Abstract
Neuropilins (Nrps) are co-receptors for class 3 semaphorins and vascular endothelial growth factors and important for the development of the nervous system and the vasculature. The extracellular portion of Nrp is composed of two domains that are essential for semaphorin binding (a1a2), two domains necessary for VEGF binding (b1b2), and one domain critical for receptor dimerization (c). We report several crystal structures of Nrp1 and Nrp2 fragments alone and in complex with antibodies that selectively block either semaphorin or vascular endothelial growth factor (VEGF) binding. In these structures, Nrps adopt an unexpected domain arrangement in which the a2, b1, and b2 domains form a tightly packed core that is only loosely connected to the a1 domain. The locations of the antibody epitopes together with in vitro experiments indicate that VEGF and semaphorin do not directly compete for Nrp binding. Based upon our structural and functional data, we propose possible models for ligand binding to neuropilins.
Keywords: neuropilin, neuroscience, tumorigenesis, vasculogenesis, X-ray crystallography
Introduction
Neuropilin-1 and -2 (Nrp1 and Nrp2), non-tyrosine kinase receptors with a molecular weight of approximately 120 kDa, were initially described as axonally expressed proteins and later recognized as binding partners for class 3 semaphorins (Chen et al, 1997; He and Tessier-Lavigne, 1997; Kolodkin et al, 1997). They further serve as isoform-specific VEGF (vascular endothelial growth factor) co-receptors on endothelial cells and are involved in vascular development and tumorigenesis (Soker et al, 1998; Gluzman-Poltorak et al, 2000; Ellis, 2006). Genetic ablation of Nrp1 or Nrp2 indicate that both homologs play critical, but nonoverlapping roles during neuronal and vascular development (Kawasaki et al, 1999; Chen et al, 2000; Giger et al, 2000; Takashima et al, 2002; Yuan et al, 2002). Upregulation of Nrp expression has been proposed to contribute to tumor formation in a number of diseases such as prostate, breast, and colon cancer (reviewed in Ellis, 2006), and has been implicated in increasing mortality in acute myeloid leukemia (Kreuter et al, 2006). The linkage between neuronal and vascular wiring along with the upregulation of neuropilin in tumors has prompted considerable efforts to understand the molecular roles of Nrps in the processes of angiogenesis, neuronal development, and tumor biology, and makes these receptors attractive targets for drug development.
Nrp1 and Nrp2 share a sequence identity of 44% (Chen et al, 1997; Kolodkin et al, 1997); they are composed of five extracellular domains followed by a single transmembrane segment and a short cytoplasmic tail. The extracellular region of Nrps contains two N-terminal CUB domains (C1r/C1s, uEGF, bone morphogenetic protein; also known as a1 and a2), two coagulation factor V/VIII homology domains (F5/8; also known as b1 and b2), and a single MAM domain (meprin, A5, and μ-phosphatase; also known as c) (Takagi et al, 1991; Beckmann and Bork, 1993). The cytoplasmic tail includes about 42–44 amino acids and does not display catalytic activity of its own, but presents a binding site for the PDZ domain of Nrp1-interacting protein (NIP or GIPC) (Cai and Reed, 1999; Wang et al, 2006). The association of Nrp and NIP may represent one pathway that is important for signaling. Additional signaling pathways are mediated by the recruitment of semaphorins and their respective co-receptors, the type A Plexins (Takahashi et al, 1999; Tamagnone et al, 1999; Yaron et al, 2005), or by binding specific isoforms of the five mammalian VEGF family members (VEGF-A, -B, -C, -D, and PlGF) and their co-receptor tyrosine kinases (VEGFR1–3) (Migdal et al, 1998; Soker et al, 1998; Makinen et al, 1999; Fuh et al, 2000; Gluzman-Poltorak et al, 2000, 2001; Karkkainen et al, 2001; Mamluk et al, 2002; Soker et al, 2002; Favier et al, 2006; Karpanen et al, 2006; Pan et al, 2007b).
Class 3 semaphorins are secreted members of the semaphorin family and interact with Nrps on two distinct sites. One binding site includes residues from the N-terminal β-propeller or ‘Sema' domain of the semaphorins and the a1a2 domains of Nrp. This site dictates the specificity for Nrp1 to bind Sema3A and Sema3C and for Nrp2 to recognize Sema3C and Sema3F (Chen et al, 1998; Giger et al, 1998; Nakamura et al, 1998; Renzi et al, 1999; Gu et al, 2002; Antipenko et al, 2003). The second binding site, formed between the carboxyl tail of the semaphorins and the b1 domain of Nrp, increases the affinity of the complexes (Giger et al, 1998; Nakamura et al, 1998; Gu et al, 2002; Chen et al, 1998; Renzi et al, 1999) but does not influence selectivity between the binding partners. The key angiogenic factor VEGF165, as well as VEGF-B and PlGF-2, binds the b1 domain and to a lesser extent the b2 domain of the Nrps via a C-terminal heparin-binding domain (Giger et al, 1998; Migdal et al, 1998; Soker et al, 1998; Makinen et al, 1999; Fuh et al, 2000; Gluzman-Poltorak et al, 2000; Gu et al, 2002; Mamluk et al, 2002; Vander Kooi et al, 2007). Shorter VEGF variants such as VEGF109, which only contain the VEGF receptor-binding domain that is responsible for binding to VEGFRs (Wiesmann et al, 1997; Fuh et al, 2000), are unable to recognize Nrps. In addition to binding VEGF, the Nrp b1b2 domains interact with heparin (Migdal et al, 1998; Fuh et al, 2000; Gluzman-Poltorak et al, 2000; Mamluk et al, 2002; Vander Kooi et al, 2007). The presence of heparin improves binding of VEGF165 to Nrp up to 100-fold (Fuh et al, 2000; Mamluk et al, 2002), suggesting that heparin or heparan sulfate containing proteoglycans can either bridge or stabilize the interface between Nrps and VEGF. VEGF-C and -D contain no known heparin-binding motif yet they are able to bind the b1b2 domain as well (Karpanen et al, 2006). Finally, the membrane-proximal c domain of Nrps appears to have no direct role in ligand binding; however, it is critical for dimerization or oligomerization (Chen et al, 1998; Giger et al, 1998; Nakamura et al, 1998; Takahashi et al, 1998; Renzi et al, 1999).
Based on the domain architecture and functional properties, the extracellular domains of Nrps are typically divided into three functional units: a1a2, b1b2, and c that represent the primary semaphorin-binding, VEGF-binding, and dimerization regions, respectively (Takagi et al, 1991; Chen et al, 1998; Giger et al, 1998; Nakamura et al, 1998; Renzi et al, 1999; Gu et al, 2002; Mamluk et al, 2002). Although structure–function studies define the overall domain requirements for neuropilin function, the molecular details of these receptor/ligand complexes remain poorly understood. To date, structural information on Nrps is limited to the b1b2 domain of Nrp1 (Lee et al, 2003; Vander Kooi et al, 2007). The crystal structure of Nrp1 b1b2 in complex with Tuftsin, a tetrapeptide homologous to the C terminus of VEGF165, provided the first structural information on the interaction of Nrp with its binding partner VEGF (von Wronski et al, 2006; Vander Kooi et al, 2007).
Here, we present the crystal structure of Nrp2 a1a2b1b2 in complex with a Fab fragment (anti-panNrpA) of an antibody that blocks Sema3 binding and function meditated through both Nrp1 and Nrp2 in vitro. We also compare and contrast structures of the a2b1b2 and b1b2 fragments of both Nrp1 and Nrp2. Finally, we illustrate the structure of the b1 domain of Nrp1 combined with a Fab fragment (anti-Nrp1B) of a recently described phage-derived antibody (Liang et al, 2007; Pan et al, 2007a) that specifically inhibits VEGF165 binding to Nrp1. In vivo, this antibody not only reduces vascular remodeling in the mouse retina, but also works as a single agent and in combination with anti-VEGF to slow tumor growth (Liang et al, 2007; Pan et al, 2007a). Together, these structures present a detailed picture of a large portion of the Nrp extracellular domain and suggest models for VEGF and semaphorin binding.
