Abstract
The vaccinia virus mRNA capping enzyme is a multifunctional heterodimeric protein associated with the viral polymerase that both catalyses the three steps of mRNA capping and regulates gene transcription. The structure of a subcomplex comprising the C-terminal N7-methyl-transferase (MT) domain of the large D1 subunit, the stimulatory D12 subunit and bound S-adenosyl-homocysteine (AdoHcy) has been determined at 2.7 Å resolution and reveals several novel features of the poxvirus capping enzyme. The structure shows for the first time the critical role played by the proteolytically sensitive N-terminus of the MT domain in binding the methyl donor and in catalysis. In addition, the poxvirus enzyme has a completely unique mode of binding of the adenosine moiety of AdoHcy, a feature that could be exploited for design of specific anti-poxviral compounds. The structure of the poxvirus-specific D12 subunit suggests that it was originally an RNA cap 2′O-MT that has evolved to a catalytically inactive form that has been retained for D1 stabilisation and MT activity enhancement through an allosteric mechanism.
Keywords: methyl-transferase, mRNA capping enzyme, poxvirus, S-adenosyl homocysteine, X-ray crystallography
Introduction
The 5′ end of eukaryotic cellular and viral mRNAs (and more generally most RNA polymerase II transcripts) is usually modified by a N7-methyl-guanosine moiety linked to the first transcribed nucleotide (N) by a 5′-5′ triphosphate bridge, resulting in the structure m7GpppN (cap 0 structure). Generally, a second methylation occurs at the 2′-O position of the ribose of the first transcribed base (cap 1 structure) and in some cases additional methyl groups are added to the first and second nucleotides (cap 2 structure). The cap structure is essential for efficient splicing, export, translation and stability of the mRNA (Cougot et al, 2004). Capping is the first step in co-transcriptional pre-mRNA processing and in many eukaryotes the capping machinery is directly bound to the phosphorylated C-terminal domain of RNA polymerase II (Cho et al, 1997; Fabrega et al, 2003). Biosynthesis of the cap 0 structure requires three consecutive enzymatic activities: hydrolysis of the 5′ triphosphate end of the nascent transcript to a diphosphate by an RNA triphosphatase; capping of the diphosphate with GMP by an RNA guanylyl-transferase, which uses GTP as a substrate and GMP covalently linked to an active site lysine as an intermediate; and finally methylation of the 5′ guanine base at the N7 position by an RNA methyl-transferase (MT) (Shuman, 2001), with corresponding conversion of the methyl donor S-adenosyl-methionine (AdoMet) to S-adenosyl-homocysteine (AdoHcy). The three-component mRNA capping apparatus is functionally conserved in all eukaryotic organisms thus far examined. However, the subunit organisation and even the folds of the catalytic domains vary considerably. For example, in yeast, the three enzymatic activities are on separate polypeptides, whereas in metazoans and plants there are only two genes, encoding respectively the bifunctional triphosphatase/guanylyl-transferase and the MT. Several DNA and RNA viruses encode their own capping enzymes, often on a single multifunctional polypeptide (Shuman, 2002).
Poxviruses are complex, enveloped DNA viruses that infect vertebrates (chordopoxviridae) or invertebrates (entomopoxviridae) and which encode about 250 proteins. Vaccinia virus, used for vaccination against the similar variola virus (the causal agent of smallpox), is most likely derived from cowpox virus. Like all poxviruses, vaccinia virus replicates in the cytoplasm, where it does not have access to the nuclear mRNA maturation machinery. Instead viral transcripts acquire the 5′ cap and 3′ poly-A tail through the action of virally encoded enzymes. The vaccinia virus capping machinery is composed of two subunits, D1 (844 residues, 97 kDa) and D12 (287 residues, 33 kDa) (Shuman et al, 1980). The RNA triphosphatase and guanylyl-transferase activities are located in the N-terminal domain of D1 (Niles and Christen, 1993; Myette and Niles, 1996), whereas the guanine N7-MT active site is in the C-terminal region of D1 (Cong and Shuman, 1992). However, the MT domain requires association with the D12 subunit for full activity, having an intrinsic activity 40–50 times less than that of the D1–D12 heterodimer (Higman et al, 1992; Mao and Shuman, 1994). The D12 subunit shows no sequence similarity with any other known protein except to homologues present in other poxviruses. The D1–D12 heterodimer also plays a role in transcription initiation of intermediate genes (Vos et al, 1991) as well as transcription termination of early genes of vaccinia virus (Shuman and Moss, 1988). Vaccinia viral mRNAs acquire the cap 1 structure by 2′-O-methylation of the first transcribed nucleotide by the VP39 protein, which is also a processivity subunit for the viral poly-A polymerase (VP55) (Schnierle et al, 1992).
