Since the stabilization of p53 occurs more rapidly than the inhibition of p53 translation, a modest degree of p53 accumulation occurs before further synthesis is shut off. With both aspects of p53 regulation intact, energy starvation of wild-type cells only initiates a limited elevation of p53, which induces cell cycle arrest and protects the cells from unfavorable conditions. However, loss of TSC1 or TSC2 results in a dramatic elevation of p53 protein because p53 translation is no longer inhibited by energy starvation. The high levels of p53, therefore, induce apoptosis in TSC cells under energy starvation and may contribute to the highly apoptotic and benign nature of TSC tumors.
The delayed effect of mTOR inhibition on p53 synthesis may be explained by p53 translation via both cap-dependent and internal ribosomal entry site (IRES)-dependent mechanisms (Ray et al, 2006; Yang et al, 2006). Although rapamycin decreases phosphorylation on S6K and 4EBP very rapidly, the effects on p53 synthesis require several hours to become apparent. In TSC1−/− MEFs, rapamycin can rapidly shut down cap-dependent p53 synthesis; however, IRES-dependent p53 translation may continue as a result of low AKT levels (Ray et al, 2006; Yang et al, 2006). After prolonged exposure to rapamycin, negative feedback on AKT via mTOR-S6K1-IRS-1 is relieved, and AKT activity is restored, which inhibits IRES-dependent p53 translation. Thus, only after rapamycin inhibits both mechanisms for p53 translation does the change in p53 synthesis become easily detectable.
Since mTOR acts upon p53 synthesis, the effect of rapamycin is ambivalent toward the initiator of p53 stabilization. It has also been reported that treatment with rapamycin reduces p53-dependent apoptosis by HIV infection (Castedo et al, 2001) and ionization radiation (Tirado et al, 2003). Consistent with mTOR's role in regulating p53 synthesis, it has also been reported that loss of PTEN, which activates mTOR by activation of AKT, also increases p53 expression and upregulation of p53 gene targets (Kim et al, 2007). Furthermore, TSC2−/− MEFs undergo early senescence through elevated p21, a transcriptional target of p53, and loss of p53 is necessary to prevent senescence (Zhang et al, 2003a). Taken together, it is possible that mTOR is critical for modulating the effects of p53 during a variety of stresses.
It has been previously shown that AMPK activation leads to direct phosphorylation of p53 Ser15 by AMPK. AMPK activation also inhibits mTOR via TSC2 phosphorylation. Inactivation of mTOR also activates the α4/PP2A phosphatase complex, which plays a role in dephosphorylation of p53 Ser15; therefore, inactivation of the PP2A phosphatase also contributes to the increase of p53 phosphorylation in an mTOR-dependent manner (Levine et al, 2006). Additionally, our studies do not elucidate whether the regulation of p53 phosphorylation by mTOR is direct; therefore, downstream effectors such as S6K may play a role in p53 phosphorylation. Further studies using the S6K1 and S6K2 double knockout cells or S6K RNAi would provide further insight into S6K's role in p53 regulation.
In addition to DNA damage and glucose starvation, activation of mTOR has also been shown to sensitize cells to cell death by other stimuli. Activation of AKT plays an important role in cell survival; however, activation of AKT is antagonized by mTOR-dependent inhibition of IRS-1 via S6K. Consequently, TSC1−/− and TSC2−/− p53−/− MEFs have also shown an increased sensitivity to serum starvation (Shah et al, 2004). Furthermore, activation of mTOR has also been shown to reduce NF-κB activation, thereby sensitizing cells to apoptosis. Through downregulation of NF-κB, TSC1−/− and TSC2−/− p53−/− MEFs are sensitized to TNFα and DNA damage (Ghosh et al, 2006).