Results and discussion
Construction and function of anti-Nrp1B and anti-panNrpA
We recently described a strategy to develop phage-derived antibodies that selectively block binding of either semaphorin or VEGF to Nrp1 (Liang et al, 2007; Pan et al, 2007a). These monoclonal antibodies were designed as research tools to discriminate between Nrp1-mediated responses to either ligand and to evaluate their potential as therapeutics in murine tumor models. Anti-Nrp1B is a VEGF-blocking antibody that binds Nrp1 with an affinity of 0.2 nM. Although this antibody blocks the interaction between VEGF and Nrp1, it does not antagonize Sema3A function (Liang et al, 2007; Pan et al, 2007a).
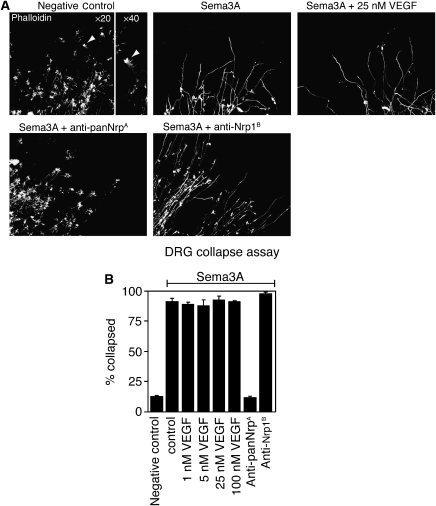
Here, we introduce an antibody (anti-panNrpA) that crossreacts with both Nrp1 and Nrp2 with affinities of 0.21 and 0.15 nM, respectively (Supplementary Figure S1). A thorough description of the phage-library construction and antibody screening will be detailed elsewhere (manuscript in preparation). In contrast to anti-Nrp1B, anti-panNrpA does not affect the binding of VEGF165 or VEGF-C to Nrp1 and Nrp2 (data not shown). We also evaluated the capacity of both antibodies to inhibit the Sema3A-mediated collapse of axon growth cones from murine dorsal root ganglia (DRG) (Figure 1). The addition of Sema3A results in the retraction of the actin processes within the DRG growth cones; this function of Sema3A is completely antagonized by anti-panNrpA, but not by anti-Nrp1B (Figure 1).
Figure 1.
VEGF does not block Sema3A-induced growth cone collapse of DRG neurons. (A) Images of axon growth cones. Untreated dorsal root ganglia (DRG) have large actin-rich growth cones (arrowheads) that are significantly reduced in number upon addition of Sema3A. Anti-Nrp antibodies were added at 50 μg/ml; Sema3A was added at 2.2 nM. Only anti-panNrpA blocks Sema3A-mediated collapse. (B) Quantification of Sema3A-induced growth cone collapse. The percentage of collapsed growth cones were calculated by counting collapsed and uncollapsed growth cones (N=4 explants per condition). Error bars represent standard error of the mean.
Crystallization of Nrp1, Nrp2, and Nrp/Fab complexes
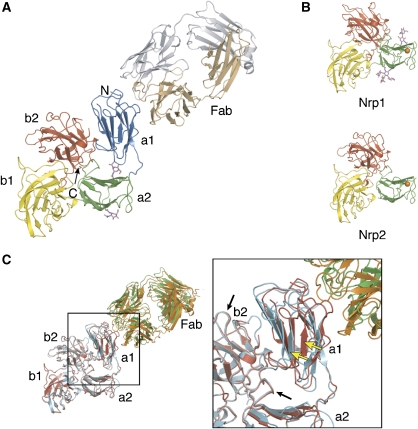
For structural studies, the smaller Nrp fragments that include the type b domains were expressed in E. coli while the larger Nrp constructs required production as secreted proteins from baculovirus-infected insect cells. An overview of the seven structures is presented in Figure 2. Crystals of the VEGF-binding portion (b1b2) of Nrp1 and Nrp2 diffracted to a maximum resolution of 1.8 and 1.95 Å, respectively. Crystals of the a2b1b2 domains of Nrp1 and Nrp2 were refined to 2.0 and 2.3 Å resolution, respectively, while the b1 domain of Nrp1 in complex with the VEGF-blocking Fab, anti-Nrp1B, diffracted to 2.2 Å resolution. Finally, the crystal structure of the Nrp2 a1a2b1b2 domains was solved in complex with the semaphorin-blocking Fab, anti-panNrpA; monoclinic (space group C2) and trigonal (space group P3221) crystal forms of this complex were identified that diffracted to 2.75 and 3.1 Å, respectively. All structures were solved by molecular replacement and are reported with final Rwork/Rfree values below 19/25% and good stereochemistry (Table I). The Nrp1 a2b1b2 structure has two N-linked glycosylation sites on opposite sides of the a2 domain (Figure 3B). The monoclinic form of the Nrp2/Fab complex also shows two glycosylation sites within the a2 domain (Figure 3A); however, these sugar moieties are not well defined in the electron density of the Nrp2 a2b1b2 structure and therefore are not modeled (Figure 3B).
Figure 2.
Summary of the neuropilin crystal structures. (A) The Nrp ectodomain is comprised of tandem CUB (a1a2), tandem coagulation factor V/VIII (b1b2), and one MAM (c) domain. Cartoon representation of the seven crystal structures presented in this report with the resolution limits listed below. Magenta spheres indicate a bound calcium ion in the a2 domain. (B) Sequence alignment of the a1a2b1b2 domains of human Nrp1 and Nrp2. Secondary structure elements refer to the Nrp2 a1a2b1b2 structure (blue, a1; green, a2; yellow, b1; red, b2) and are named according to conventions adopted for the spermadhesin CUB domain (Romero et al, 1997) and the coagulation factor V C2 domain (Macedo-Ribeiro et al, 1999). Residues boxed in blue and yellow delineate the antibody epitopes for anti-panNrpA and anti-Nrp1B, respectively. Amino acids highlighted in orange indicate the Ca2+-binding site in a2, while residues in red represent a putative Ca2+-binding site in the a1 domain. Residues shaded in green highlight the positions of amino acid that when substituted disrupt interactions between Sema3A and Nrp1 (Gu et al, 2002). This alignment was produced with EsPript (Gouet et al, 2003).
Table 1.