Here, we report the crystal structure of the heterodimer of the N7-MT domain of vaccinia virus D1 with bound AdoHcy in complex with its stimulatory D12 subunit. The catalytic domain possesses a class I AdoMet-dependent MT fold (Martin and McMillan, 2002), similar to that of the monomeric N7-MT of the microsporidian parasite Encephalitozoon cuniculi (denoted Ecm1) (Fabrega et al, 2004) with both structures exhibiting a deep cleft containing the AdoMet/AdoHcy-binding site. However, in Ecm1, electron density for the N-terminal 40 residues of the enzyme is absent and as residues 30–40 are essential for activity (Fabrega et al, 2004), critical structural aspects of the enzyme mechanism are likely to have been missed. In comparison, the vaccinia structure shows the catalytically important N-terminal extension to the MT domain folded back over the AdoHcy-binding site, burying the ligand, in a remarkably similar conformation to that previously observed in some small molecule MTs. Other unexpected features of the D1CTD-D12-AdoHcy ternary complex are a unique conformation for the adenosine moiety of the bound AdoHcy and a structural similarity between the D12 stimulatory subunit and cap 2′O-MTs, perhaps indicative of the evolutionary origin of this subunit.
Results
Structure determination and general features of the D1CTD-D12 MT subcomplex
Taking into account the previous attempts to optimise the N-terminus of the MT transferase domain for expression, solubility and activity (Higman et al, 1994; Mao and Shuman, 1994; Saha and Shuman, 2001), we coexpressed, purified and crystallised a catalytically active subcomplex containing residues 545–844 of D1 (denoted D1CTD) and the full-length D12 subunit. Crystals were obtained only in the presence of the ligands AdoMet or AdoHcy and m7GDP although electron density was only observed for AdoHcy. Three different crystal forms, one of native heterodimer (space-group P1, diffraction to 2.7 Å resolution) and two grown in the presence of trace amounts of trypsin (space-groups C2 and P6522, diffracting to, respectively, 2.8 and 3.1 Å resolution) were used to determine the structure (see Materials and methods and Table I). Briefly, the structure was solved in the hexagonal form using seleno-methionine (SeMet)-substituted protein and an initial model built that was improved using molecular replacement into the monoclinic crystal form. Finally, a full model was obtained in the untrypsinated triclinic form, which allowed completion of some loops previously ill-defined, notably the N-terminal residues 545–564 of D1CTD (Supplementary Figure S1). The final R-factor (Rfree) is 22.3 (26.5). The four independent copies of the subcomplex in the P1 asymmetric unit are very similar with one heterodimer showing better order of the N-terminal peptide (D1 chain C and D12 chain D).
Table 1.
X-ray data collection and refinement statistics
| D1ctd+D12+AdoHcy | D1ctd+D12+AdoHcy | D1ctd+D12+AdoHcy SeMet-labelled | |
|---|---|---|---|
| Data collection | |||
| Beamline | ID29 | ID29 | ID14-EH4 |
| Wavelength | 0.97626 | 0.97626 | 0.97932 |
| Space group | P1 | C2 | P6522 |
| Cell dimensions | |||
| a, b, c (Å) | 60.9, 61.0, 225.8 | 158.8, 78.7, 61.1 | 137.0, 137.0, 178.3 |
| α, β, γ (deg) | 94.2, 92.9, 108.3 | 90.0, 93.1, 90.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 48.4–2.7 (2.8–2.7)a | 30–2.8 (2.85–2.8)a | 40–3.1 (3.2–3.1)a |
| Rsym | 0.090 (0.486)a | 0.078 (0.406)a | 0.072 (0.560)a |
| I/σI | 7.31 (1.94)a | 9.14 (2.24)a | 10.08 (2.14)a |
| Completeness (%) | 89.7 (79.6)a | 98.1 (98.9)a | 97.3 (97.4)a |
| Redundancy | 1.87 (1.83)a | 2.04 (2.04)a | 2.46 (2.48)a |
| Refinement | |||
| Resolution (Å) | 48.4–2.70 (2.70–2.77)a | ||