The majority of these studies have been performed in TSC1−/− and TSC2−/− cells, which assumes that the effect of TSC1 and TSC2 loss is constitutive mTOR activation. It is possible that TSC1 and TSC2 have functions in addition to inhibiting mTOR. However, inhibition of mTOR by rapamycin rescued glucose deprivation-induced apoptosis. Furthermore, overexpression of Rheb sensitized cells to stress-induced apoptosis. Our data are consistent with Rheb-mTOR playing a major role in the hypersensitivity of TSC mutant cells in response to stress, although we cannot exclude the involvement of mTOR-independent function in this apoptotic response.
From our experiments, we have identified a novel mechanism by which mTOR regulates p53 to maintain cell viability. This provides new insights into the proapoptotic role of mTOR and may help to explain the benign nature of many hamartoma syndromes, including TSC (Inoki et al, 2005). Immunohistochemical staining of TSC tumors showed concurrent staining by VEGF and p53, which indicates that in tumors lacking TSC, p53 levels are substantially elevated under stress conditions (Figure 7A and B). These data indicate that our model of overaccumulation of p53 by disruption of the mTOR pathway is not limited to just cell culture but may also play a role in vivo. Given that the TP53 gene encodes the most commonly mutated tumor suppressor in human cancers, our study suggests that p53 status may be important for determining the effect of mTOR inhibitors against various malignant and benign neoplasms. However, recent studies have shown that mTOR inhibitors enhance the effects of chemotherapeutics in A549 human non-small-cell lung carcinomas as well as ovarian cancer cells (Beuvink et al, 2005; Treeck et al, 2006). It is possible that the protective effect of rapamycin against DNA damage agents may be limited to benign tumors such as tuberous sclerosis.
Materials and methods
Antibodies and materials
Anti-caspase 12, anti-caspase 9, anti-caspase 3, anticleaved caspase 3, anti-p53, anti-phospho p53 (S6, S9, S15, S20, S392), anti-AMPK, and anti-phospho AMPK (T172) antibodies were obtained from Cell Signaling (Beverly, MA). Anti-Actin, anti-β-tubulin, and anti-caspase 8 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Horseradish peroxidase-conjugated IgG secondary antibodies were obtained from Amersham (Buckinghamshire, UK).
The AMPK inhibitor, commonly known as compound C, was obtained from Merck (Whitehouse Station, NJ) and was described previously (Zhou et al, 2001). Cells were treated with 10 μM of the compound suspended in DMSO. Rapamycin was purchased from Cell Signaling, suspended in methanol, and used at 20 nM. The caspase 9 inhibitor Z-LEHD-FMK and pan-caspase inhibitor Z-VAD-FMK were purchased at R&D Systems and used at 20 μM 1 h prior to and during glucose starvation. The calpain inhibitors, ALLN and ALLM, were purchased at EMD Biosciences, and treatment with 10 μM started 1 h prior to and during glucose starvation. The proteosome inhibitor MG132 was obtained from Sigma and used at 20 μM. Protein stability was assayed with cycloheximide from Sigma and used at 50 ng/ml. Etoposide was purchased from Sigma and used at 6 μg/ml. MMS was also purchased from Sigma.
Cell culture and transfection
MEF cells were cultured in DMEM (Invitrogen) containing 10% fetal bovine serum (Invitrogen) and 50 μg/ml penicillin/streptomycin (P/S). TSC2−/− LExF2 cells (LEF) were maintained in DMEM/F12 (Invitrogen) containing 10% FBS and 50 μg/ml P/S. Glucose starvation was performed with glucose-free DMEM (Invitrogen) containing 25 mM HEPES, 10% dialyzed FBS (Invitrogen), and 50 μg/ml P/S.
RNAi used to knock TSC2 (smart pool) was purchased from Dharmacon.
Annexin V/PI staining
Annexin V/PI double staining was carried out with Annexin V and PI (BD Biosciences) as per the manufacturer's protocol, and samples were analyzed via BD FACScalibur (BD Biosciences).