Data collection and refinement statistics
| Nrp1 b1b2 | Nrp1 a2b1b2 | Nrp1 b1/Fab | Nrp2 b1b2 | Nrp2 a2b1b2 | Nrp2 a1a2b1b2/Fab | Nrp2 a1a2b1b2/Fab | |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| PDB code | 2QQI | 2QQM | 2QQN | 2QQJ | 2QQO | 2QQK | 2QQL |
| Space group | P212121 | P21 | H3 | P212121 | P21 | C2 | P3221 |
| Cell dimensions | |||||||
| a, b, c (Å) | 65.9, 67.0, 74.7 | 53.2, 68.2, 66.6 | 214, 214, 45.5 | 36.5, 70.5, 122 | 50.1, 193, 66.2 | 148, 106, 92.4 | 121, 121, 203 |
| α, β, γ (deg) | 90, 90, 90 | 90, 102, 90 | 90, 90, 120 | 90, 90, 90 | 90, 90.1, 90 | 90, 98.8, 90 | 90, 90, 120 |
| Resolution (Å) | 50–1.8 | 50–2.0 | 50–2.2 | 50–1.95 | 50–2.3 | 50–2.75 | 50–3.1 |
| No. of reflections | 30 653 | 30 901 | 39 195 | 23 691 | 52 659 | 36 321 | 31 929 |
| Completeness (%) | 97.6 (99.3) | 97.5 (88.7) | 99.0 (98.0) | 99.8 (99.9) | 94.5 (85.6) | 99.6 (100) | 99.6 (98.9) |
| Redundancy | 4.6 (4.6) | 3.5 (3.0) | 4.4 (3.6) | 5.3 (5.2) | 4.2 (4.1) | 4.2 (4.2) | 3.8 (3.6) |
| Rsym | 5.0 (51.5) | 8.6 (32.2) | 9.6 (48.5) | 8.9 (53.0) | 5.6 (34.4) | 10.4 (52.7) | 6.1 (51.5) |
| I/σI | 26.9 (3.2) | 13.8 (2.4) | 14.3 (2.2) | 16.8 (3.1) | 20.7 (3.7) | 10.9 (3.3) | 18.3 (2.6) |
| Refinement | |||||||
| Resolution (Å) | 20–1.8 | 20–2.0 | 20–2.2 | 20–1.95 | 20–2.3 | 30–2.75 | 30–3.1 |
| No. of reflections | 29 018 | 29 213 | 37 085 | 22 404 | 49 667 | 34 487 | 30 265 |
| Rwork/Rfree | 0.159/0.201 | 0.183/0.243 | 0.159/0.207 | 0.172/0.235 | 0.185/0.239 | 0.186/0.243 | 0.188/0.232 |
| No. of atoms | |||||||
| Protein | 2549 | 3459 | 4540 | 2509 | 6813 | 7593 | 7740 |
| Ligand/ion | 6 | 25 | 12 | 6 | 34 | 0 | 0 |
| Sugar | 0 | 76 | 0 | 0 | 0 | 28 | 0 |
| Water | 282 | 194 | 249 | 239 | 365 | 0 | 0 |
| B-factors (Å2) | 26.6 | 36.3 | 33.2 | 22.5 | 31.8 | 74.9 | 97.1 |
| R.m.s. deviations | |||||||
| Bond lengths (Å) | 0.014 | 0.012 | 0.013 | 0.013 | 0.013 | 0.011 | 0.010 |
| Bond angles (deg) | 1.5 | 1.5 | 1.4 | 1.5 | 1.4 | 1.3 | 1.3 |
| One crystal was used for data collection for each structure. Values in parentheses refer to the highest-resolution shell. | |||||||
Figure 3.
Overall domain architecture of neuropilins. (A) Domain organization of Nrp2 (blue, a1; green, a2; yellow, b1; red, b2) in complex with the Fab fragment of anti-panNrpA (tan, heavy chain; gray, light chain). N-glycosylated residues are indicated in magenta. (B) Ribbon representation of the Nrp a2b1b2 structures; the orange spheres highlight a bound calcium ion. (C) Superposition of the Nrp2/Fab complex from two different crystal forms based on the a2b1b2 domains. Note the poor superposition of the a1 domains (yellow arrows) in comparison to the a2b1b2 region (black arrows). Structure figures were produced with PyMol (http://www.pymol.org).
The overall domain architecture of the Nrp ectodomain
The Nrp extracellular region is often divided into three units that describe the primary semaphorin-binding (a1a2), VEGF-binding (b1b2), and dimerization (c) domains. The crystal structure of rat Nrp1 b1b2 identified a large interface between the b1 and b2 domains and, as the residues buried between domains are conserved, this b1b2 domain arrangement was recognized as a general feature of the Nrp family (Vander Kooi et al, 2007). We have solved four crystal structures of Nrp that include the a2, b1, and b2 domains (Figures 2 and 3) and were crystallized from a variety of conditions (Supplementary Table S1). Two models include the a1 domains bound to Fab fragments, while two others include calcium ions in the a2 domains. Despite these differences, the three domains (a2, b1, and b2) share the same arrangement in all crystal structures and pack tightly around a pseudo-three-fold axis (Figure 3). The Nrp1 and Nrp2 a2b1b2 structures are very similar and superimpose with a root mean square deviation (r.m.s.d.) of 1.2 Å over 317 Cα atoms. As predicted (Vander Kooi et al, 2007), the interface between b1 and b2 is conserved between both neuropilins, burying about 1200–1500 Å2 of solvent accessible surface. Notably, the interface between the a2 domain and the b1b2 is conserved in all structures as well and buries more than 2000 Å2 of solvent accessible surface, indicating that all three domains form a rigid structural unit (Figure 3).
In contrast, the domain arrangement between a1 and a2b1b2 is not preserved between the two crystal forms of the Nrp2/anti-panNrpA–Fab complex (Figure 3C). When superimposing the a2b1b2 cores of the receptors, the a1 domains are displaced by about 7 Å with respect to each other. The interface between a1 and a2b1b2 is small (approximately 800 Å2 buried surface area) and not conserved. The lack of strong interactions identifies flexibility of the a1 domain, suggesting that it can undergo conformational changes with respect to the remainder of Nrp in solution or upon receptor binding.
Nrp CUB domains include calcium- and semaphorin-binding sites
The a1 and a2 domains of Nrp1 and Nrp2 are CUB domains (Takagi et al, 1991). In the CUB domain prototype, spermadhesin (Romero et al, 1997), the fold comprises two five-stranded β-sheets that form a β-sandwich. Strands β2, β4, β9, β6, and β7 form one β-sheet with the other sheet containing strands β1, β3, β10, β5, and β8; however, strands β1 and β2 are frequently absent in CUB domains from several complement family proteins (Feinberg et al, 2003; Gregory et al, 2003, 2004). Similar to these proteins, the a1 and a2 domains of Nrps lack the β1 strand (Figure 4). Overall, CUB domains display a high degree of similarity. For example, the Nrp2 a2 domain is structurally similar to the Nrp1 a2 domain (r.m.s.d. of 0.9 Å; 108 Cα atoms), the Nrp2 a1 domain (r.m.s.d. of 0.9 Å; 96 Cα atoms), and the CUB domain from spermadhesin (r.m.s.d. of 1.4 Å; 86 Cα atoms).
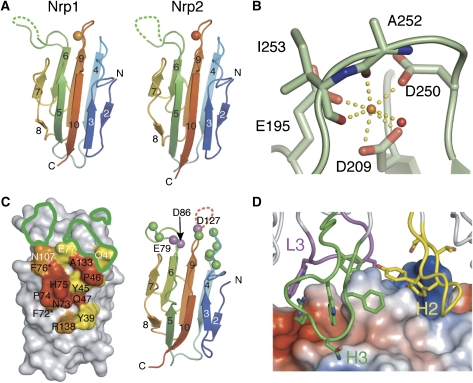
Figure 4.
Molecular details of the neuropilin CUB domains. (A) In the a2b1b2 structures, the a2 domain includes a bound calcium ion (orange). The Nrp a1 (panel C) and a2 domains lack the β1 strand found in spermadhesin. (B) The calcium ion of the Nrp1 a2 domain is coordinated by three negatively charged amino acids, two main-chain carbonyl oxygens, and a water molecule. These interactions are highly conserved among Nrps (Supplementary Figures S2 and S3). (C) (left panel) Amino acids are colored according to the percentage of solvent-accessible surface that is buried at the Nrp2/Fab interface (red, 75–100%; orange, 50–74%; yellow, 25–49%). Anti-panNrpA is crossreactive for Nrp1 and Nrp2, and 11 of the 14 residues in the structural epitope are identical (black text, identical; white text, nonconserved; asterisks indicate residues in which the side chain points toward the a1 protein core). Residues outlined in green indicate the position of amino acids that when replaced were shown to abrogate binding between Nrp1 and Sema3A (Gu et al, 2002). (right panel) The Cα atoms of these amino-acid substitutions are indicated as green spheres. Cα atoms shaded in purple represent a putative calcium-binding site. (D) The anti-panNrpA/Nrp2 interface. Nrp2 is shown as a molecular surface with local concentrations of charged residues colored blue (basic) or red (acidic) as calculated by PyMol. The antibody contact residues are dominated by aromatic resides from CDRs H2, H3, and L3.