| No. of reflections in work set (free set) | 68 062 (3818) | ||
| Rwork | 22.3 (32.9)a | ||
| Rfree | 26.5 (39.2)a | ||
| Total non-hydrogen atoms | 18 796b | ||
| Ligand (AdoHcy) | 4 × 26 | ||
| Solvent molecules | 11 sulphate | ||
| Average B-factor (Å2) | 40.3 | ||
| Wilson B-factorc | 43.4 | ||
| RMSD | |||
| Bond lengths (Å) | 0.011 | ||
| Bond angles (deg) | 1.258 | ||
| aHighest resolution shell is shown in parentheses. | |||
| bFour complexes in the asymmetric unit. | |||
| cFrom XDS output. | |||
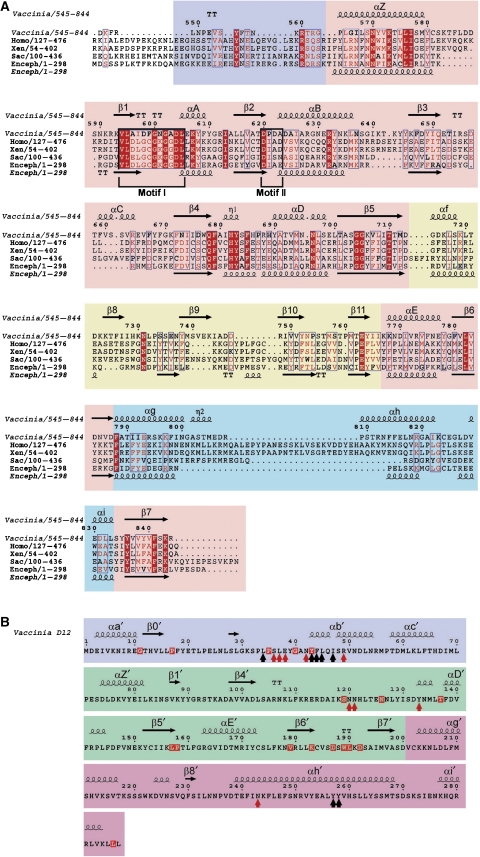
The structure of D1CTD (Figure 1) exhibits the characteristic core fold of the class I MT family comprising a seven-stranded β-sheet with three helices each side (Martin and McMillan, 2002). A search of the PDB using DALI (Holm and Sander, 1993) confirmed a high structural similarity of the D1CTD subunit with Ecm1 (PDB entry 1RI1)(Fabrega et al, 2004) with a 2.3 Å root-mean square deviation (RMSD) for 241 aligned Cα positions (19% sequence identity, Dali Z-score: 26.6). Nevertheless, some differences between the two structures are evident, such as the presence of extra helices (αC, η2 and αh), β-strand (β8) or larger loops (mainly involved in D1–D12 complex formation). A major difference is that the cleft between the core-fold and the smaller domain comprising strands β6–β9 and helix α6 is filled by the N-terminal peptide (residues 545–562), which is notably absent in Ecm1 structure (Figures 2A and 3). Amongst the next highest similarity hits were small molecule MTs, such as glycine N-MT (GNMT) (PDB entry 1NBH, Z=17.1) and phenylethanolamine N-MT (Z=16.6) and the cap RNA MT-2 from the reovirus λ2 turret complex (PDB entry 1EJ6, Z=14.5) (Reinisch et al, 2000).
Figure 1.
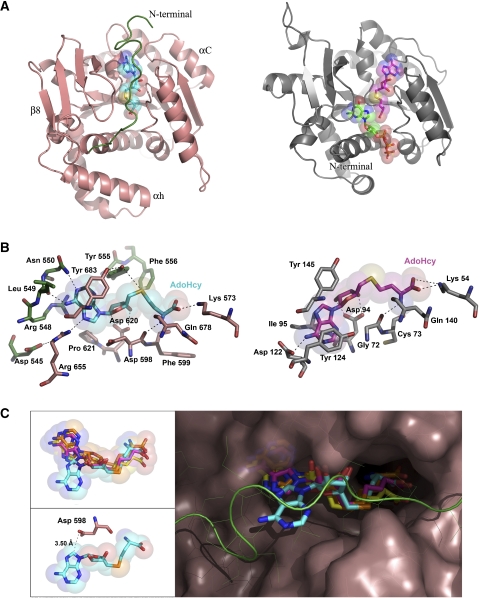
Structure of the vaccinia virus N7-MT subcomplex. (A) Two orthogonal views of the overall structure of the D1CTD(pink)-D12(green) subcomplex in ribbon representation. AdoHcy is shown in cyan sticks. Oxygen, nitrogen and sulphur atoms are in red, blue and orange, respectively. (B) Surface and ribbon representation of the D1CTD–D12 subcomplex coloured by domains as in Figure 1A. AdoHcy is shown in cyan sticks. (C) Schematic diagram of the topology of the D1CTD and D12 subunit coloured by domains as in (B). Some of the regions involved in complex formation are shown by dotted lines.
Figure 2.
Structure based sequence alignments of D1CTD and D12. (A) Sequence alignment of representative mRNA cap guanine N7 MTs from vaccinia virus (D1CTD, residues 545–844), Homo sapiens (Hcm1, residues 127–476), Xenopus laevis (residues 54–402), Saccharomyces cerevisiae (Abd1, residues 100–436) and E. cuniculi (Ecm1). Residues 100% conserved are in solid red boxes and those with homology of 70% or higher are depicted in red type. The secondary structure of vaccinia virus D1CTD is shown above and that of Ecm1 below the alignment (α, α-helices, η, 310-helices, β, β-strands). The class I MT core of D1CTD is highlighted with a pink background and conserved motifs I and II are boxed, whereas extra elements such as the N-terminal peptide, the internal α/β domain or the C-terminal domain are shown with violet, yellow and cyan backgrounds, respectively. (B) Sequence and secondary structure of vaccinia virus D12. Red and black arrowheads indicate positions involved in the major polar and nonpolar interactions with D1CTD, respectively. The truncated MT core of D12 is highlighted with a green background, whereas the N- and the C-terminal domains are shown with violet and magenta backgrounds, respectively. Residues conserved in all known D12 homologues are in red boxes (Supplementary Figure S3).
