Abstract
Previous studies in human cells indicate that sister telomeres have distinct requirements for their separation at mitosis. In cells depleted for tankyrase 1, a telomeric poly(ADP-ribose) polymerase, sister chromatid arms and centromeres separate normally, but telomeres remain associated and cells arrest in mitosis. Here, we use biochemical and genetic approaches to identify proteins that might mediate the persistent association at sister telomeres. We use immunoprecipitation analysis to show that the telomeric proteins, TRF1 (an acceptor of PARsylation by tankyrase 1) and TIN2 (a TRF1 binding partner) each bind to the SA1 ortholog of the cohesin Scc3 subunit. Sucrose gradient sedimentation shows that TRF1 cosediments with the SA1–cohesin complex. Depletion of the SA1 cohesin subunit or the telomeric proteins (TRF1 and TIN2) restores the normal resolution of sister telomeres in mitosis in tankyrase 1-depleted cells. Moreover, depletion of TRF1 and TIN2 or SA1 abrogates the requirement for tankyrase 1 in mitotic progression. Our studies indicate that sister telomere association in human cells is mediated by a novel association between a cohesin subunit and components of telomeric chromatin.
Keywords: cohesins, sister chromatids, tankyrase 1, telomeres, TRF1
Introduction
Human telomeres are specialized nucleoprotein complexes comprised of TTAGGG repeats and a six-subunit protein complex termed shelterin that is required for the protection and replication of chromosome ends, reviewed by de Lange (2005). Shelterin contains two double-stranded DNA-binding proteins TRF1 (Chong et al, 1995) and TRF2 (Bilaud et al, 1997; Broccoli et al, 1997), that mediate the replication and protective functions, respectively (van Steensel and de Lange, 1997; van Steensel et al, 1998). TRF1 and TRF2 do not interact directly, but they do have a common binding partner, TIN2 (Kim et al, 1999, 2004; Houghtaling et al, 2004; Liu et al, 2004a; Ye et al, 2004a). TIN2 binds to TPP1 (Houghtaling et al, 2004; Liu et al, 2004b; Ye et al, 2004b), that in turn recruits POT1, which binds to the single-stranded 3′ extension at chromosome ends (Baumann and Cech, 2001; Loayza and de Lange, 2003). The sixth subunit, RAP1 binds mostly to TRF2 (Li et al, 2000). While shelterin can be detected as a single complex, subcomplexes containing either TRF1 or TRF2 along with other subunits predominate in lysed cells (Houghtaling et al, 2004; Kim et al, 2004; Liu et al, 2004a; Ye et al, 2004a).
Tankyrase 1 is a poly(ADP-ribose) polymerase (PARP) that regulates TRF1 function at telomeres, reviewed by Hsiao and Smith (2007). Tankyrase 1 poly(ADP-ribosyl)ates (PARsylates) TRF1 in vitro inhibiting its binding to telomeric DNA (Smith et al, 1998). Overexpression of tankyrase 1 removes TRF1 from telomeres (Smith and de Lange, 2000; Cook et al, 2002), resulting in TRF1 ubiquitination and degradation by the proteasome (Chang et al, 2003). Long-term overexpression of tankyrase 1 leads to telomere elongation (Smith and de Lange, 2000; Cook et al, 2002), dependent on the catalytic PARP activity of tankyrase 1 and on telomerase (Cook et al, 2002; Chang et al, 2003). Conversely, long-term partial knockdown of tankyrase 1 in telomerase-positive cells leads to telomere shortening (Donigian and de Lange, 2007). Together these data indicate tankyrase 1 as a positive regulator of telomere length. Tankyrase 1 PARP activity at telomeres is regulated by TIN2 (Ye and de Lange, 2004). TIN2 does not bind directly to tankyrase, but rather forms a ternary complex with TRF1 and tankyrase 1, where it protects TRF1 from PARsylation by tankyrase 1 (Ye and de Lange, 2004).
Depletion of tankyrase 1 in HeLa cells by siRNA led to a mitotic arrest (Dynek and Smith, 2004). This phenotype could be rescued by wild-type tankyrase 1, but not a PARP dead mutant, indicating a requirement for PARsylation (Dynek and Smith, 2004). Tankyrase 1-depleted mitotic cells were unable to resolve their telomeres despite separation of sister chromatid arms and centromeres. Live cell imaging showed that in tankyrase 1-depleted cells chromosomes congressed normally to the metaphase plate, but then cells underwent a struggle to segregate their chromosomes (Dynek and Smith, 2004). These and other observations led to the hypothesis that in the absence of tankyrase 1, cells arrest in early anaphase with unresolved sister telomeres (Dynek and Smith, 2004). A subsequent study observed a similar mitotic arrest in tankyrase 1-depleted cells, but observed fully paired sister chromatids and proposed a pre-anaphase arrest (Chang et al, 2005b).
Sister chromatids are held together from the time of their replication in S phase until their separation in mitosis by cohesin, a multisubunit ring-shaped complex (Haering et al, 2002; Gruber et al, 2003). Cohesin consists of a heterodimer of Smc1 and Smc3 bound to Scc1 and Scc3, reviewed by Losada and Hirano (2005) and Nasmyth and Haering (2005). Vertebrate cells contain two orthologs of Scc3, termed SA1 and SA2 (Losada et al, 2000; Sumara et al, 2000), with SA2 the major ortholog in HeLa cells (Losada et al, 2000; Sumara et al, 2000). Cohesin complexes contain either SA1 or SA2, but not both. In human cells, cohesins are removed in a two-step process involving post-translational modification of the Scc3 and Scc1 subunits; cohesin is removed from chromosome arms in prophase by phosphorylation of the Scc3 variant SA2 (Waizenegger et al, 2000; Losada et al, 2002; Sumara et al, 2002; Hauf et al, 2005) and from centromeres at the metaphase to anaphase transition by proteolytic cleavage of Scc1 (Hauf et al, 2001).
While studies indicate distinct mechanisms for removal of arm and centromere cohesion, very little is not known about removal of cohesion from telomeres. Previous studies used chromosome specific fluorescent in situ hybridization (FISH) to analyze the timing of sister chromatid resolution at human telomeric regions and found that sister telomeres (like arms) were fully resolved by the time cells reached metaphase (Ofir et al, 2002; Yalon et al, 2004). Interestingly, however, as cells approached senescence they displayed persistent sister telomere (but not arm) associations in metaphase, leading to the speculation that in human cells telomeric regions could be sites of persistent cohesion (Ofir et al, 2002; Yalon et al, 2004). Similarly, our studies indicate that in the absence of tankyrase 1 sister chromatid associations persist at telomeres in mitosis. Our observation that these associations are not due to a gross block in telomere replication or to covalent linkage (Dynek and Smith, 2004) suggest that protein–protein interactions might mediate the persistent sister telomere associations in tankyrase 1-depleted cells.
Here, we take a biochemical and a genetic approach to identify proteins required for the persistent telomere associations observed in tankyrase 1-depleted cells. We show that the telomeric proteins TRF1 and TIN2 each bind to the Scc3 cohesin ortholog SA1, but not the closely related SA2. We demonstrate that depletion of TRF1 and TIN2 or SA1 can abrogate the requirement for tankyrase 1 in sister telomere resolution and mitotic progression. Our studies suggest a novel mode of association between cohesins and telomeric chromatin in human cells.