The crystal structures of CUB domains from the complement family proteins C1s and MASP-2 (Gregory et al, 2003, 2004) contain a calcium-binding site at one end of the sandwich. The a2 domain from the Nrp1 and Nrp2 a2b1b2 structures also contain a bound ion at an identical location (Figures 3B and 4A). In the Nrp1 structure, the ion is clearly observed in the electron density (Supplementary Figure S2) even though no divalent cations were included during crystallization (Supplementary Table S1). In Nrp1, the calcium ion is coordinated by three negatively charged side chains (Glu195, Asp209, and Asp250), two carbonyl oxygens (Ala252 and Ile253), and a water molecule (Figure 4B). Similarly, calcium coordination involves three negatively charged side chains in Nrp2 (Supplementary Figure S2), and these three amino acids are absolutely conserved in the a2 domain from 12 distantly related species (Supplementary Figure S3).
A sequence alignment of the a1 and a2 domains of Nrps (Supplementary Figures S2 and S3) shows that this triad of charged residues is also strictly conserved within the a1 domain of the Nrps. However, in the Nrp2/anti-panNrpA–Fab complex, there is no indication of a bound ion in the a1 domain. The loops with the residues that define the putative a1 Ca2+-binding site are poorly ordered, suggesting that Ca2+ plays a role in stabilizing the fold of the domain. Based on the high degree of sequence conservation, it is likely that calcium binding represents a shared feature of Nrp CUB domains.
Neuropilins function as co-receptors for select members of the class 3 family of semaphorins. The semaphorin N terminus contains the signature ‘Sema' domain, a seven-bladed β-propeller (Antipenko et al, 2003), which is required for binding the a1a2 domains of neuropilins. Anti-panNrpA blocks Sema3A-induced growth cone collapse mediated through Nrp1 (Figure 1) and Sema3F-mediated growth cone collapse mediated through Nrp2, but does not affect VEGF binding (data not shown). In the structure of the Nrp2/anti-panNrpA–Fab complex (Figure 3A), the Fab fragment contacts only the a1 domain of Nrp2 on the β8-5-10-3 face of the sandwich, burying approximately 1400 Å2 of the solvent accessible surface (Figure 4C). The interface recognized by this antibody is well conserved between Nrp1 and Nrp2. Eleven of the 14 Nrp2 residues that have more than 25% of their accessible surface buried in the interface are identical between the two receptors (Figure 4C). This recognition of an epitope that is conserved between Nrp1 and Nrp2 explains the ability of the antibody to bind both receptors with affinities in the sub-nanomolar range (Supplementary Figure S1).
On the antibody side, 11 side chains contact Nrp2 from complementary-determining regions (CDRs) L3, H2, and H3, seven of which are aromatic in character (Figure 4D). Previous site-directed mutagenesis studies (Gu et al, 2002) outlined the semaphorin-binding site of Nrp1. Based on a model of spermadhesin, putatively solvent-exposed residues on the surface of the a1 domain were selected for amino-acid substitutions. Using this approach (Gu et al, 2002), a number of mutants were identified that disrupt the interactions between Sema3A and Nrp1 and when mapped onto Nrp1 a1, they are located on the loops at one pole of the a1 β-sandwich (Figure 4C). Substitutions at the other end of the domain had no effect on Sema3A binding (Gu et al, 2002). The mutations disrupting Sema3A binding are adjacent to the epitope recognized by the Sema3A-blocking Fab (Figure 4C), strongly suggesting that the sema domain binds the loops and the β8-5-10-3 face of the sandwich within the Nrp2 a1 domain. The location of the semaphorin-binding site is also adjacent to the putative calcium-binding site of the a1 domain (Figure 4C), suggesting that calcium binding may play a role in the interactions between the ligand and receptor.
To test this hypothesis, we examined the effects of the Ca2+ chelator EGTA in vitro. While Sema3A results in the collapse of actin-rich growth cones in DRG, pre-incubation with EGTA blocks this activity of Sema3A (Supplementary Figure S4). These observations coupled with the high conservation of the amino acids that coordinate the calcium ion suggest that calcium may play an important role for Sema3/Nrp interactions.
Nrp type b domains contain the heparin- and VEGF-binding sites
The b domains from the human neuropilin b1b2 structures (Supplementary Figure S5) share significant homology with the phospholipid-binding (type C2) modules from coagulation factors V and VIII (Takagi et al, 1991; Macedo-Ribeiro et al, 1999; Pratt et al, 1999; Lee et al, 2003; Vander Kooi et al, 2007), and to the galactose-binding domain of bacterial sialidase (Gaskell et al, 1995). Collectively, these domains define the discoidin fold that is topologically classified as a distorted jelly-roll β-barrel composed of eight core β-strands. One pole of the domain contains three extended ‘spikes' or loops (Supplementary Figure S5) that typically constitute the ligand-binding site for discoidin family members (Gaskell et al, 1995; Macedo-Ribeiro et al, 1999; Pratt et al, 1999; Vander Kooi et al, 2007). The b1b2 fragment of Nrp1 and Nrp2 share 50% sequence identity and superimpose with an r.m.s.d. of 2.3 Å over 307 Cα atoms. While the b1 domains of Nrp1 and Nrp2 are nearly indistinguishable (r.m.s.d=0.6 Å), the b2 domains superimpose less well (r.m.s.d.=2.7 Å) with the differences largely in the conformations of the ‘spikes' (Supplementary Figure S5). As these spikes frequently define binding sites within the discoidin family (Gaskell et al, 1995; Macedo-Ribeiro et al, 1999; Pratt et al, 1999; Vander Kooi et al, 2007), the dissimilarity between Nrp1 and Nrp2 may represent flexibility in this region, and thus a way to recognize multiple-binding partners.
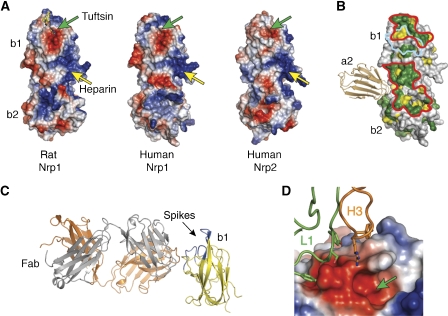
Despite the differences between the b2 domains of the Nrp1 and Nrp2, the b domains are tightly packed and form a rigid scaffold (Figure 5). The interdomain junction forms a deep cleft that runs approximately perpendicular to the long axis of the two b domains. This cleft, created by the β4:β5 loop of b1 and the β5:β6 loop of b2, is surrounded by a number of positively charged residues, which based on mutagenesis experiments on rat Nrp1, represents the heparin-binding site (Vander Kooi et al, 2007). The electropositive patch is present in the structure of both human Nrps as well (Figure 5A). The C-terminal domain of VEGF165 (also known as VEGF55) also contains a heparin-binding site (Fairbrother et al, 1998). As heparin increases the affinity of b1b2 for VEGF165 up to 100-fold (Fuh et al, 2000; Mamluk et al, 2002), it is likely that Nrps use heparin to recruit VEGF165 to this region.
Figure 5.