Figure 3.
Comparison of vaccinia virus cap MT structure and mode of AdoHcy binding with that of E. cuniculi. (A) Comparison of the global fold of D1CTD from vaccinia virus (left, pink ribbons) and Ecm1 from E. cuniculi (right, grey ribbons, PDB entry 1RI1) cap MTs. Extra elements present in D1CTD are labelled. The N-terminal peptide of D1CTD is highlighted in green and AdoHcy in cyan. In Ecm1, AdoHcy and GTP are shown in magenta and green sticks, respectively. (B) The AdoHcy-binding sites in D1CTD (left) and Ecm1 (right). The AdoHcy molecules are depicted in cyan for D1CTD and magenta for Ecm1. Residues of D1CTD involved in ligand binding are in pink, except those from the N-terminal peptide, which are in green. Residues of Ecm1 are in grey. Likely hydrogen bonds and salt bridges are represented as dashed lines. (C) Comparison of the AdoMet/AdoHcy conformations after superimposing the ligand-bound structures of the glycine N-MT from rat (yellow, PDB entry 1NBH), VP39 from vaccinia (orange, PDB entry 1VP3), Ecm1 (magenta) and D1CTD (cyan). The D1CTD protein surface is shown in pink with the exception of the N-terminal peptide, which is shown in green. The upper-left inset shows the same view for just the ligands, whereas the lower-left inset shows the AdoHcy molecule and residue Asp598 in D1CTD.
The topology of the stimulatory subunit D12 unexpectedly also reveals a class I MT like core, however, with a truncated AdoMet-binding domain (Figure 1C and 2B), consistent with its lack of MT activity (Mao and Shuman, 1994). Thus, strand β1′ connects directly with β4′, and strands β2′ and β3′ as well as helices A′ to C′ are absent. Using DALI or Dali-lite, we found that the two closest structural homologues of D12 are the cap 2′-O-MT (MT-1) of the reovirus λ2 subunit (RMSD 3.2 Å over 165 residues, Z=8.2) and the cap 2′-O-MT domain of the flavivirus RNA polymerase (PDB entry 1L9K, RMSD 3.3 Å over 152 residues, Z=7.2) (Egloff et al, 2002) as shown in Supplementary Figure S2A. Third is spermidine synthetase and fourth the vaccinia virus cap 2′-O-MT VP39 (PDB entry 3MAG, Z=3.8). Although D1CTD and D12 are structurally homologous to, respectively, MT-2 and MT-1 of reovirus λ2, their relative orientation is not the same (Supplementary Figure S2B). Implications of the unexpected structural homology of D12 with cap 2′-O-MTs are discussed below.
A unique mode of AdoHcy binding to the vaccinia virus D1 N-MT
Crystals of the D1CTD–D12 subcomplex only appeared in the presence of AdoMet or AdoHcy and the structure solved contains AdoHcy. The ligand is located at the C-termini of strands β1 and β2 and tightly bound in the general configuration described for most AdoMet-dependent MTs (Martin and McMillan, 2002). The most conserved residues in the AdoMet/AdoHcy-binding pocket are the glycine-rich loop between β1 and α2 (the so-called Motif I: Gln/Asp-X-Gly-X-Gly-X-Gly), which mainly interacts with the amino-acid portion of the methyl donor, and the acidic loop between β2 and αB (Motif II), which interacts with the ribose hydroxyls (Martin and McMillan, 2002; Figure 2A). In D1CTD, the adenosine moiety is stacked between Tyr-683 (equivalent in sequence but not functionally to Tyr-145 in Ecm1 from E. cuniculi) and Arg-548; in Ecm1, the base is stacked between Tyr-124 (for which there is no equivalent in D1CTD) and Ile-95 (Figure 3B). Polar contacts also exist between the adenine N6 and the carbonyl group of Leu-549, N7 with side chain of Arg-655 (which in turn is stabilised by the side-chain of Asp-545) and N1 with side-chain of Asn-550. The 2′- and 3′-OH of the adenosine ribose, are coordinated by the carboxylate of Asp-620 (Asp-94 in Ecm1) and the hydrophobic face of the ribose packs against the aromatic ring of Tyr-555. The amino group of the homocysteine moiety makes two hydrogen bonds to the main-chain carbonyl oxygens of Gln-678 and Asp-598 (Gln-140 and Gly-72 in Ecm1) and the carboxylate group interacts with Lys-573 (Lys-54) from helix α1. Thus, compared to the E. cuniculi cap-MT, the vaccinia virus enzyme only shares a few conserved AdoHcy-binding residues and differs notably in the involvement of several residues from the N-terminal peptide (545–550, 555), resulting in the almost complete burial of the ligand (Figure 3).