Results
Tankyrase 1-depleted cells have persistent sister telomere associations
We showed previously using a combination of immunofluorescence analysis (FISH) and chromosome spread analysis that sister chromatids were separated at centromeres and arms, but remained associated at telomeres (Dynek and Smith, 2004). A subsequent study called these results into question. Using electron microscopy these authors observed fully paired sister chromatids in misaligned mitotic cells and thus suggested that sister chromatid cohesion was intact in tankyrase 1 siRNA cells (Chang et al, 2005b). To resolve this discrepancy, we repeated and extended our analysis of sister chromatid associations using more quantitative and conventional approaches.
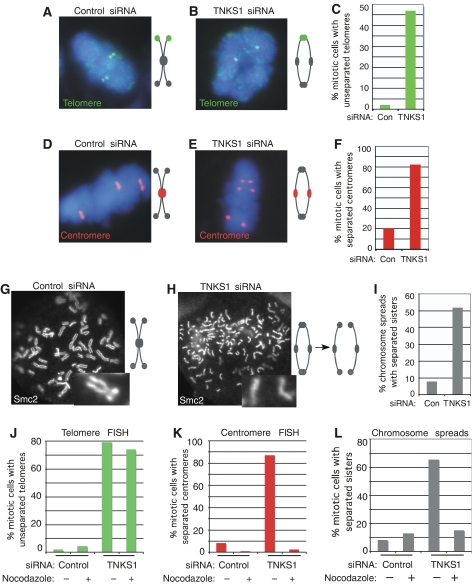
First, we used chromosome-specific FISH to analyze the association of sister chromatids in tankyrase 1-depleted cells. HelaI.2.11 cells were treated for 48 h with tankyrase 1 siRNA, isolated by mitotic shake-off, fixed, and analyzed by FISH. As shown in Figure 1A, and previously (Dynek and Smith, 2004), in control mitotic cells sister telomeres appear as doublets indicating resolution of sister telomeres. By contrast, in tankyrase 1 siRNA cells telomeric regions appear as singlets (Figure 1B), indicating a block in resolution of sister telomeres. Quantification of this analysis shows a greater than 10-fold increase in unresolved telomeres in tankyrase 1-depleted versus control cells (Table I; Figure 1C). We next addressed the cohesion status of centromeres. Note that the centromere probe used here detects a locus that is trisomic in HeLa cells and therefore labels three (rather than two) pairs of sister chromatids. In control mitotic cells, sister centromeres are tightly associated and thus appear as a closely opposed doublets (Figure 1D). By contrast, in tankyrase 1 siRNA cells, there is a clear separation of sister centromeres, indicating a loss in centromeric cohesion (Figure 1E and F; Table II).
Figure 1.
Sister telomeres remain associated in tankyrase 1 siRNA cells. (A–F) Chromosome-specific FISH analysis of HeLaI.2.11 cells collected by mitotic shake-off at 48 h after treatment with control (GFP) or tankyrase 1 siRNA. Cells were fixed directly in methanol-acetic acid without hypotonic swelling and hybridized to a telomere probe 16pter (green) (A, B) or a centromere probe 6cen (red) (D, E). DNA was stained with DAPI (blue). (C, F) Histograms showing the percentage of mitotic cells with unseparated telomeres (C) or separated centromeres (F); at least 100 mitotic cells were scored for each sample. (G–I) Chromosome spread analysis of HelaI.2.11 cells collected after 48 h of treatment with control (GFP) or tankyrase 1 siRNA, swollen in hypotonic buffer and fixed in paraformaldehyde. Cells were treated with colcemide 90 min before harvesting. Chromosome arms were visualized by staining with antibodies to the condensin subunit Smc2. (I) Histogram showing % mitotic cells with single sisters; at least 200 spreads were scored for each sample. (J–L) Histograms showing analysis of nocodazole-arrested cells. HeLaI.2.11 cells were treated with control (GFP) or tankyrase 1 siRNA for 16 h and then incubated without (−) or with (+) nocodazole for an additional 12 h, harvested and processed for telomere FISH (J), centromere FISH (K), and chromosome spreads (L) as described above. Approximately 100 mitotic cells or more were scored for each sample.
Table 1.
Chromosome-specific FISH analysis of telomere cohesion using a 16pter probe on HeLaI.2.11 cells following 48 h of transfection with the indicated siRNAs
| 1st siRNA | 2nd siRNA | % Two singlets | % Two doublets | % Other | No. of cells examined |
|---|---|---|---|---|---|
| GFP | — | 2 | 71 | 27 | 128 |
| TNKS1 | — | 47 | 30 | 23 | 356 |
| TNKS1 | GFP | 60 | 17 | 24 | 153 |
| GFP | SA1.a | 6 | 76 | 18 | 143 |
| TNKS1 | SA1.a | 11 | 67 | 22 | 115 |
| GFP | SA2.a | 9 | 78 | 14 | 103 |
| TNKS1 | SA2.a | 60 | 15 | 25 | 125 |
| GFP | TRF1.b | 11 | 69 | 20 | 100 |
| TNKS1 | TRF1.b | 45 | 32 | 24 | 110 |
| GFP | TIN2.a | 10 | 70 | 29 | 100 |
| TNKS1 | TIN2.a | 13 | 47 | 40 | 100 |
Table 2.
Chromosome-specific FISH analysis of centromere cohesion using a 6cen probe on HeLaI.2.11 cells following 48 h of transfection with the indicated siRNAs
| 1st siRNA | 2nd siRNA | % Three separated | % Three together | % Other | No. of cells examined |
|---|---|---|---|---|---|
| GFP | — | 20 | 71 | 9 | 128 |
| TNKS1 | — | 82 | 13 | 5 | 263 |
| TNKS1 | GFP | 59 | 39 | 2 | 105 |
| GFP | SA1.a | 16 | 80 | 3 | 116 |
| TNKS1 | SA1.a | 27 | 72 | 1 | 134 |
| GFP | TIN2.a | 12 | 85 | 4 | 137 |
| TNKS1 | TIN2.a | 15 | 82 | 3 | 118 |
In a second approach, we used chromosome spread analysis to analyze sister chromatid cohesion. As shown in Figure 1G, in control spreads sister chromatids are resolved along their arms, but remain associated at their centromeres. By contrast, in tankyrase 1 siRNA cells chromosomes appear as separated sister chromatids, consistent with progression to anaphase and a loss in centromeric cohesion (Figure 1H and I; Table III).
Table 3.
Chromosome spread analysis of HeLaI.2.11 cells following 48 h of transfection with the indicated siRNAs
| 1st siRNA | 2nd siRNA | % Sisters separated | % Sisters together | No. of cells examined |
|---|---|---|---|---|
| GFP | — | 8 | 92 | 922 |
| TNKS1 | — | 52 | 48 | 215 |
| TNKS1 | GFP | 26 | 74 | 814 |
| GFP | SA1.a | 9 | 911 | 243 |
| TNKS1 | SA1.a | 10 | 90 | 768 |
| GFP | TIN2.a | 6 | 94 | 203 |
| TNKS1 | TIN2.a | 7 | 93 | 970 |
We note that the telomere associations observed by FISH analysis (Figure 1B) do not survive chromosome spread preparation (Figure 1H). Spread preparations differ from FISH analysis in two ways, either of which could influence sister telomere associations. First, spread preparations include a 90 min colcemide treatment. Studies have shown that spindle poisons can promote separation of sister arms (Rieder and Cole, 1999). Thus, we asked if sister chromatids in tankyrase 1-depleted cells would separate even in the absence of colcemide. As shown in Supplementary Figure S1, sister chromatids are separated in spreads prepared without colcemide treatment from tankyrase 1-depleted cells, indicating that the loss in telomere association is not due to colcemide per se. The second difference between FISH analysis and chromosome spread preparations, is the inclusion of hypotonic treatment in spread preparations. This treatment, which was developed to enhance visualization of sister chromatid arms, may also release chromosomal proteins (Ohnuki, 1968). To determine if hypotonic treatment releases persistent sister telomere associations, cells were subjected to hypotonic treatment prior to FISH analysis. As shown in Supplementary Figure S2, persistent sister telomere associations are resolved by hypotonic treatment, suggesting that these associations do not survive chromosome spread analysis due to the hypotonic treatment.