Features of the Nrp VEGF- and heparin-binding domains. (A) The molecular surface of the rat (PDB code 2ORZ) (Vander Kooi et al, 2007) and human b1b2 crystal structures are colored as described in Figure 4D. Green arrows indicate an acidic groove that is formed by the ‘spikes' in the b1 domain (Supplementary Figure S5); this feature forms the Tuftsin-binding site of rat Nrp1. Yellow arrows indicate the approximate location of the heparin-binding patch. (B) The sequence conservation (green, 100%; yellow, ⩾75%) of the b1b2 domains among 12 Nrps (Supplementary Figure S3) was mapped onto the surface of the human Nrp1 b1b2 structure. Two highly conserved patches are delineated in red. Residues outlined in cyan indicate those residues that contact the Fab in the anti-Nrp1B-Fab/b1 complex. The a2 domain (tan) is shown by using a superposition of the b1b2 and a2b1b2 structures from Nrp1. (C) Ribbon representation of the anti-Nrp1B–Fab/b1 complex (yellow, b1; orange, heavy chain; gray, light chain). (D) The anti-Nrp1B/b1 interface. The b1 domain is depicted as a molecular surface with a green arrow indicating the Tuftsin/VEGF tail-binding groove. Only CDRs H3 and L1 contact b1.
In addition to this heparin-mediated interaction between exon 7 of VEGF165 and Nrps, the C-terminal tail of VEGF (CDKPRRCOOH) encoded by exon 8 may bind directly to Nrps. In the crystal structure of rat Nrp1 b1b2 in complex with Tuftsin (Vander Kooi et al, 2007), a tetrapeptide mimetic (TKPRCOOH) of the VEGF tail (von Wronski et al, 2006), the C-terminal arginine is tucked tightly into an acidic groove created by residues from the conserved ‘spikes' of the b1 domain. In several of our structures, including all three Fab complexes, the C-terminal residue (a histidine) from a symmetry-related molecule occupies the same acidic pocket as the Tuftsin peptide (Supplementary Figure S6).
To further detail potential residues involved in VEGF binding, we examined the conservation of Nrp residues on the b1b2 surface (Figure 5B). Two contiguous patches are conserved among 12 Nrp1 and Nrp2 proteins (Supplementary Figure S3). One of these sites maps directly to the Tuftsin-binding site, while the second site includes residues on the edge of the heparin-binding patch, suggesting an additional area for interactions between Nrp and VEGF.
In the crystal structure of the Nrp1 b1/anti-Nrp1B–Fab complex (Figure 5), the Fab contacts an epitope located between these two putative VEGF-binding sites and partially overlaps with the Tuftsin-binding cleft. The epitope of this VEGF-blocking antibody is unusual, involving only residues from CDRs L1 and H3. In general, Fab/antigen interfaces bury an average of approximately 1700 Å2 of solvent-exposed surface (Lo Conte et al, 1999); however, only 900 Å2 are shielded in the Nrp1 b1/Fab interface. Despite the small interface, anti-Nrp1B binds tightly to Nrp1 with an affinity of 0.2 nM (Liang et al, 2007; Pan et al, 2007a).
VEGF and Sema3A do not compete for Nrp binding
Several studies have reported that VEGFs and semaphorins compete for binding on the cell surface and that this competition involves a partially overlapping binding site on the b1 domain (Miao et al, 1999; Gu et al, 2002; Narazaki and Tosato, 2006).
As the carboxyl tail of VEGF165 and class 3 semaphorins are both rich in basic residues, it was suggested that these tails might compete for the electronegative groove created by the ‘spikes' in the b1 domain (Lee et al, 2003; Vander Kooi et al, 2007). The recent b1b2/Tuftsin crystal structure indicated that the VEGF165 tail occupies this binding site. However, the C terminus of Sema3A does not end with the highly conserved arginine of VEGF165 or Tuftsin that is tucked in the VEGF-binding groove on the b1b2 structure (Vander Kooi et al, 2007); instead, it contains a hydrophobic valine, rendering it unlikely that the C terminus of semaphorins occupy the same binding pocket. We have previously shown that anti-Nrp1B does not antagonize Sema3A-mediated collapse of axons from DRG (Figure 1) (Liang et al, 2007; Pan et al, 2007a). As the VEGF-blocking antibody does not affect Sema3 function, we examined whether VEGF165 might antagonize Sema3A-mediated collapse of axonal growth cones from DRGs. In this assay, even at concentrations up to 100 nM, VEGF165 did not reduce the ability of Sema3A (at 2.2 nM) to cause the retraction of actin processes (Figure 1). In addition, saturating concentrations of Sema3A do not alter VEGF-induced endothelial cell migration (Pan et al, 2007a). To test whether VEGF and Sema3A exclude each other from binding to Nrps, we studied the interaction between immobilized Nrp1 and its ligands VEGF165 and Sema3A using surface plasmon resonance. Both Nrp-binding partners show specific binding to Nrp1-Fc, although they display different binding modes as illustrated in the differences between their association and dissociation phases (Supplementary Figure S7). When VEGF165 and Sema3A are coinjected at equimolar amounts, the resulting binding sensorgram is almost identical to the sum of the two single binding sensorgrams, as would be expected for simultaneous binding (Supplementary Figure S7). Antibodies that have been shown to block binding of Sema3A and VEGF165 to Nrp1 (Liang et al, 2007; Pan et al, 2007a) also completely block binding in this in vitro assay (Supplementary Figure S7). Furthermore, in our Nrp/Fab crystal structures, the binding epitopes blocking VEGF and Sema3 binding are separated by 65 Å and located on opposite sites of the Nrp (Supplementary Figure S8). The large distance between the interfaces for VEGF- and Sema3-blocking Fabs supports the notion that both ligands, semaphorin and VEGF, can bind Nrps simultaneously and do not compete for each other.
Previous competition experiments (Miao et al, 1999; Gu et al, 2002; Narazaki and Tosato, 2006), indicating that VEGF and semaphorins compete for Nrp binding, employed VEGFs and semaphorins with heterologous tags (such as alkaline phosphatase) at the C termini of these proteins. It might be possible that the observed competition is a result of steric clashes with the tags rather than the result of direct competition between the VEGF and the Sema3 tail.
Models for neuropilin dimerization
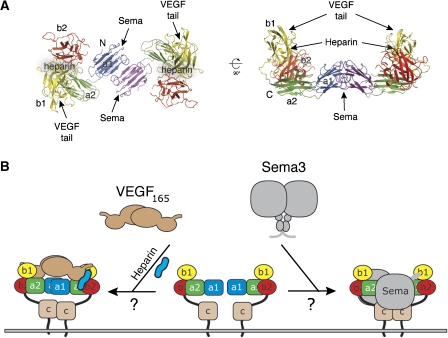
Nrp1 and Nrp2 form homo- or hetero-multimers even in the absence of ligand (Chen et al, 1998; Nakamura et al, 1998; Takahashi et al, 1998). Data have demonstrated that the c domain is important for multimerization; however, since truncation mutants lacking this domain can self-associate, the c domain might not be the only domain involved in Nrp oligomerization (Chen et al, 1998; Giger et al, 1998; Nakamura et al, 1998; Renzi et al, 1999). Nrp constructs that include the a1a2 and b1b2 domains have higher affinity for VEGFs than constructs that span only the b1b2 domains (Gu et al, 2002; Mamluk et al, 2002; Karpanen et al, 2006) even though the a1a2 domains do not bind VEGF. It is therefore possible that the a1a2 domains contribute directly to Nrp dimerization and strengthen interactions between the Nrp dimer and the VEGF dimer by stabilizing the 2:2 complex.