35S labeling
35S Pulse/Chase labeling was carried out with 0.2 mCi/ml of 35S-Met/Cys Trans label in DMEM (−Met/−Cys) containing 10% dialyzed FBS for 1 h prior to chase with DMEM containing 10% dialyzed FBS, 18 mg L-Cys/100 ml media, and 9 mg L-Met/100 ml media. p53 was immunoprecipitated with anti-p53 antibodies and resolved by SDS–PAGE.
35S Pulse labeling for assay of p53 synthesis was performed with labeling media as described earlier but incubated for only 8 min prior to immunoprecipitation by anti-p53 antibodies and resolution by SDS–PAGE.
qRT–PCR
Dishes (10 cm) of TSC1−/− or TSC1+/+ MEFs were glucose starved or rapamycin treated for 6 h prior to lysis with 1 ml Trizol (Sigma), and the aqueous layer was collected after addition of 200 μl chloroform. mRNA was precipitated with 1 volume isopropanol. The isopropanol was removed with a 70% ethanol wash and the RNA pellet air dried. Reverse transcription was performed with the Superscript First Strand Synthesis System for RT–PCR (Invitrogen) as per the manufacturer's protocol.
Quantitative PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems). p53 was amplified using the forward primer 5′-AACCGCCGACCTATCCTTAC-3′ and the reverse primer 5′-CTTCTGTACGGCGGTCTCTC-3′. HPRT was amplified using the forward primer 5′-TCATTATGCCGAGGATTTGGA-3′ and the reverse primer 5′-GCACACAGAGGGCCACAAT-3′. Actin was amplified using the forward primer 5′-CCGGGAGAAGATGACTCAAA-3′ and the reverse primer 5′-CCAGAATCCAACACGATGC-3′. Samples were done in triplicate to calculate averages and standard deviations.
Polysome fractionation
Mouse embryonic fibroblasts were cultured in DMEM, supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). MEFs were seeded in 150 mm Petri dishes (5 × 106 cells/dish) and collected 24 h later. Prior to harvesting, cells were treated with cycloheximide (100 μg/ml) for 10 min. Cells were then washed twice with 5 ml of PBS (containing 100 μg/ml), collected by scraping and pelleted at 500 g for 5 min. Cells were lysed in 0.8 ml of extraction buffer (5 mM Tris (pH 7.5), 2.5 mM MgCl2, 1.5 mM KCl, 100 μg/ml cycloheximide, 2 mM DTT, 0.5% Triton-X 100, and 0.5% sodium deoxycholate). Extracts were cleared by centrifugation at 13 000 g for 2 min and then loaded on 11 ml sucrose gradients (10–50%) buffered in 20 mM HEPES (pH 7.6), 100 mM KCl, 5 mM MgCl2. Gradients were subjected to centrifugation using a Beckman SW40Ti Rotor at 38 000 r.p.m. for 2.2 h at 4°C. Gradients were then fractionated (from the lightest to the heaviest fraction) into 24 fractions (12 drops per fraction; approximately 0.5 ml) while monitoring the optical density at 254 nm. Adjacent fractions were pooled to yield a total of 12 fractions for qPCR.
Immunohistochemistry
Sections (4 μm) were deparaffinized in xylene and rehydrated in a gradient series of ethanol. For antigen retrieval, sections were boiled in Citric Buffer (10 mM sodium citrate-trisodium salt dehydrate, Sigma, St Louis, MO), pH 6.0, for 10 min. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in methanol for 15 min at room temperature. Nonspecific background was eliminated by incubating the tissue with normal goat serum for 10 min at room temperature (Zymed, San Francisco, CA). The sections were then incubated in a humidified chamber with mouse monoclonal antibody against p53 (1C12), dilution 1:100 (Cell Signaling Technology, Beverly, MA) or prediluted rabbit monoclonal antibody against VEGF (SP28) (Abcam Inc., Cambridge, MA) overnight at 4°C. The slides were then washed, incubated with biotinylated affinity-purified secondary antibodies (Zymed, San Francisco, CA) for 10 min at room temperature, then washed and incubated with enhanced horseradish peroxidase-conjugated streptavidin (Zymed) for 10 min at room temperature. After washing, the slides were developed using AEC Chromogen Solution (Zymed), lightly counterstained with hematoxylin (Biomeda, Foster City, CA), and mounted using GelMount (Biomeda).