As described above, in tankyrase 1-depleted cells centromeres separate, but telomeres remain associated. Based on these and previous results (Dynek and Smith, 2004), we hypothesize that tankyrase 1-depleted cells proceed normally through metaphase, centromeres separate, but cells arrest in early anaphase with unresolved sister telomeres. To determine if centromere separation in tankyrase 1-depleted cells results from normal mitotic progression, we blocked progression by incubation with the spindle poison nocodazole, which arrests cells in prometaphase. For these experiments, to capture tankyrase 1-depleted cells before they undergo mitotic arrest, cells were treated with tankyrase 1 siRNA for a short time (16 h) and then nocodazole was added for an additional 12 h incubation to allow cells to accumulate in prometaphase. First, as shown by FISH in Figure 1J, we observe persistent sister telomere associations in tankyrase 1-depleted cells in the presence or absence of nocodazole, indicating that prometaphase arrest (or long-term treatment with spindle poison) does not influence sister telomere association. Strikingly, however, sister centromeres do not separate in tankyrase 1-depleted cells arrested in prometaphase, as measured by FISH (Figure 1K) and chromosome spread analysis (Figure 1L), indicating that when tankyrase 1-depleted cells are prevented from progressing through mitosis, sister centromeres do not separate. These data are consistent with the notion that centromere separation in tankyrase 1-depleted cells results from progression to anaphase.
TRF1 associates with the SA1–cohesin complex
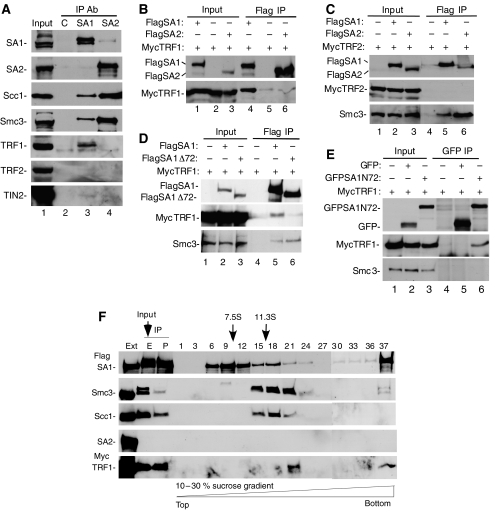
As described above persistent telomere associations in tankyrase 1-depleted cells are sensitive to hypotonic treatment. This observation, combined with our previous analysis showing that persistent telomere associations are not due to incomplete DNA replication or to covalent ligation, suggests that telomeres remain associated through proteinaceous bridges. We thus asked what might be mediating the persistent association at telomeres. Previous studies indicated a critical role for the Scc3-SA2 cohesin subunit in loss of cohesion along chromosome arms (Hauf et al, 2005). We thus tested for interaction between the Scc3 subunit and telomeric proteins. Immunoprecipitations were performed with lysates from logarithmically growing HeLa cells using antibodies directed against the two variants of Scc3 (SA1 or SA2) and analyzed by immunoblotting with antibodies to cohesin subunits (SA1, SA2, Scc1, or Smc3) or shelterin subunits (TRF1, TRF2, or TIN2). As shown in Figure 2A, lane 3, SA1 immunoprecipitates (IPs) contained the cohesin subunits Scc1 and Smc3. The SA1 IPs did not contain SA2, as expected since cohesin complexes contain only SA1 or SA2. Of note, SA1 IPs also contained the telomeric protein TRF1. By contrast, SA2 IPs (which, due to the greater abundance of SA2 in HeLa cells, contained more Scc1 and Smc3 than SA1 IPs) did not contain TRF1 (Figure 2A, lane 4). TIN2 was only weakly detected in the SA1 IP (see below), but not in the SA2 IP, and TRF2 was not detected in SA1 or SA2 IPs. These data demonstrate that endogenous SA1 (not SA2) binds to TRF1 (not TRF2) in human cells.
Figure 2.
TRF1 binds to the SA1 cohesin complex. (A) Endogenous TRF1 is co-immunoprecipitated by SA1 not SA2. HeLaI.2.11 cells were lysed and immunoprecipitated (IP) with anti-Flag as a control (C), anti-SA1 (BL143G), or anti-SA2. Proteins were fractionated on SDS–PAGE and analyzed by immunoblotting with antibodies against SA1, SA2, Scc1, Smc3, TRF1, TRF2, or TIN2. (B, C) Exogenous TRF1 not TRF2 is co-immunoprecipitated by SA1 not SA2. 293T cells were transfected with FlagSA1 or FlagSA2 and (B) MycTRF1 or (C) MycTRF2. Cell lysates were immunoprecipitated with anti-Flag beads and analyzed by immunoblotting with antibodies against Flag, Myc, or Smc3. (D) TRF1 is not co-immunoprecipitated by SA1Δ72. 293T cells were transfected with FlagSA1 or FlagSA1Δ72 and MycTRF1. Cell lysates were immunoprecipitated with anti-Flag beads and analyzed by immunoblotting with antibodies against Flag, Myc, or Smc3. (E) The N-terminal 72 amino acids of SA1 is sufficient for binding to TRF1. 293T cells were transfected with GFP or GFPSA1N72 and MycTRF1. Cell lysates were immunoprecipitated with anti-GFP antibody and analyzed by immunoblotting with antibodies against GFP, Myc, or Smc3. (A–E) Input indicates 4% of extract. (F) TRF1 forms a complex with SA1–cohesin. 293T cells were transfected with FlagSA1 and MycTRF1. Cell extracts (ext; 1% of total) were immunoprecipitated with anti-Flag beads. The immunocomplex was eluted (E; 2.5% of total) from the beads (P, pellet; 2.5% of total) with Flag peptide and separated by sucrose density gradient sedimentation. Fractions (numbered 1–37) were analyzed by immunoblotting with antibodies against Flag, Smc3, Scc1, SA2, or TRF1. The sedimentation positions of aldolase (7.5S) and catalase (11.3S) are indicated.
We note that only a small fraction of endogenous TRF1 associates with SA1. While the frequency of cohesin binding to human chromosomes is not known, studies in yeast suggest that cohesins are space roughly at 10 kb intervals (Blat and Kleckner, 1999; Laloraya et al, 2000). Thus, we might anticipate only a few cohesin complexes per telomere. As telomeres are coated with TRF1 protein along their length, we predict only a small fraction of TRF1 to be complexed with SA1.
We next sought to confirm these interactions using exogenously expressed proteins. SA1 and SA2 alleles containing triple Flag epitope tags at their N termini were generated and each cotransfected into human 293T cells with Myc epitope tagged TRF1 or TRF2. Protein complexes were immunoisolated using anti-Flag antibody agarose beads and analyzed by immunoblot. As shown in Figure 2B, lane 4, FlagSA1 but not FlagSA2 (lane 6) co-immunoprecipitated MycTRF1. By contrast, MycTRF2 was not detected in the FlagSA1 or FlagSA2 IPs (Figure 2C, lanes 5 and 6), whereas, the cohesin subunit Smc3 (which serves as a positive control for the immunoprecipitation) was contained in the FlagSA1 and SA2 IPs. These data confirm the specific interaction between SA1 and TRF1. This association is independent of DNA as it occurs in the presence of 10 μg/ml ethidium bromide (data not shown).