Interestingly, the crystal structures of a1a2b1b2 domains from two Nrp2 crystal forms (Table I) and contain a conserved, crystallographic interface that is mediated by a1. In this dimer, the a1 domains align in an approximately antiparallel arrangement along the β7 and β8 strands. The interface buries approximately 1200 Å2 of solvent accessible surface area and is dominated by hydrophobic interactions (Supplementary Figure S8). Notably, similar dimers have been observed for other CUB domain family members (Romero et al, 1997). Other features of the crystallographic dimer are less supportive that the observed a1 interface is biologically relevant. For one, the amino acids along the interface are only moderately conserved (Supplementary Figure S8); in addition, the shape complementarity of the interface is poor with an Sc value of only 0.4 (Lawrence and Colman, 1993). Therefore, we examined the molecular mass of three Nrp2 constructs (a2b1b2, a1a2b1b2, and a1a2b1b2c) by multiangle light scattering, but in this experiment none of the constructs formed dimers even when the c domain was present. Although immunoprecipitation experiments have demonstrated that Nrp self-associates (Chen et al, 1998; Giger et al, 1998; Nakamura et al, 1998; Takahashi et al, 1998, 1999), there is no data that rigorously show the formation of Nrp dimers in solution. This underscores the difficulty in designing experiments that directly measure dimerization of wild-type or mutant Nrps that might disrupt the a1 interface.
Although Nrp2 a1a2b1b2 is monomeric by dynamic light scattering, the orientation of crystallographic Nrp2 dimer suggests a model for VEGF binding. The a1 interface creates a saddle-shaped dimer with a width of approximately 70 Å (Figure 6), large enough to accommodate a VEGF109-dimer with dimensions of about 35 by 60 Å. Importantly, the Tuftsin-binding sites and the heparin-binding patches are found on the inner surface of the saddle. Heparin could stabilize interactions between the heparin-binding sites of Nrp and VEGF and further facilitate the association of the VEGF tail with the ‘spikes' of b1. This arrangement would also be able to accommodate VEGFR binding via the VEGF receptor-binding domain for downstream signaling. Therefore, although the a1 domains do not directly engage VEGF, they could enhance VEGF binding of Nrps by facilitating Nrp dimerization. Alternatively, it has been suggested that heparin binding induces dimerization of Nrp1 b1b2 (Vander Kooi et al, 2007) and that this activity could explain how Nrps dimerize in the absence of the MAM domain.
Figure 6.
A Nrp2 crystallographic dimer suggests models for VEGF and Sema3 binding. (A) Nrp2 forms a saddle-shaped dimer in both crystal forms of the Nrp2/Fab complex. This figure highlights the Nrp2 a1a2b1b2 domains from the monoclinic crystal form of the Fab complex. The putative VEGF tail-, heparin-, and semaphorin-binding sites are indicated. (B) Potential models of VEGF/Nrp and semaphorin/Nrp complexes based on the Nrp a1-mediated dimer in the crystal structures.
This dimer could further accommodate Sema3 binding (Figure 6). In the Sema3A crystal structure, two ‘sema' domains pack tightly together at an interface. Upon Nrp binding, the sema domains of Sema3A dissociate to allow interactions with the primary binding site on a1a2 (Antipenko et al, 2003). In our model of the a1-mediated Nrp dimer, the two putative Sema3A-binding sites are located on opposite sides, and distant enough to accommodate the large β-propellers of two bound Sema3 molecules (Figures 6 and Supplementary S5).
Our structural analysis provides the first detailed picture of both the VEGF-binding (b1b2) and semaphorin-binding (a1a2) portions of the Nrp ECD. The antibody complexes along with previous mutagenesis studies (Gu et al, 2002; Vander Kooi et al, 2007) delineate the Nrp-binding sites for the Sema domain of semaphorins, and the heparin-binding domain of VEGF. These structures provide a basis for future experiments and provide strategies to elucidate the structures of Nrps in complex with their ligands (VEGFs and semaphorins) and signaling receptors (VEGFRs and plexins).
Materials and methods
Functional assays and antibody-binding affinities
Collapse assays and determination of anti-Nrp antibody-binding affinities were performed as previously described (He and Tessier-Lavigne, 1997; Liang et al, 2007; Pan et al, 2007a).
Protein expression and purification
Nrp1 b1, Nrp1 b1b2, and Nrp2 b1b2 (Supplementary Table S1) were cloned into pET15b (Novagen) and expressed in E. coli following induction at 37°C (Nrp1 b1 and b1b2) or 16°C (Nrp2 b1b2). All neuropilin type b fragments are expressed as soluble proteins without the need for a refolding protocol. Following cell lysis, proteins were purified using nickel-nitrilotriacetic acid (Ni-NTA) resin in 50 mM Tris (pH 8.0), 300–500 mM NaCl, and 20 mM imidazole, and eluted in the same buffer plus 250 mM imidazole. The his6-tags were removed with thrombin, and samples were further purified using a Superdex-75 column equilibrated in 25 mM Tris (pH 8.0) and 150 mM NaCl.
Recombinant baculoviruses were generated to facilitate the secretion of Nrp a2b1b2, a1a2b1b2, and full-length ECD constructs (Supplementary Table S1). Nrp2 a1a2b1b2 and the full-length Nrp2-ECD were subcloned with the Nrp2 native secretion signal and a C-terminal His6-tag into pENTR/D-TOPO (Invitrogen) and recombined into pDEST8 (Invitrogen) to generate a viral bacmid. Nrp1 and Nrp2 a2b1b2 were cloned into pAcGP67B (Clonetech). Following infection, the culture media were collected and supplemented with 50 mM Tris (pH 8.0), 5 mM CaCl2, and 1 mM NiCl2; proteins were purified with Ni-NTA and gel filtration chromatography as described for bacterial-expressed constructs.
The Fab fragments for anti-Nrp1B (YW107.4.87) (Liang et al, 2007; Pan et al, 2007a) and anti-panNrpA (YW68.11.26) were expressed in E. coli, captured on a Protein G column equilibrated in PBS, and eluted with 0.58% acetic acid. Protein fractions were further purified by ion exchange chromatography (SP-sepharose) in 20 mM MES (pH 5.5) and eluted with a gradient from 0 to 250 mM NaCl. Fab/Nrp complexes were typically mixed at 1:1 molar ratio and further purified using a Superdex-200 column equilibrated in 25 mM Tris–HCl (pH 7.5) and 200 mM NaCl. For crystallization, all unbound neuropilin and Nrp/Fab complex samples were concentrated as detailed in Supplementary Table S1.
Crystallization, structure determination, and refinement
All crystals were obtained by vapor diffusion at 19°C by mixing equal volumes of protein plus well solution (Supplementary Table S1). Crystals were cryoprotected as described in Supplementary Table S1. Data sets were collected at the Advanced Light Source (beam lines 5.0.1 or 5.0.2) or the Stanford Synchrotron Radiation Laboratory (beam lines 9-2 or 11-1) and processed with the HKL package (Otwinowski and Minor, 1997).
All crystal structures were solved by molecular replacement using Phaser (McCoy et al, 2005). The Nrp1 b1b2 structure was solved using the Nrp1 b1 crystal structure (pdb code 1KEX) (Lee et al, 2003), and the refined coordinates of the Nrp1 b1b2 structure served as the search probe for the Nrp2 b1b2 structure. The monoclinic form of the Nrp2 a1a2b1b2/anti-panNrpA–Fab complex was solved using the Nrp2 b1b2 structure and Fab fragments containing either the variable domains (VH/VL) or the constant domains (CH1/CL) from the B20-4/VEGF complex (pdb code 2FJH) (Fuh et al, 2006). The a2 domain was identified by molecular replacement using the N-terminal CUB domain from MASP-2 (pdb code 1NT0) (Feinberg et al, 2003); the a1 domain was placed manually within the electron density. The Nrp1 b1/Fab complex was solved using the Nrp1 b1 crystal structure and the B20-4 Fab fragments described above. All remaining structures were solved using the refined coordinates of the b1b2 structures and the a2 CUB domain from the Nrp2 a1a2b1b2/Fab complex. Atomic models were built using Coot (Emsley and Cowtan, 2004) and refined with Refmac (Murshudov et al, 1997).
Supplementary Material
Supplementary Figure S1–S8
Acknowledgments
We thank members of the structure group for assistance with data collection. Data were collected at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the US Department of Energy at Lawrence Berkeley National Laboratory and at the Stanford Synchrotron Radiation Laboratory, which is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. The atomic coordinates and structure factors have been deposited in the Protein Data Bank (pdb codes 2QQI, 2QQJ, 2QQK, 2QQL, 2QQM, 2QQN, and 2QQO).