These interactions stabilise a highly atypical, indeed unique, syn-conformation of the adenine base, not previously seen in AdoMet-dependent MTs (Figure 3C). A key correlate for this conformation is the occurrence of the sequence 596-Ala-Ile-Asp-Phe-Gly-Asn-Gly in the Motif I of all poxvirus cap MTs, where Asp-598 is substituted for the normally highly conserved glycine at this position. The side chain of Asp-598 is close to the base (the OD1 of Asp-598 is 3.5 Å from the adenine C8) and sterically incompatible with the adenosine adopting the usual anti-conformation. Importantly, the rarer syn-conformation of the adenine is stabilised by stacking interactions, notably with Tyr-683, and several hydrogen bonds to the base by residues from the N-terminal peptide (Figure 3B). Indeed, the presence of Asp-598 is apparently not essential, as an Asp-598 to Ala mutation in D1(498–844)–D12 only reduces MT activity by a factor of two (Mao and Shuman, 1996). Whereas Asp-598 is conserved in the capping enzyme of all poxviruses so far sequenced, this position is the usual glycine in the related large DNA virus such as African swine fever virus (ASFV) and mimivirus (Supplementary Figure S3), which may thus have the more usual anti-conformation of the adenine base.
The role of the N-terminal extension in mRNA cap MT activity
Observations on several other MTs, for instance the thiopurine MT (Scheuermann et al, 2004) and GNMT (Konishi and Fujioka, 1988; Takata et al, 2003; Pakhomova et al, 2004) have shown firstly, that binding of AdoMet can induce protein conformational changes protecting the enzyme against proteolytic degradation and loss of activity and secondly, that a protease sensitive N-terminal peptide can be functionally important for enzyme activity. Similar observations have been made on RNA cap MTs, all of which have an N-terminal extension to the core MT fold (Figure 2; Supplementary Figure S4), which tolerates only limited truncations before activity is severely affected. For Ecm1, N-terminal deletions less than 30 residues do not affect the enzyme's ability to complement a yeast cap MT deletion strain, whereas a deletion of 30 residues led to reduced activity and the −40 deletion was unable to complement (Fabrega et al, 2004). Thus, residues 30–40 are essential for activity but not visualised in the crystal structure. For the vaccinia virus cap MT, similar complementation experiments have shown that whereas D1 residues before 540 are dispensable, constructs starting at 560 or 580 are lethal (Schwer and Shuman, 2006). Thus, residues between 540 and 560 are required for activity, consistent with our observations on the mode of AdoHcy binding. Interestingly, this region also corresponds to the protease-sensitive region of D1 separating the N-terminal triphosphatase and guanylyl-transferase activities from the MT activity (Shuman, 1989).
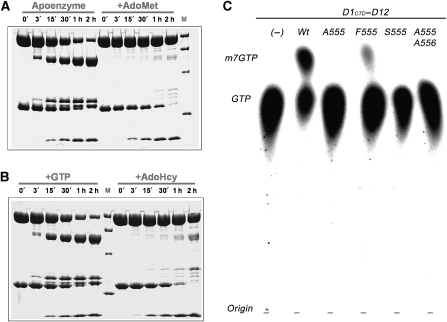
These results prompted us to re-investigate the role of the protease-sensitive linker in the D1 subunit of vaccinia virus capping enzyme. We performed limited proteolysis experiments with the complete capping complex in the absence or presence of AdoMet, AdoHcy or GTP ligands. In the absence of ligands, trypsin digestion showed, as expected (Shuman, 1989), that D1 is degraded into two major fragments corresponding to the N- and C-terminal domains of, respectively, 60 and 35 kDa (Figure 4A). The D12 subunit is also degraded into two principal fragments of 20 and 10 kDa (C- and N-terminus, respectively). However, when the enzyme is pre-incubated with 3 mM of AdoMet or AdoHcy, the D1 subunit, but not D12, was protected from trypsination, whereas 3 mM GTP alone did not protect D1 (Figure 4A and B). These observations suggest that the proposed flexible linker between the N- and C-terminal domains of D1 undergoes a conformational change upon binding AdoMet or AdoHcy, protecting it from proteolysis. This is consistent with our structure which shows that residues 545–560 are intimately involved in AdoHcy binding (closed conformation), but are presumably loosely bound or unbound in the absence of ligand (open conformation), making this region susceptible to proteolysis.
Figure 4.
Functional importance of the N-terminal peptide of D1CTD. (A) Time course of limited proteolysis in the presence or absence of ligands. Full-length vaccinia virus capping enzyme was incubated at room temperature with trypsin (1/500 w/w) in the absence (left) and presence (right) of 3 mM S-adenosyl-methionine (AdoMet). Aliquots were taken every 0, 3, 15, 30, 60 and 120 min, denatured in denaturing buffer (125 mM Tris pH 6.8, 260 mM DTT, 30% glycerol, 10% SDS and 0.025% Coomassie Blue) and loaded onto a 12.5% SDS polyacrylamide gel. Protein marker (M) is the Low Range marker (BioRad) corresponding to 97, 66, 45 and 31 kDa. (B) As in panel (A), limited proteolysis experiments were performed in the presence of 3 mM GTP (left) and 3 mM AdoHcy (right). (C) Methyl-transfer assays using the enzymes corresponding to wild-type D1CTD–D12, single mutants of Tyr 555 to Ala (A555), Phe (F555) or Ser (S555) and double mutant Tyr 555 and Phe 556 to alanines (A555/A556). Substrates used were AdoMet and [α-32P]GTP, which were incubated with the corresponding enzymes at 37oC for 60 min (see Materials and methods).