SA1 and SA2 are conserved along their length, but diverge at their N termini. SA1 has a unique 72 amino-acid domain. As TRF1 bound exclusively to SA1 and not SA2, we wondered if this unique domain might underlie the specificity in binding. We thus generated a deletion construct lacking the N-terminal 72 amino acids (FlagSA1Δ72) and used it in immunoprecipitation analysis. As shown in Figure 2D, whereas MycTRF1 was readily detected in the FlagSA1 IP (lane 5), it was greatly diminished in the FlagSA1Δ72 IP (lane 6), suggesting that the N-terminal domain of SA1 was required for TRF1 binding. As a control we show that Smc3 was detected in both FlagSA1 and FlagSA1Δ72 IPs (lanes 5 and 6). To determine if the 72 amino-acid domain of SA1 was sufficient for binding to TRF1, we fused this domain to GFP to generate GFPSA1N72. As shown in Figure 2E, GFPSA1N72 (lane 6) but not GFP (lane 5) co-immunoprecipitated MycTRF1. Together these data indicate that the 72 amino-acid N-terminal domain of SA1 is necessary and sufficient for TRF1 binding. A recent report described a form of SA2 that contains an additional 69 amino acids at its N terminus (Hauf et al, 2005). This sequence bears no homology to the 72 amino-acid domain of SA1 and when fused to GFP does not co-immunoprecipitate MycTRF1 (data not shown).
Finally, to determine if TRF1 incorporated into the cohesin complex, the FlagSA1-MycTRF1 IPs were subjected to sucrose gradient sedimentation. Previous studies indicated that the cohesin subunits cofractionated with a sedimentation coefficient of approximately 14S. The FlagSA1-MycTRF1 complex was immunoisolated using anti-Flag agarose as described above. The complex was eluted with Flag peptide, fractionated by sucrose gradient sedimentation, and the fractions analyzed by immunoblot. As shown in Figure 2F, FlagSA1 cosedimented with the other cohesin subunits at greater than 11S (in fractions 15, 18 and 21). MycTRF1 was detected in the heaviest fraction (fraction 21) where it cosedimented with FlagSA1, Scc1 and Smc3. These data indicate that TRF1 can incorporate into the SA1-containing cohesin complex.
TIN2 binds to SA1
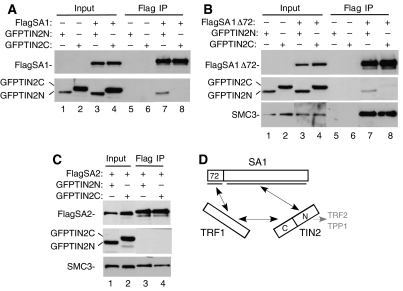
The studies described above show that TRF1 but not TRF2 associates with the endogenous SA1–cohesin complex. TIN2, which binds to both TRF1 and TRF2, was detected but at very low levels (Figure 2A). The presence of TIN2 in the SA1 IP could be due to an indirect association (via TRF1) and/or to an independent association with SA1. We thus asked if TIN2 might bind to SA1. TIN2 is at the center of the shelterin complex serving as a scaffold for interaction with three shelterin subunits (TRF1, TRF2, and TPP1). TIN2 is a modular protein; the N-terminal half (TIN2N) binds to TPP1 and TRF2, whereas the C-terminal half (TIN2C) binds to TRF1 (see schematic in Figure 3D). We used GFP–TIN2 fusions to test for interaction between TIN2N or TIN2C and SA1. FlagSA1 was cotransfected with GFPTIN2N or GFPTIN2C into 293T cells and protein complexes immunoisolated on anti-Flag beads. As shown in Figure 3A, lane 7, GFPTIN2N, but not GFPTIN2C (lane 8) was detected in the FlagSA1IPs. Unlike TRF1, TIN2 did not require the N-terminal domain of SA1 for binding; FlagSA1Δ72 co-immunoprecipitated GFPTIN2N (Figure 3B, lane 7). Similar to TRF1, TIN2 binding was specific for SA1; FlagSA2 did not co-immunoprecipitate GFPTIN2N or GFPTIN2C (Figure 3C, lanes 3 and 4).
Figure 3.
TIN2 binds to SA1. (A–C) TIN2N not TIN2C is co-immunoprecipitated by SA1 or SA1Δ72, not SA2. 293T cells were transfected with GFPTIN2N or GFPTIN2C and (A) FlagSA1, (B) FlagSA1Δ72, or (C) FlagSA2. Cell lysates were immunoprecipitated with anti-Flag beads. Proteins were fractionated on SDS–PAGE and analyzed by immunoblotting with antibodies against Flag, GFP, or Smc3. Input indicates 4% of extract. (D) Schematic representation of the interactions between SA1 and TRF1 and TIN2.
Together these data indicate that SA1 can bind to either TRF1 or TIN2 and it does so using distinct domains; the N-terminal domain binds to TRF1 and the C-terminal domain binds to TIN2 (Figure 3D). Binding of TIN2 to SA1 appears to be efficient in the transfected cell extracts (Figure 3A and B), whereas association between the endogenous proteins is much weaker (Figure 2A). There may only be a small amount of TIN2 associated with cohesin in vivo. Alternatively, and consistent with our previous cell fractionation studies (W Chang, S Smith, unpublished data), the stability of endogenous TIN2 in nuclear complexes may be very susceptible to cell lysis conditions.
Depletion of SA1 or TIN2 and TRF1 rescues the mitotic arrest observed in tankyrase 1-depleted cells
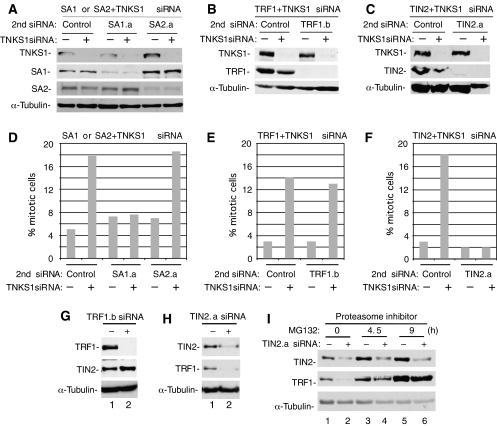
The biochemical analyses described above raise the possibility that the association of TRF1 and TIN2 with SA1 might mediate the persistent telomere association, and hence the mitotic arrest, observed in tankyrase 1-depleted cells. If so, removal of these proteins should allow sister telomeres to separate and allow normal mitotic progression, even in the absence of tankyrase 1. To address this question, we established conditions for double siRNA of tankyrase 1 with control siRNA, cohesin subunits (SA1 or SA2), or telomeric proteins (TRF1 or TIN2). For these experiments, we used a constant amount of tankyrase 1 siRNA in each sample, along with an equal amount of the second siRNA. As shown by immunoblot analysis (Figure 4A–C), we achieved efficient knockdown of tankyrase 1 along with each of the other proteins. Importantly, immunoblot analysis (Figure 4A–C) and immunofluorescence analysis (data not shown) demonstrate that the second siRNA oligo has no effect on depletion of tankyrase 1 protein.
Figure 4.