Supplementary Material
Supplementary Figures
Supplementary Figure Legends
Acknowledgments
We are indebted to Dr RS Yeung for providing the LExF2 cell line; Dr DJ Kwiatkowski for TSC1−/−, +/+ and TSC2−/−,+/+ p53−/− MEFs; Dr G Zhou for the AMPK inhibitor. We also thank Jeffery Lee, Grant Yang, Eunjung Kim, Qian Yang, and Xiaoming Zhao for their critical reading and comments on the manuscript, and this study was funded by grants from the NIH and Department of Defense (KLG).
References
- Ali SM, Sabatini DM (2005) Structure of S6 kinase 1 determines whether raptor-mTOR or rictor-mTOR phosphorylates its hydrophobic motif site. J Biol Chem 280: 19445–19448 [DOI] [PubMed] [Google Scholar]
- Beranek DT (1990) Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res 231: 11–30 [DOI] [PubMed] [Google Scholar]
- Beuvink I, Boulay A, Fumagalli S, Zilbermann F, Ruetz S, O'Reilly T, Natt F, Hall J, Lane HA, Thomas G (2005) The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell 120: 747–759 [DOI] [PubMed] [Google Scholar]
- Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG Jr (2003) TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell 4: 147–158 [DOI] [PubMed] [Google Scholar]
- Castedo M, Ferri KF, Blanco J, Roumier T, Larochette N, Barretina J, Amendola A, Nardacci R, Metivier D, Este JA, Piacentini M, Kroemer G (2001) Human immunodeficiency virus 1 envelope glycoprotein complex-induced apoptosis involves mammalian target of rapamycin/FKBP12-rapamycin-associated protein-mediated p53 phosphorylation. J Exp Med 194: 1097–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro AF, Rebhun JF, Clark GJ, Quilliam LA (2003) Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J Biol Chem 278: 32493–32496 [DOI] [PubMed] [Google Scholar]
- Duckett DR, Bronstein SM, Taya Y, Modrich P (1999) hMutSalpha- and hMutLalpha-dependent phosphorylation of p53 in response to DNA methylator damage. Proc Natl Acad Sci USA 96: 12384–12388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hashemite N, Walker V, Zhang H, Kwiatkowski DJ (2003) Loss of Tsc1 or Tsc2 induces vascular endothelial growth factor production through mammalian target of rapamycin. Cancer Res 63: 5173–5177 [PubMed] [Google Scholar]
- Feng Z, Zhang H, Levine AJ, Jin S (2005) The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA 102: 8204–8209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G (2003) Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 11: 1457–1466 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Tergaonkar V, Rothlin CV, Correa RG, Bottero V, Bist P, Verma IM, Hunter T (2006) Essential role of tuberous sclerosis genes TSC1 and TSC2 in NF-kappaB activation and cell survival. Cancer Cell 10: 215–226 [DOI] [PubMed] [Google Scholar]
- Hardie DG, Carling D, Carlson M (1998) The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem 67: 821–855 [DOI] [PubMed] [Google Scholar]
- Henske EP, Neumann HP, Scheithauer BW, Herbst EW, Short MP, Kwiatkowski DJ (1995) Loss of heterozygosity in the tuberous sclerosis (TSC2) region of chromosome band 16p13 occurs in sporadic as well as TSC-associated renal angiomyolipomas. Genes Chromosomes Cancer 13: 295–298 [DOI] [PubMed] [Google Scholar]
- Hresko RC, Mueckler M (2005) mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem 280: 40406–40416 [DOI] [PubMed] [Google Scholar]
- Inoki K, Corradetti MN, Guan KL (2005) Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet 37: 19–24 [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL (2003a) Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 17: 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL (2002) TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4: 648–657 [DOI] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL (2003b) TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590 [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 6: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB (2005) AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 18: 283–293 [DOI] [PubMed] [Google Scholar]