Mechanism of methyl-transfer
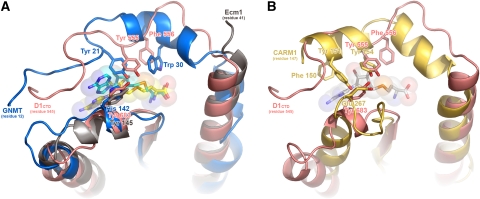
In the light of the previous discussion, the alignment of the structure of the rat liver GNMT (Takata et al, 2003) with D1CTD, both with bound AdoMet or AdoHcy, is particularly instructive (Figure 5A). The N-terminal extension of GNMT follows the same path as that of D1CTD, largely burying the ligand and making numerous interactions with both sides of the cleft. Strikingly, the side chains of Tyr-21 and Trp-30 in GNMT closely overlap with, respectively, that of Tyr-555 and Phe-556 in D1CTD, with the hydroxyl of Tyr-555 at 4 Å from the sulphur atom (SD) of AdoHcy (Figure 5A). Tyr-21 has been implicated in catalysis by GNMT with the proposed role of forming a charge–dipole interaction with the positively charged SD of the AdoMet thus favouring methyl-transfer (Takata et al, 2003). A strikingly similar situation has been observed in recent structures of another N-MT, the mammalian protein arginine MT (PRMT) CARM1 (Troffer-Charlier et al, 2007; Yue et al, 2007). In this case, the region 144–164 folds over bound AdoHcy, with Tyr-154, part of a conserved YFxxF motif in PRMTs, occupying exactly the same position as Tyr-555 in vaccinia D1CTD (Figure 5B).
Figure 5.
Conformation of the N-terminal peptide of D1CTD compared to two other N-methyltransferases. (A) Superimposed N-terminal regions of D1CTD from vaccinia virus (pink ribbons and AdoHcy ligand in cyan sticks), glycine N-methyltransferase (blue ribbons and AdoHcy ligand in yellow sticks, PDB entry 1NBH) and Ecm1 from E. cuniculi (grey ribbons, AdoHcy not shown, PDB entry 1RI1). Note the close correspondence between Tyr-555 and Phe-556 of D1CTD with Tyr-21 and Trp-30 of GNMT. (B) Superimposed N-terminal regions of D1CTD (pink ribbons, AdoHcy not shown) and the MT domain of CARM1 (gold ribbons and AdoHcy ligand in white sticks, PDB entry 2V74). As for D1CTD and GNMT (A), there is a similar close correspondence between Tyr-555 of D1CTD and conserved Tyr-154 of CARM1. Another conserved residue of PRMTs essential for activity, Glu-267, superimposes with Tyr683 of D1CTD.
To test whether Tyr-555 and Phe-556 play a significant role in vaccinia cap MT, we mutated both positions to alanine residues (Ala-555/Ala-556) or just Tyr-555 to alanine (Ala-555), serine (Ser-555) or phenylalanine (Phe-555) residues. The mutated subcomplexes were purified and assayed for MT activity, using GTP as methyl acceptor. For the Ala-555/Ala-556, Ala-555 and Ser-555 mutants, in vitro methylase activity was abolished when compared with the wild-type construct under the same conditions, whereas the more conservative Phe-555 change still showed methylase activity, but at a reduced level (Figures 4C; Supplementary Figure S5).