Depletion of SA1 or TIN2 (and TRF1) rescues the mitotic arrest phenotype induced by tankyrase 1 siRNA. (A–C) Immunoblot analysis of extracts from HeLaI.2.11 cells transfected for 48 h with tankyrase 1 siRNA and a 2nd siRNA: (A) control (GFP), SA1.a, or SA2.a siRNA (B) control (scramble) or TRF1.b siRNA, and (C) control (scramble) or TIN2.a siRNA. (D–F) Histograms showing the percentage of cells in mitosis following 48 h of treatment with (+) or without (−) tankyrase 1 siRNA and the indicated 2nd siRNA. Approximately 1000 cells were scored for each sample by immunofluorescence analysis of cells stained with anti-α-tubulin antibody and DAPI. Control siRNAs; (D) GFP (E, F) scramble. (G, H) Immunoblot analysis of cell extracts from HeLaI.2.11 cells transfected with (G) TRF1.b siRNA or (H) TIN2.a siRNA. (I) TRF1 is degraded by the proteasome in TIN2 siRNA cells. Immunoblot analysis of cell extracts from HeLaI.2.11 cells transfected with TIN2.a siRNA for 48 h. Before harvesting, cells were treated with (+) or without (−) proteasome inhibitor MG132 (12.5 μM) for the indicated times.
The double siRNA-treated cells were analyzed by immunofluorescence analysis to determine the number of cells in mitosis. As described previously, treatment with tankyrase 1 siRNA alone showed that approximately 40% of the cells arrest in mitosis (Dynek and Smith, 2004). When tankyrase 1 siRNA was combined with a control GFP oligo a similar mitotic arrest was observed, although reduced by about half, as half the amount of tankyrase 1 siRNA is used in the double siRNA experiments (Figure 4D). When tankyrase 1 siRNA was combined with SA1 siRNA the mitotic index was restored to control levels (Figure 4D). This rescue of the mitotic arrest was not observed with SA2 siRNA (Figure 4D). A combination of TRF1 with tankyrase 1 siRNA did not rescue the mitotic arrest (Figure 4E), but when tankyrase 1 siRNA was combined with TIN2 siRNA, the mitotic index was restored to control levels (Figure 4F).
We were surprised to see that TIN2 siRNA, but not TRF1 siRNA rescued the telomere cohesion defect in tankyrase 1-depleted cells. We thus investigated the levels of TRF1 and TIN2 in the siRNA cells. As shown in Figure 4G, lane 2, in TRF1 siRNA cells, TIN2 protein levels remain the same or slightly elevated. By contrast, in TIN2 siRNA cells TRF1 is dramatically reduced (Figure 4H, lane 2). Our previous studies indicated that PARsylation of TRF1 releases TRF1 from telomeres, rendering it susceptible to ubiquitination and degradation by the proteasome (Chang et al, 2003). In addition, studies have shown that in the absence of TIN2, TRF1 becomes sensitive to PARsylation by tankyrase and is released from telomeres (Ye and de Lange, 2004). We thus wondered if the TIN2 siRNA-induced loss of TRF1 protein was due to protein degradation. As shown in Figure 4I, lanes 4 and 6, incubation of TIN2 siRNA cells with the proteasome inhibitor MG132 restored TRF1 (but not TIN2) protein levels, indicating that TIN2 siRNA induces proteasome-mediated degradation of TRF1 protein. These data suggest that the depletion of both TIN2 and TRF1 (that occurs with TIN2 siRNA) may be required to rescue the mitotic arrest in tankyrase 1-depleted cells.
The data described above show that SA1 or TIN2 (and TRF1) depletion allows tankyrase 1-depleted cells to proceed normally through mitosis. As controls we show that depletion of SA1 or TIN2 alone does not induce a mitotic arrest (Figure 4D and F). Furthermore, analysis of the distribution of the different stages of mitosis in SA1 or TIN2-depleted cells shows that it is similar to control cells (Supplementary Figure S3).
SA1 and TIN2 are required for persistent sister telomere association in tankyrase 1-depleted cells
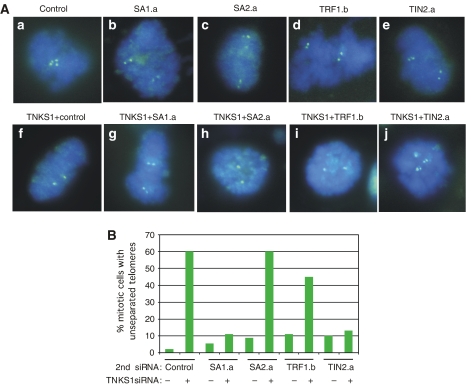
Our previous studies suggested that tankyrase 1-depleted cells arrest in early anaphase with persistent sister telomere associations. As described above, depletion of SA1 or TIN2 rescues this mitotic arrest. We thus wondered if the persistent sister telomere association phenotype was also rescued in the doubly depleted cells. As described previously, and shown in Figure 1B, treatment with tankyrase 1 siRNA alone shows unresolved sister telomeres. When tankyrase 1 siRNA was combined with a control GFP oligo a similar phenotype is observed (Figure 5Af). By contrast, when tankyrase 1 siRNA was combined with SA1 (Figure 5Ag) but not SA2 (Figure 5Ah) siRNA, sister telomeres resolved normally, similar to control cells. Combining TRF1 siRNA with tankyrase 1 had only a minor effect (Figure 5Ai), but when TIN2 siRNA was combined with tankyrase 1, sister telomeres resolved normally, similar to control cells. These data indicate that depletion of SA1 or TIN2 (and TRF1) abrogates the requirement for tankyrase 1 in resolving sister telomere association (Figure 5A and B; Table I).
Figure 5.
Depletion of SA1 or TIN2 rescues the persistent telomere associations in tankyrase 1-depleted cells. (A) Chromosome-specific FISH analysis of HeLaI.2.11 cells collected by mitotic shake-off at 48 h after treatment with (a) control (GFP), (b) SA1.a, (c) SA2.a, (d) TRF1.b, or (e) TIN2.a siRNA without (a–e) or with (f–j) tankyrase 1 siRNA. Cells were fixed directly in methanol-acetic acid without hypotonic swelling and hybridized to a telomere probe 16pter (green). DNA was stained with DAPI (blue). (B) Histogram showing percentage of mitotic cells with unseparated telomeres; at least 100 mitotic cells were scored for each sample.
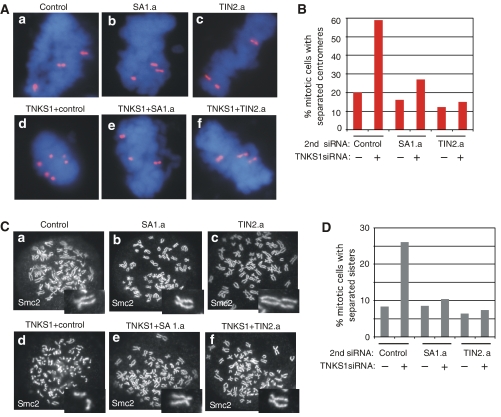
Finally, according to our hypothesis, the separated sister centromeres observed in tankyrase 1-depleted cells are not due to a premature loss in centromere cohesion, but rather to the fact that tankyrase 1-depleted cells proceed normally to early anaphase, but then arrest due to persistent telomere associations. Thus, we predict that in the cells doubly depleted for tankyrase 1 and SA1 or TIN2 (that are no longer arrested in mitosis with persistent telomere associations) centromere cohesion should be normal. We thus used centromere FISH and chromosome spread analysis to determine if normal centromere cohesion was restored in these double siRNA cells. When tankyrase 1 siRNA was combined with a control (GFP) siRNA, cells retained separated centromeres as evidenced by widely spaced doublets (Figure 6Ad). By contrast, double siRNA with tankyrase 1 and SA1 or TIN2 restored normal centromere cohesion, as evidenced by the closely opposed doublets (Figure 6Ae, f and B; Table II). Similar results were obtained by chromosome spread analysis. When tankyrase 1 siRNA was combined with a control (GFP) siRNA, sister chromatids remained separated (Figure 6Cd). By contrast, double siRNA with tankyrase 1 and SA1 or TIN2 rescued the separated sister phenotype of tankyrase 1 siRNA cells and restored the normal configuration of sister chromatids (Figures 6Ce, f and D; Table III). As controls we show that single depletions of TIN2 or SA1 are similar to control depletions, and have no effect on sister telomere associations (Figure 5; Table I), sister centromere associations (Figure 6; Tables II and III), or on mitotic progression (Figure 4; Supplementary Figure S3).