- Kahn BB, Alquier T, Carling D, Hardie DG (2005) AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab 1: 15–25 [DOI] [PubMed] [Google Scholar]
- Kapoor M, Hamm R, Yan W, Taya Y, Lozano G (2000) Cooperative phosphorylation at multiple sites is required to activate p53 in response to UV radiation. Oncogene 19: 358–364 [DOI] [PubMed] [Google Scholar]
- Karbowniczek M, Yu J, Henske EP (2003) Renal angiomyolipomas from patients with sporadic lymphangiomyomatosis contain both neoplastic and non-neoplastic vascular structures. Am J Pathol 162: 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee C, Bonifant CL, Ressom H, Waldman T (2007) Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Mol Cell Biol 27: 662–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Inoki K, Guan KL (2007) mTOR pathway as a target in tissue hypertrophy. Annu Rev Pharmacol Toxicol 47: 443–467 [DOI] [PubMed] [Google Scholar]
- Levine AJ, Feng Z, Mak TW, You H, Jin S (2006) Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev 20: 267–275 [DOI] [PubMed] [Google Scholar]
- Li Y, Inoki K, Guan KL (2004) Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol Cell Biol 24: 7965–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN (2002) Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10: 457–468 [DOI] [PubMed] [Google Scholar]
- Ray PS, Grover R, Das S (2006) Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep 7: 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14: 1296–1302 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 22: 159–168 [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307: 1098–1101 [DOI] [PubMed] [Google Scholar]
- Shah OJ, Wang Z, Hunter T (2004) Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol 14: 1650–1656 [DOI] [PubMed] [Google Scholar]
- Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC (2004) The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6: 91–99 [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J (2003) Tuberous sclerosis complex gene products, tuberin and hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 13: 1259–1268 [DOI] [PubMed] [Google Scholar]
- Tirado OM, Mateo-Lozano S, Sanders S, Dettin LE, Notario V (2003) The PCPH oncoprotein antagonizes the proapoptotic role of the mammalian target of rapamycin in the response of normal fibroblasts to ionizing radiation. Cancer Res 63: 6290–6298 [PubMed] [Google Scholar]
- Treeck O, Wackwitz B, Haus U, Ortmann O (2006) Effects of a combined treatment with mTOR inhibitor RAD001 and tamoxifen in vitro on growth and apoptosis of human cancer cells. Gynecol Oncol (E-pub ahead of print) [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M, Nellist M, Nagelkerken B, Cheadle J, Snell R, van den Ouweland A, Reuser A, Sampson J, Halley D, van der Sluijs P (1998) Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum Mol Genet 7: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Wataya-Kaneda M, Kaneda Y, Hino O, Adachi H, Hirayama Y, Seyama K, Satou T, Yoshikawa K (2001) Cells derived from tuberous sclerosis show a prolonged S phase of the cell cycle and increased apoptosis. Arch Dermatol Res 293: 460–469 [DOI] [PubMed] [Google Scholar]
- Yang D, Halaby M, Zhang Y (2006) The identification of an internal ribosomal entry site in the 5-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene 25: 4613–4619 [DOI] [PubMed] [Google Scholar]
- Young J, Povey S (1998) The genetic basis of tuberous sclerosis. Mol Med Today 4: 313–339 [DOI] [PubMed] [Google Scholar]
- Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, Vazquez F, Carpenter CL, Kwiatkowski DJ (2003a) Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest 112: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D (2003b) Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol 5: 578–581 [DOI] [PubMed] [Google Scholar]
- Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, Wu M, Ventre J, Doebber T, Fujii N, Musi N, Hirshman MF, Goodyear LJ, Moller DE (2001) Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest 108: 1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Figure Legends