The inactive mutant enzymes were further biochemically characterised to ascertain whether the mutations affected substrate binding or the catalytic step. The Ala-555 and Ala-555/Ala-556 mutations were introduced into the full-length capping enzyme and purified mutant D1–D12 complexes subjected to trypsin digestion in the presence or absence of AdoMet. Degradation of the mutant D1 subunits was inhibited by the presence of the methyl-donor in a qualitatively similar fashion to that previously found for the wild-type enzyme, although protection was less effective, particularly for the double mutation (Supplementary Figure S6). In addition, direct binding of AdoHcy to wild-type and mutant MT subcomplexes was assayed by isothermal titration calorimetry experiments. These measurements clearly show AdoHcy binding to the Ala-555 and Ala-555/Ala-556 mutant subcomplexes with comparable binding constants to the wild type (data not shown). This is consistent with the fact that in some of our crystal structures, bound AdoHcy is observed even though the N-terminal extension is poorly ordered. Taken together, these results show that mutations at positions 555–556 do not significantly affect methyl-donor binding, nor the ability of the enzyme to undergo an AdoMet-induced conformational change. The inactivity of the Ser-555, Ala-555 and Ala-555/Ala-556 mutants is thus most likely due to a defect in the catalytic step of the reaction. Although we have not measured precise kinetic parameters, the slightly reduced catalytic efficiency of the Phe-555 mutation without affecting AdoMet binding, is similar to that observed upon mutation of structurally equivalent Tyr-21 to Phe in the case of GNMT (Takata et al, 2003). The roles of the two tyrosines thus seem to be analogous. However, our additional results showing that mutation to Ser or Ala totally inactivates the enzyme (such mutations were not performed for GNMT) suggest that the major function of this residue is probably optimal positioning of the methyl-donor through aromatic interactions (stacking against the ribose) rather than the charge–dipole interaction the hydroxyl group of the tyrosine with the SD of AdoMet. We note that in the AdoHcy bound vaccinia enzyme, the hydroxyl of Tyr-555 forms hydrogen bond with that of Tyr-683 (Figure 6). This interaction may stabilise the binding of the N-terminal extension to the catalytic site and the precise positioning of the methyl-donor; however, it has been shown by mutation of Tyr-683 to Phe, Ala or Ser (Mao and Shuman, 1996) that, again, it is actually the aromatic nature of this residue that is essential for function (stacking against the adenine of AdoMet) rather than the hydroxyl group.
Figure 6.
Schematic diagram of the active site of vaccinia D1CTD with putative interactions with AdoMet and cap (GTP) during the methyl-transfer reaction. Residues labelled with * correspond to those modelled on the basis of the corresponding Ecm1 structure (PDB entry 1RI3). Dashed lines indicate potential hydrogen bonds. Residues labelled in green correspond to those located in the N-terminal peptide (compare Figure 8 of Schwer et al, 2006).
We have unfortunately been unsuccessful in visualising the substrate or product of the methylation reaction bound to the enzyme despite extensive soaking/cocrystallisation trials with GTP, m7-GDP or m7GpppG. This is possibly due to the use of sulphate ions in our crystallisation conditions, which compete with phosphate-binding sites. However, superimposition of the vaccinia enzyme on Ecm1 (with bound cap analogue and AdoHcy) suggests a very similar mode of substrate binding due to the conservation of most of the residues that contact the guanosine. These include Asn-570 in D1 (Asn-51 in Ecm1), Phe-679 (Phe-141), His-682 (His-144), Glu-763 (Glu-225) and Tyr-836 (Tyr-284) (Figure 6). The triphosphate of a modelled GTP, which bridges to the first transcribed nucleotide, is probably directly contacted by Lys-607 and Arg-632 (conserved as Lys-81 and Arg-106 in Ecm1) as well as Arg-560 and Arg-562 (equivalent basic residues in this region are not visible in the in Ecm1 structure), with electron density for a putative sulphate ions being observed in the corresponding position.
The D1–D12 interface
D12 has no significant sequence homology with any other known protein apart from the corresponding homologues in the poxviridae family. Even viruses distantly related to poxvirus-encoding homologues of the D1 subunit, such as ASFV and mimivirus, lack a recognisable D12 subunit. D12 is essential for vaccinia replication (Carpenter and DeLange, 1991) but lacks any MT activity of its own (Mao and Shuman, 1994) and instead stimulates the weak intrinsic MT activity of D1 by 30–50 fold (Higman et al, 1994; Mao and Shuman, 1994; Saha and Shuman, 2001). Early cross-linking studies showed that D12 does not affect the extent of substrate binding to the catalytic subunit (Higman and Niles, 1994), suggesting that there was no direct interaction of D12 with the active site. Recent studies propose that D12 has an allosteric role with heterodimer formation leading to an increased affinity for both ligands, especially the substrate, and a modest increase in the turnover number (Schwer et al, 2006). An additional role of D12 is to structurally stabilise D1 leading to increased thermal stability of the heterodimer compared to the catalytic domain alone (Schwer et al, 2006).