Figure 6.
Depletion of SA1 or TIN2 restores normal centromere cohesion in tankyrase 1-depleted cells. (A) Chromosome-specific FISH analysis of HeLaI.2.11 cells collected by mitotic shake-off at 48 h after treatment with (a) control (GFP), (b) SA1.a, or (c) TIN2.a siRNA without (a–c) or with (d–f) tankyrase 1 siRNA. Cells were fixed directly in methanol-acetic acid without hypotonic swelling and hybridized to a centromere probe 6cen (red). DNA was stained with DAPI (blue). (B) Histogram showing percentage of mitotic cells with separated centromeres; at least 100 mitotic cells were scored for each sample. (C) Chromosome spread analysis of HelaI.2.11 cells collected after 48 h of treatment with (a) control (GFP), (b) SA1.a, or (c) TIN2.a siRNA without (a–c) or with (d–f) tankyrase 1 siRNA. Cells were swollen in hypotonic buffer and fixed in paraformaldehyde. Cells were treated with colcemide for 90 min before harvesting. Chromosome arms were visualized by staining with antibodies to the condensin subunit Smc2. (D) Histogram showing % mitotic cells with separated sisters; at least 200 mitotic cells were scored for each sample.
Discussion
Previous studies suggested that sister telomeres uniquely require tankyrase 1 for their resolution at mitosis. Here, we demonstrate a novel association between cohesins and telomeric chromatin via the SA1 cohesin subunit and the telomeric proteins TRF1 and TIN2. We show that depletion of these components abrogates the requirement for tankyrase 1 in sister telomere resolution and mitotic progression. Our studies suggest that sister telomere associations are mediated via a novel SA1–TRF1/TIN2 association that requires tankyrase 1 for its resolution at mitosis.
Interactions between cohesins and telomeric chromatin
We demonstrate that endogenous SA1-containing (but not SA2-containing) cohesin complexes associate with TRF1 in HeLa cells. This distinction between SA1- and SA2-containing cohesin complexes was unexpected. SA1 and SA2 are very similar in their primary structure and are believed to have similar function. In fact, it is not known why vertebrate cells contain two orthologs of the Scc3 cohesin subunit. However, the observation that the ratio of SA1 versus SA2 can vary dramatically in different cell types (Losada et al, 2000) suggests that they may have different functions. These functions could be mediated by unique protein-binding partners (such as TRF1 and TIN2) that can target cohesin complexes to specific chromatin domains (such as telomeres), as we have shown here.
We show that SA1 binds to the shelterin subunit TRF1 but not TRF2. While shelterin can exist as an intact six-subunit complex, in lysed cells it is found predominantly in subcomplexes containing either TRF1 or TRF2 (Houghtaling et al, 2004; Kim et al, 2004; Liu et al, 2004a; Ye et al, 2004a). TRF2 plays a critical role in the protection of chromosome ends, most likely mediated by its association with the TIN2/TPP1/POT1 complex. When TRF2 is removed from telomeres (by a dominant negative allele), TRF1 remains at telomeres, but it is not sufficient to protect chromosome ends (van Steensel et al, 1998). By contrast, when TRF1 is removed from telomeres by tankyrase 1 or a TRF1 dominant negative allele, TRF2 remains and telomeres remain protected (van Steensel and de Lange, 1997; Smith and de Lange, 2000). Association of cohesin specifically with TRF1 could provide a mechanism to allow for removal of cohesin (via tankyrase 1 release of TRF1) without the concomitant loss of the essential TRF2-mediated protective function at telomeres.
In addition to TRF1, SA1 binds to TIN2, the central subunit of the shelterin complex. SA1 binds to the TIN2 N-terminal domain, which is the same domain that binds to TPP1 and TRF2 (Figure 3D). By binding to the N-terminal domain of TIN2, SA1 may compete with TPP1 and/or TRF2, thereby creating a sub complex existing exclusively of TRF1 and TIN2 bound to cohesin. Previous studies have shown that TIN2 can modulate tankyrase 1 PARsylation of TRF1 (Ye and de Lange, 2004). Thus, TIN2 could play a regulatory role here, preventing removal of telomeric cohesin by tankyrase 1 until the appropriate time in the cell cycle.
Mitotic arrest in tankyrase 1-depleted cells
Based on a number of observations, we previously hypothesized that tankyrase 1-depleted cells proceed normally through metaphase and arrest in early anaphase with unresolved sister telomeres (Dynek and Smith, 2004). First, live cell imaging showed that chromosomes congressed normally to the metaphase plate, but then underwent a struggle to segregate. Second, the SCC1 subunit of cohesin was cleaved, centromeres were separated and bipolar spindles were retracted in anaphase configuration. These observations lead us to hypothesize that in tankyrase 1 siRNA cells chromosomes line up on the metaphase plate and centromeres separate but then cells cannot proceed through mitosis due to persistent telomere associations. A subsequent study observed a similar mitotic arrest in tankyrase 1-depleted cells, but observed intact sister chromatid cohesion and suggested a pre-anaphase arrest (Chang et al, 2005b). One possible explanation for the discrepancy may be the different assays used to analyze sister chromatid cohesion. In the subsequent study, thin section electron microscopy was used to analyze sister chromatids in tankyrase 1-depleted cells and the authors noted that when single whole chromosomes could be distinguished they were fully paired and thus they concluded that sister chromatid cohesion was intact (Chang et al, 2005b). However, only a few chromosomes were indicated and the results were not quantified. In our previous work (Dynek and Smith, 2004) and in the present study, we use standard techniques to measure sister chromatid cohesion (chromosome specific FISH and chromosome spread analysis) that permit quantifiable analysis of sister chromatid associations. We show that sister centromeres are separated in the majority of mitotic cells following tankyrase 1 depletion and further that this separation depends on mitotic progression, consistent with progression to anaphase.
Our data indicate that tankyrase 1 is required for separation of sister telomeres and mitotic progression. We note, however, that tankyrase 1 may have additional activities that could influence mitosis. In addition to its telomeric localization, tankyrase 1 localizes to Golgi-associated membranes (Chi and Lodish, 2000) and to spindle poles (Smith and de Lange, 1999), reviewed by Hsiao and Smith (2007). Indeed, spindle pole defects were detected in tankyrase 1-depleted cells (Chang et al, 2005b). However, our observation that depletion of shelterin or cohesin subunits abrogates the requirement for tankyrase 1 in mitotic progression support the notion that persistent telomere associations contribute to the mitotic arrest phenotype.
A special mechanism for sister telomere cohesion
Our work here is in line with previous studies suggesting that specialized chromatin domains use distinct mechanisms for sister chromatid cohesion, reviewed by Losada (2007). For example, studies with fission yeast show that cohesin is recruited to heterochromatic regions (centromeres and mating type loci) via an association between the heterochromatin protein Swi6/HP1 and the Scc3 subunit of cohesin (Bernard et al, 2001; Nonaka et al, 2002; Partridge et al, 2002). A second example comes from recent work in budding yeast, where it was found that cohesin's association with the silent HMR mating-type locus was mediated by the Sir proteins and silent chromatin (Chang et al, 2005a).