References
- Antipenko A, Himanen JP, van Leyen K, Nardi-Dei V, Lesniak J, Barton WA, Rajashankar KR, Lu M, Hoemme C, Puschel AW, Nikolov DB (2003) Structure of the semaphorin-3A receptor binding module. Neuron 39: 589–598 [DOI] [PubMed] [Google Scholar]
- Beckmann G, Bork P (1993) An adhesive domain detected in functionally diverse receptors. Trends Biochem Sci 18: 40–41 [DOI] [PubMed] [Google Scholar]
- Cai H, Reed RR (1999) Cloning and characterization of neuropilin-1-interacting protein: a PSD-95/Dlg/ZO-1 domain-containing protein that interacts with the cytoplasmic domain of neuropilin-1. J Neurosci 19: 6519–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Bagri A, Zupicich JA, Zou Y, Stoeckli E, Pleasure SJ, Lowenstein DH, Skarnes WC, Chedotal A, Tessier-Lavigne M (2000) Neuropilin-2 regulates the development of selective cranial and sensory nerves and hippocampal mossy fiber projections. Neuron 25: 43–56 [DOI] [PubMed] [Google Scholar]
- Chen H, Chedotal A, He Z, Goodman CS, Tessier-Lavigne M (1997) Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III. Neuron 19: 547–559 [DOI] [PubMed] [Google Scholar]
- Chen H, He Z, Bagri A, Tessier-Lavigne M (1998) Semaphorin–neuropilin interactions underlying sympathetic axon responses to class III semaphorins. Neuron 21: 1283–1290 [DOI] [PubMed] [Google Scholar]
- Ellis LM (2006) The role of neuropilins in cancer. Mol Cancer Ther 5: 1099–1107 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fairbrother WJ, Champe MA, Christinger HW, Keyt BA, Starovasnik MA (1998) Solution structure of the heparin-binding domain of vascular endothelial growth factor. Structure 6: 637–648 [DOI] [PubMed] [Google Scholar]
- Favier B, Alam A, Barron P, Bonnin J, Laboudie P, Fons P, Mandron M, Herault JP, Neufeld G, Savi P, Herbert JM, Bono F (2006) Neuropilin-2 interacts with VEGFR-2 and VEGFR-3 and promotes human endothelial cell survival and migration. Blood 108: 1243–1250 [DOI] [PubMed] [Google Scholar]
- Feinberg H, Uitdehaag JC, Davies JM, Wallis R, Drickamer K, Weis WI (2003) Crystal structure of the CUB1-EGF-CUB2 region of mannose-binding protein associated serine protease-2. EMBO J 22: 2348–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuh G, Garcia KC, de Vos AM (2000) The interaction of neuropilin-1 with vascular endothelial growth factor and its receptor flt-1. J Biol Chem 275: 26690–26695 [DOI] [PubMed] [Google Scholar]
- Fuh G, Wu P, Liang WC, Ultsch M, Lee CV, Moffat B, Wiesmann C (2006) Structure–function studies of two synthetic anti-vascular endothelial growth factor Fabs and comparison with the Avastin Fab. J Biol Chem 281: 6625–6631 [DOI] [PubMed] [Google Scholar]
- Gaskell A, Crennell S, Taylor G (1995) The three domains of a bacterial sialidase: a beta-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure 3: 1197–1205 [DOI] [PubMed] [Google Scholar]
- Giger RJ, Cloutier JF, Sahay A, Prinjha RK, Levengood DV, Moore SE, Pickering S, Simmons D, Rastan S, Walsh FS, Kolodkin AL, Ginty DD, Geppert M (2000) Neuropilin-2 is required in vivo for selective axon guidance responses to secreted semaphorins. Neuron 25: 29–41 [DOI] [PubMed] [Google Scholar]
- Giger RJ, Urquhart ER, Gillespie SK, Levengood DV, Ginty DD, Kolodkin AL (1998) Neuropilin-2 is a receptor for semaphorin IV: insight into the structural basis of receptor function and specificity. Neuron 21: 1079–1092 [DOI] [PubMed] [Google Scholar]
- Gluzman-Poltorak Z, Cohen T, Herzog Y, Neufeld G (2000) Neuropilin-2 is a receptor for the vascular endothelial growth factor (VEGF) forms VEGF-145 and VEGF-165. J Biol Chem 275: 29922. [DOI] [PubMed] [Google Scholar]
- Gluzman-Poltorak Z, Cohen T, Shibuya M, Neufeld G (2001) Vascular endothelial growth factor receptor-1 and neuropilin-2 form complexes. J Biol Chem 276: 18688–18694 [DOI] [PubMed] [Google Scholar]
- Gouet P, Robert X, Courcelle E (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31: 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory LA, Thielens NM, Arlaud GJ, Fontecilla-Camps JC, Gaboriaud C (2003) X-ray structure of the Ca2+-binding interaction domain of C1s. Insights into the assembly of the C1 complex of complement. J Biol Chem 278: 32157–32164 [DOI] [PubMed] [Google Scholar]
- Gregory LA, Thielens NM, Matsushita M, Sorensen R, Arlaud GJ, Fontecilla-Camps JC, Gaboriaud C (2004) The X-ray structure of human mannan-binding lectin-associated protein 19 (MAp19) and its interaction site with mannan-binding lectin and L-ficolin. J Biol Chem 279: 29391–29397 [DOI] [PubMed] [Google Scholar]
- Gu C, Limberg BJ, Whitaker GB, Perman B, Leahy DJ, Rosenbaum JS, Ginty DD, Kolodkin AL (2002) Characterization of neuropilin-1 structural features that confer binding to semaphorin 3A and vascular endothelial growth factor 165. J Biol Chem 277: 18069–18076 [DOI] [PubMed] [Google Scholar]
- He Z, Tessier-Lavigne M (1997) Neuropilin is a receptor for the axonal chemorepellent Semaphorin III. Cell 90: 739–751 [DOI] [PubMed] [Google Scholar]
- Karkkainen MJ, Saaristo A, Jussila L, Karila KA, Lawrence EC, Pajusola K, Bueler H, Eichmann A, Kauppinen R, Kettunen MI, Yla-Herttuala S, Finegold DN, Ferrell RE, Alitalo K (2001) A model for gene therapy of human hereditary lymphedema. Proc Natl Acad Sci USA 98: 12677–12682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpanen T, Heckman CA, Keskitalo S, Jeltsch M, Ollila H, Neufeld G, Tamagnone L, Alitalo K (2006) Functional interaction of VEGF-C and VEGF-D with neuropilin receptors. FASEB J 20: 1462–1472 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Kitsukawa T, Bekku Y, Matsuda Y, Sanbo M, Yagi T, Fujisawa H (1999) A requirement for neuropilin-1 in embryonic vessel formation. Development 126: 4895–4902 [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty DD (1997) Neuropilin is a semaphorin III receptor. Cell 90: 753–762 [DOI] [PubMed] [Google Scholar]
- Kreuter M, Woelke K, Bieker R, Schliemann C, Steins M, Buechner T, Berdel WE, Mesters RM (2006) Correlation of neuropilin-1 overexpression to survival in acute myeloid leukemia. Leukemia 20: 1950–1954 [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM (1993) Shape complementarity at protein/protein interfaces. J Mol Biol 234: 946–950 [DOI] [PubMed] [Google Scholar]
- Lee CC, Kreusch A, McMullan D, Ng K, Spraggon G (2003) Crystal structure of the human neuropilin-1 b1 domain. Structure 11: 99–108 [DOI] [PubMed] [Google Scholar]
- Liang WC, Dennis MS, Stawicki S, Chanthery Y, Pan Q, Chen Y, Eigenbrot C, Yin J, Koch AW, Wu X, Ferrara N, Bagri A, Tessier-Lavigne M, Watts RJ, Wu Y (2007) Function blocking antibodies to neuropilin-1 generated from a designed human synthetic antibody phage library. J Mol Biol 366: 815–829 [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J (1999) The atomic structure of protein–protein recognition sites. J Mol Biol 285: 2177–2198 [DOI] [PubMed] [Google Scholar]
- Macedo-Ribeiro S, Bode W, Huber R, Quinn-Allen MA, Kim SW, Ortel TL, Bourenkov GP, Bartunik HD, Stubbs MT, Kane WH, Fuentes-Prior P (1999) Crystal structures of the membrane-binding C2 domain of human coagulation factor V. Nature 402: 434–439 [DOI] [PubMed] [Google Scholar]
- Makinen T, Olofsson B, Karpanen T, Hellman U, Soker S, Klagsbrun M, Eriksson U, Alitalo K (1999) Differential binding of vascular endothelial growth factor B splice and proteolytic isoforms to neuropilin-1. J Biol Chem 274: 21217–21222 [DOI] [PubMed] [Google Scholar]
- Mamluk R, Gechtman Z, Kutcher ME, Gasiunas N, Gallagher J, Klagsbrun M (2002) Neuropilin-1 binds vascular endothelial growth factor 165, placenta growth factor-2, and heparin via its b1b2 domain. J Biol Chem 277: 24818–24825 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61: 458–464 [DOI] [PubMed] [Google Scholar]
- Miao HQ, Soker S, Feiner L, Alonso JL, Raper JA, Klagsbrun M (1999) Neuropilin-1 mediates collapsin-1/semaphorin III inhibition of endothelial cell motility: functional competition of collapsin-1 and vascular endothelial growth factor-165. J Cell Biol 146: 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migdal M, Huppertz B, Tessler S, Comforti A, Shibuya M, Reich R, Baumann H, Neufeld G (1998) Neuropilin-1 is a placenta growth factor-2 receptor. J Biol Chem 273: 22272–22278 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, AAVagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nakamura F, Tanaka M, Takahashi T, Kalb RG, Strittmatter SM (1998) Neuropilin-1 extracellular domains mediate semaphorin D/III-induced growth cone collapse. Neuron 21: 1093–1100 [DOI] [PubMed] [Google Scholar]
- Narazaki M, Tosato G (2006) Ligand-induced internalization selects use of common receptor neuropilin-1 by VEGF165 and semaphorin3A. Blood 107: 3892–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pan Q, Chanthery Y, Liang WC, Stawicki S, Mak J, Rathore N, Tong RK, Kowalski J, Yee SF, Pacheco G, Ross S, Cheng Z, Le Couter J, Plowman G, Peale F, Koch AW, Wu Y, Bagri A, Tessier-Lavigne M, Watts RJ (2007a) Blocking neuropilin-1 function has an additive effect with anti-VEGF to inhibit tumor growth. Cancer Cell 11: 53–67 [DOI] [PubMed] [Google Scholar]
- Pan Q, Chanthery Y, Wu Y, Rahtore N, Tong RK, Peale F, Bagri A, Tessier-Lavigne M, Koch AW, Watts RJ (2007b) Neuropilin-1 binds to VEGF121 and regulates endothelial cell migration and sprouting. J Biol Chem 282: 24049–24056 [DOI] [PubMed] [Google Scholar]
- Pratt KP, Shen BW, Takeshima K, Davie EW, Fujikawa K, Stoddard BL (1999) Structure of the C2 domain of human factor VIII at 1.5 A resolution. Nature 402: 439–442 [DOI] [PubMed] [Google Scholar]
- Renzi MJ, Feiner L, Koppel AM, Raper JA (1999) A dominant negative receptor for specific secreted semaphorins is generated by deleting an extracellular domain from neuropilin-1. J Neurosci 19: 7870–7880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero A, Romao MJ, Varela PF, Kolln I, Dias JM, Carvalho AL, Sanz L, Topfer-Petersen E, Calvete JJ (1997) The crystal structures of two spermadhesins reveal the CUB domain fold. Nat Struct Biol 4: 783–788 [DOI] [PubMed] [Google Scholar]
- Soker S, Miao HQ, Nomi M, Takashima S, Klagsbrun M (2002) VEGF165 mediates formation of complexes containing VEGFR-2 and neuropilin-1 that enhance VEGF165-receptor binding. J Cell Biochem 85: 357–368 [DOI] [PubMed] [Google Scholar]
- Soker S, Takashima S, Miao HQ, Neufeld G, Klagsbrun M (1998) Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 92: 735–745 [DOI] [PubMed] [Google Scholar]
- Takagi S, Hirata T, Agata K, Mochii M, Eguchi G, Fujisawa H (1991) The A5 antigen, a candidate for the neuronal recognition molecule, has homologies to complement components and coagulation factors. Neuron 7: 295–307 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Fournier A, Nakamura F, Wang LH, Murakami Y, Kalb RG, Fujisawa H, Strittmatter SM (1999) Plexin–neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 99: 59–69 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Nakamura F, Jin Z, Kalb RG, Strittmatter SM (1998) Semaphorins A and E act as antagonists of neuropilin-1 and agonists of neuropilin-2 receptors. Nat Neurosci 1: 487–493 [DOI] [PubMed] [Google Scholar]
- Takashima S, Kitakaze M, Asakura M, Asanuma H, Sanada S, Tashiro F, Niwa H, Miyazaki Ji J, Hirota S, Kitamura Y, Kitsukawa T, Fujisawa H, Klagsbrun M, Hori M (2002) Targeting of both mouse neuropilin-1 and neuropilin-2 genes severely impairs developmental yolk sac and embryonic angiogenesis. Proc Natl Acad Sci USA 99: 3657–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L, Artigiani S, Chen H, He Z, Ming GI, Song H, Chedotal A, Winberg ML, Goodman CS, Poo M, Tessier-Lavigne M, Comoglio PM (1999) Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 99: 71–80 [DOI] [PubMed] [Google Scholar]
- Vander Kooi CW, Jusino MA, Perman B, Neau DB, Bellamy HD, Leahy DJ (2007) Structural basis for ligand and heparin binding to neuropilin B domains. Proc Natl Acad Sci USA 104: 6152–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wronski MA, Raju N, Pillai R, Bogdan NJ, Marinelli ER, Nanjappan P, Ramalingam K, Arunachalam T, Eaton S, Linder KE, Yan F, Pochon S, Tweedle MF, Nunn AD (2006) Tuftsin binds neuropilin-1 through a sequence similar to that encoded by exon 8 of vascular endothelial growth factor. J Biol Chem 281: 5702–5710 [DOI] [PubMed] [Google Scholar]
- Wang L, Mukhopadhyay D, Xu X (2006) C terminus of RGS-GAIP-interacting protein conveys neuropilin-1-mediated signaling during angiogenesis. FASEB J 20: 1513–1515 [DOI] [PubMed] [Google Scholar]
- Wiesmann C, Fuh G, Christinger HW, Eigenbrot C, Wells JA, de Vos AM (1997) Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell 91: 695–704 [DOI] [PubMed] [Google Scholar]
- Yaron A, Huang PH, Cheng HJ, Tessier-Lavigne M (2005) Differential requirement for Plexin-A3 and -A4 in mediating responses of sensory and sympathetic neurons to distinct class 3 semaphorins. Neuron 45: 513–523 [DOI] [PubMed] [Google Scholar]
- Yuan L, Moyon D, Pardanaud L, Breant C, Karkkainen MJ, Alitalo K, Eichmann A (2002) Abnormal lymphatic vessel development in neuropilin 2 mutant mice. Development 129: 4797–4806 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1–S8