Our studies suggest a distinct mode of binding between SA1–cohesin complexes and telomeric chromatin via TIN2 and TRF1. A distinct mode of binding may be necessary to accommodate the specialized chromatin structure and function of telomeres. Tethering of cohesins to telomeres could be required to prevent cohesins from sliding off chromosome ends. Tankyrase 1 would be needed to dissociate the complex, most likely through PARsylation of TRF1 (or as yet unidentified targets). As tankyrase 1 is phosphorylated at mitosis (Chang et al, 2005c), it may be a target for regulatory kinases. Future experiments will be required to determine if tankyrase 1 is controlled by the same (or different) pathways that regulate removal of arm and centromere cohesion.
Materials and methods
Plasmids
The SA1 cDNA (amino acids 1–1258) (Carramolino et al, 1997; Losada et al, 2000) was obtained from The I.M.A.G.E Consortium. The clone (accession # AAH64699) contained an internal deletion from amino acids 1150–1186. The SA2 cDNA (amino acids 1–1162) (Carramolino et al, 1997; Losada et al, 2000) was a gift from L Carramolino. Full-length SA1, SA1 containing amino acids 73–1258, and full-length SA2 were cloned into the vector p3XFLAG-CMV-10 (Sigma) to generate FlagSA1, FlagSA1Δ72, and FlagSA2, respectively. MycTRF1 (pLPCTRF1) (Chang et al, 2003) and MycTRF2 (pcDNA3hTRF2) (a gift from Dominique Broccoli) contain N-terminal Myc-epitope tags. GFP constructs contained GFP followed by the following: amino acids 1–72 of SA1 (GFPSA1N72); 1–180 of TIN2 (GFPTIN2N); or 180–354 of TIN2 (GFPTIN2C) cloned into the pLEGFP-C1 vector (Clontech).
Whole-cell extracts
siRNA-transfected HeLaI.2.11 cells were resuspended in four volumes of buffer C (20 mM HEPES-KOH (pH 7.9), 420 mM KCl, 25% glycerol, 0.1 mm EDTA, 5 mM MgCl2, 0.2% NP40, 1 mM dithiothreitol, and 2.5% protease inhibitor cocktail (Sigma) and incubated for 1 h on ice. Suspensions were pelleted at 8000 g for 10 min. A 25 μg (determined by Biorad protein assay) portion of the supernatant proteins was fractionated by SDS–PAGE and analyzed by immunoblotting.
Immunoprecipitation
Cells were lysed in 0.5 ml (per one 15-cm-diameter dish) TNE buffer (10 mM Tris (pH7.8), 1% Nonidet P-40, 0.15 M NaCl, 1 mM EDTA, and protease inhibitor cocktail (Sigma)) on ice for 1 h, then pelleted at 8000 g for 10 min. Supernatants were precleared with rabbit immunoglobulin (IgG) and protein G-Sepharose (GE Healthcare) rotating at 4°C for 30 min. Nonspecific antibody complexes and protein aggregates were removed by centrifugation, and the supernatant was used for immunoprecipitation analysis or fractionated directly on SDS–PAGE (indicated as input, approximately 4% of the amount used in the immunoprecipitation). Supernatants were incubated with 1.0 μg of goat anti-Flag (Bethyl Laboratories Inc.), goat anti-SA1 BL143G (Bethyl Laboratories Inc.), goat anti-SA2 BL143G (Bethyl Laboratories Inc.) or rabbit anti-GFP (Abcam) at 4°C with rocking for 2 h 30 min. Antigen–antibody complexes were collected on protein G beads at 4°C with rocking for 30 min. For Flag IPs, supernatants were pre-cleared with protein G and then incubated with 35 μl of mouse anti-Flag-agarose (Sigma) at 4°C with rocking for 3 h. Immunocomplexes were then washed three times with 1.0 ml TNE buffer and processed for sucrose gradients (described below) or suspended in Laemmli buffer. Samples were fractionated on 7.5 or 10% SDS–PAGE gels and processed for immunoblotting as described below.
Sucrose density gradient centrifugation
Flag immunocomplexes from four 15-cm-diameter dishes (prepared as described above) were eluted from anti-Flag agarose by incubation with 200 μl TNE buffer containing 50 μg/ml Flag peptide (Sigma) for 1 h at room temperature and separated in a 2 ml 10–30% sucrose gradient (prepared in TNE buffer) by centrifugation at 50 000 r.p.m. for 12 h at 4°C in a TLS-55 rotor (Beckman), as described (Tanese, 1997). Fractions (60 μl) were collected by pipetting from the top of the gradient and analyzed by immunoblotting.
Immunoblotting
Proteins were transferred to nitrocellulose electrophoretically and blocked in 5% milk in PBS containing 0.1% Tween 20. Blots were incubated with the following primary antibodies: goat anti-SA1 BL140G (1 μg/ml) (Bethyl Laboratories Inc.); goat anti-SA1 BL143G (1 μg/ml) (Bethyl Laboratories Inc.); goat anti-SA2 BL146G (1 μg/ml) (Bethyl Laboratories Inc.); rabbit anti-Scc1 (2 mg/ml, Bethyl Laboratories Inc.); rabbit anti-Smc3 (0.2 mg/ml) (Calbiochem); rabbit anti-TRF1 415 (1 μg/ml) (Cook et al, 2002); rabbit anti-tankyrase1 609 (1 μg/ml)(Cook et al, 2002); mouse monoclonal anti-TRF2 (2.0 μg/ml) (Imgenex), rabbit anti-Myc (0.8 μg/ml) (Santa Cruz Biotechnologies); mouse monoclonal anti-Flag M2 (4.3 μg/ml) (Sigma); rabbit anti-TIN2 701 (0.5 μg/ml) (Houghtaling et al, 2004); mouse anti-a-tubulin ascites (1:50 000) (Sigma); or rabbit anti-GFP serum (1:2500) (Abcam), followed by horseradish peroxidase-conjugated donkey anti-rabbit (Amersham), anti-mouse (Amersham), or anti-goat IgG (Bethyl Laboratories Inc.) (1:2500). Bound antibody was detected with Super Signal West Pico (Pierce).
Transient transfections
Plasmid transfections for immunoprecipitations were performed in 293T cells with Lipofectamine 2000 regeant (Invitrogen) for 24 h according to the manufacturer's protocol.
siRNA transfections were performed in HeLaI.2.11 cells, a HeLa-derived clonal cell line (van Steensel et al, 1998) with Oligofectamine (Invitrogen) for 48 h according to the manufacturer's protocol. The final concentration of siRNA was 100 nM. For the prometaphase arrest experiments, cells were treated with siRNA for 16 h and nocodazole (1.5 μg/ml) was added for an additional 12 h. For double siRNA experiments, each oligo was present at 50 nM. Thus, the double siRNA reactions indicted as (−) TNKS1, contain 50 nM of control siRNA (GFP or scramble). The following siRNAs (synthesized by Dharmacon Research Inc.) were used: TNKS1 (5′-AACAAUUCACCGUCGUCCUCU-3′) (Dynek and Smith, 2004); TRF1.b (5′-AAUGCCAGGAACUGCUCGAGU-3′); TIN2.a(5′-AACGCCUUUGUAUGGGCCUAA-3′); SA1.a (5′-GUGAUGCCUUCCUAAAUGA-3′); SA2.a (5′-GUACGGCAAUGUCAAUAUA-3′); Scramble II Duplex (Dharmacon); or GFP Duplex I (Dharmacon).
Chromosome spread analysis
For chromosome spread analysis, siRNA-transfected HeLaI.2.11 cells were collected by trypsinization (0.5 mg/ml colcemide was added 90 min before harvest), swollen in buffer containing 10 mM Tris–HCl (pH 7.4), 10 mM NaCl, 5 mM MgCl2 for 10 min at 37°C and sedimented onto coverslips for 15 s at 1000 r.p.m. in a Sorval RT7. Cells were fixed with 2% paraformaldehyde and stained with rabbit anti-Smc2 (0.4 μg/ml; Bethyl Laboratories Inc.). Primary antibodies were detected with fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit antibodies (1:100) (Jackson Laboratories). Images were acquired on a Zeiss Axioplan 2 microscope with a Photometrix SenSyn camera. Photographs were processed and merged using IPLab software.
Chromosome specific FISH
siRNA-transfected HeLaI.2.11 cells were collected by mitotic shake-off, fixed and processed exactly as described previously (Dynek and Smith, 2004), using the following directly labeled (FITC or TRITC) DNA probes from Cytocell: the 16pter subtelomeric-specific probe or the chromosome 6-specific alpha-satellite centromere probe. DNA was stained with 4,6-diamino-2-phenylindole (DAPI) (0.2 μg/ml).
Supplementary Material
Supplementary Figures and Legends
Acknowledgments
We thank Jose Luis Barbero for the Stag2 plasmid. We are grateful to Tom Meier, Susan Hsiao, and members of the Smith lab for critical comments on the manuscript and helpful discussions. SC was supported by a NYS DOH Breast Cancer Research Grant (C020914). BRH and JND were supported by a Predoctoral Training Program in Cell and Molecular Biology (GM07238). WGC was supported by a Predoctoral Training Program in Molecular Oncology and Immunology (CA09161). This work was supported by a grant from the Charlotte Geyer Foundation and grants from the NIH (RO1 CA95099) and (R01 CA116352).
References
- Baumann P, Cech TR (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542 [DOI] [PubMed] [Google Scholar]
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E (1997) Telomeric localization of TRF2, a novel human telobox protein. Nat Genet 17: 236–239 [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N (1999) Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98: 249–259 [DOI] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 17: 231–235 [DOI] [PubMed] [Google Scholar]
- Carramolino L, Lee BC, Zaballos A, Peled A, Barthelemy I, Shav-Tal Y, Prieto I, Carmi P, Gothelf Y, Gonzalez de Buitrago G, Aracil M, Marquez G, Barbero JL, Zipori D (1997) SA-1, a nuclear protein encoded by one member of a novel gene family: molecular cloning and detection in hemopoietic organs. Gene 195: 151–159 [DOI] [PubMed] [Google Scholar]
- Chang CR, Wu CS, Hom Y, Gartenberg MR (2005a) Targeting of cohesin by transcriptionally silent chromatin. Genes Dev 19: 3031–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P, Coughlin M, Mitchison TJ (2005b) Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol 7: 1133–1139 [DOI] [PubMed] [Google Scholar]
- Chang W, Dynek JN, Smith S (2003) TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev 17: 1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Dynek JN, Smith S (2005c) NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem J 391: 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NW, Lodish HF (2000) Tankyrase is a Golgi-Associated MAP Kinase Substrate that Interacts with IRAP in GLUT4 vesicles. J Biol Chem 275: 38437–38444 [DOI] [PubMed] [Google Scholar]
- Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T (1995) A human telomeric protein. Science 270: 1663–1667 [DOI] [PubMed] [Google Scholar]
- Cook BD, Dynek JN, Chang W, Shostak G, Smith S (2002) Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres. Mol Cell Biol 22: 332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Donigian JR, de Lange T (2007) The role of the poly(ADP-ribose) polymerase tankyrase1 in telomere length control by the TRF1 component of the shelterin complex. J Biol Chem 282: 22662–22667 [DOI] [PubMed] [Google Scholar]
- Dynek JN, Smith S (2004) Resolution of sister telomere association is required for progression through mitosis. Science 304: 97–100 [DOI] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K (2003) Chromosomal cohesin forms a ring. Cell 112: 765–777 [DOI] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9: 773–788 [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM (2005) Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol 3: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters JM (2001) Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293: 1320–1323 [DOI] [PubMed] [Google Scholar]
- Houghtaling BR, Cuttonaro L, Chang W, Smith S (2004) A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol 14: 1621–1631 [DOI] [PubMed] [Google Scholar]
- Hsiao SJ, Smith S (2007) Tankyrase function at telomeres, spindle poles, and beyond. Biochimie (10.1016/j.biochi.2007.07.012) [DOI] [PubMed] [Google Scholar]
- Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J (2004) TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem 279: 43799–43804 [DOI] [PubMed] [Google Scholar]
- Kim SH, Kaminker P, Campisi J (1999) TIN2, a new regulator of telomere length in human cells (see comments). Nat Genet 23: 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloraya S, Guacci V, Koshland D (2000) Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol 151: 1047–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Oestreich S, de Lange T (2000) Identification of human Rap1: implications for telomere evolution. Cell 101: 471–483 [DOI] [PubMed] [Google Scholar]
- Liu D, O'Connor MS, Qin J, Songyang Z (2004a) Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem 279: 51338–51342 [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z (2004b) PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol 6: 673–680 [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 423: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Losada A (2007) Cohesin regulation: fashionable ways to wear a ring. Chromosoma 116: 321–329 [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T (2002) Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev 16: 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano T (2005) Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev 19: 1269–1287 [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T (2000) Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol 150: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74: 595–648 [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y (2002) Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4: 89–93 [DOI] [PubMed] [Google Scholar]
- Ofir R, Yalon-Hacohen M, Segev Y, Schultz A, Skorecki KL, Selig S (2002) Replication and/or separation of some human telomeres is delayed beyond S-phase in pre-senescent cells. Chromosoma 111: 147–155 [DOI] [PubMed] [Google Scholar]
- Ohnuki Y (1968) Structure of chromosomes. I. Morphological studies of the spiral structure of human somatic chromosomes. Chromosoma 25: 402–428 [DOI] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC (2002) cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol 12: 1652–1660 [DOI] [PubMed] [Google Scholar]
- Rieder CL, Cole R (1999) Chromatid cohesion during mitosis: lessons from meiosis. J Cell Sci 112 (Part 16): 2607–2613 [DOI] [PubMed] [Google Scholar]
- Smith S, de Lange T (1999) Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci 112: 3649–3656 [DOI] [PubMed] [Google Scholar]
- Smith S, de Lange T (2000) Tankyrase promotes telomere elongation in human cells. Curr Biol 10: 1299–1302 [DOI] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, de Lange T (1998) Tankyrase, a poly(ADP-ribose) polymerase at human telomeres (see comments). Science 282: 1484–1487 [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM (2000) Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 151: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters JM (2002) The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell 9: 515–525 [DOI] [PubMed] [Google Scholar]
- Tanese N (1997) Small-scale density gradient sedimentation to separate and analyze multiprotein complexes. Methods 12: 224–234 [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T (1997) Control of telomere length by the human telomeric protein TRF1. Nature 385: 740–743 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM (2000) Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103: 399–410 [DOI] [PubMed] [Google Scholar]
- Yalon M, Gal S, Segev Y, Selig S, Skorecki KL (2004) Sister chromatid separation at human telomeric regions. J Cell Sci 117: 1961–1970 [DOI] [PubMed] [Google Scholar]
- Ye JZ, de Lange T (2004) TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat Genet 36: 618–623 [DOI] [PubMed] [Google Scholar]
- Ye JZ, Donigian JR, Van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, De Lange T (2004a) TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem 279: 47264–47271 [DOI] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T (2004b) POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev 18: 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Legends