Our structure enables both these roles of D12 to be rationalised. Firstly, it shows that the D1–D12 subunit interface is on the opposite side to the ligand-binding pockets of D1 (Figure 1), confirming there is no direct role for D12 in substrate binding and catalysis. The closest approach of D12 to the site of methylation is 21 Å by the side chain of Phe-44. The extensive interface between D1 and D12 is typical of a high-affinity protein–protein interaction, with about 3000 Å2 of buried surface area with both polar (about 35%) and non-polar/hydrophobic (about 65%) character (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html). From D12, significant contacts with D1 involve residues from the N-terminal 31–54, central 119–130 and C-terminal regions 239–265 consistent with the fact that N- (by 60 residues) and C- (by 48 residues) terminally truncated D12 no longer heterodimerises (Cong and Shuman, 1992; Saha and Shuman, 2001). In D1, the main contact regions are residues 571–589 and 783–815. There are salt bridges between Asp-587 and Glu-806 from D1 with, respectively, Lys-31 and Arg-49 from D12. Intermolecular hydrogen bonds exist between Asn-42 from D12 with Ser-795 and Tyr-571 from D1, Glu-38 with Asp-784 and Asn-785, and finally Glu-38 and Asn-243 with Arg-794. Previously, on the basis of the biochemical characterisation or in vivo yeast complementation assays of dialanine substitutions in conserved regions of D12, it has been proposed that the residues 16–53 (notably 42-NY) and the motifs 156-KLPT, 176-FK and 189-WLKDS might comprise part of the docking site for the D1 subunit (Saha and Shuman, 2001; Saha et al, 2003; Schwer and Shuman, 2006). As discussed below, the N-terminal residues 37–47 are indeed critically involved at the interface and functionally significant for activity. On the other hand, none of the other motifs are close to the interface, but involved in maintaining the fold of D12. For instance, Lys-156 makes strong intramolecular salt bridges with both Asp-72 and Asp-74, and Phe-176 is buried in the hydrophobic core. The extended motif 183-KCVSDSWLKDS is particularly interesting as the five bold-typed residues are absolutely conserved in all known D12 homologues, there being only 17/289 such fully conserved residues (Figures 2B; Supplementary Figure S3). This motif forms a highly structured turn between strands β6 and β7, which through a network of further interactions links together several distant segments of the polypeptide chain and thus is plausibly important for maintaining, if not nucleating, tertiary folding. Within the motif, Trp-189 stacks on Lys-183, which makes a salt bridge to Asp-187 as well as hydrogen bonding to the main-chain oxygens of Ser-188 and Ser-193. Amongst the integrating interactions emanating from the motif, Lys-191 makes a salt bridge with Asp-72 as well as interacting with the carbonyl oxygens of Ala-105 and Lys-108; Asp-192 carboxylate makes polar interactions with Tyr-253, Thr-159, Leu-160 and Ser-131 and the main chain oxygen of Leu-190 hydrogen bonds with the side chain of His-260.
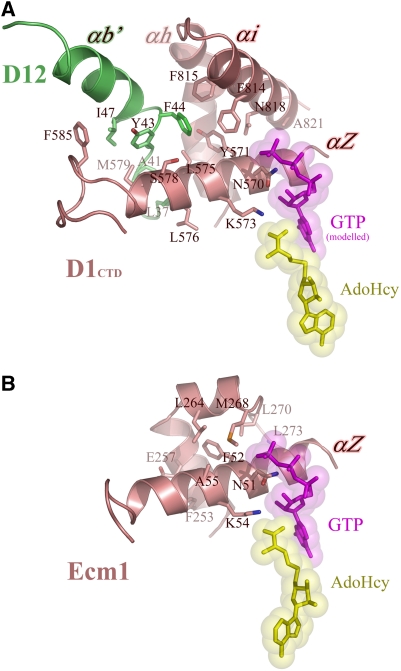
Structural basis for allosteric effect of D12 on MT activity
The proposed allosteric effect of D12 on substrate binding is based on Km values obtained for GTP and GpppA binding (in the presence of 50 μM AdoMet) and for AdoMet binding (in the presence of 5 mM GTP) measured for the MT catalytic domain alone compared to the heterodimer with D12 (Schwer et al, 2006). The results show for GTP, GpppA and AdoMet an apparent increase in affinity by a factor of 14, 10 or 6, respectively. For GTP methylation, the kcat increases by a factor of 4. As a structural explanation for this effect, we propose that there is indirect stabilisation of the ligand-binding sites by D12 via intersubunit contacts with the N-terminal helix αZ of D1 (residues 563–581). This helix carries two critical and conserved ligand-binding residues, one Asn-570, which interacts with the ribose of the GTP (based on the model derived from the Ecm1 structure, see above) and another Lys-573, which interacts with the carboxylate of AdoMet/AdoHcy (Figure 7A). In the monomeric Ecm1, the helix αZ (residues 44–61) is braced by two short helices from the region 250–270 with notably Phe-253, Tyr-254, Glu-257, Met-268, Leu-264 and Leu-270 making contact. In the vaccinia virus enzyme, the same function is shared between the equivalent region of D1 (residues 788–799 and 815–821), packing against the N-terminal part of helix αZ (residues 564–572) and the N-terminal region of D12 between residues 37–47, which interacts with the C-terminal part of helix αZ (residues 575–585). It is noteworthy that the side chain of Phe-44 of D12 nearly superimposes with that of Leu-264 of Ecm1, both being just the other side of helix αZ from Lys-573 (or Lys-54 in Ecm1), which interacts with the carboxylate of AdoMet/AdoHcy (Figure 7B). As modelling suggests that the side chains of Asn-570 and Lys573 could simultaneously hydrogen bond to each other as well as to their respective substrate and methyl donor ligands, stabilisation of helix αZ by D12 is likely to enhance affinity for both ligands as observed (Schwer et al, 2006). Also in agreement with our structure is the fact that the D12 dipeptide 42-NY to alanine substitution was highlighted in previous experiments as being defective in MT stimulation independently of hetero-dimerisation, with the conclusion that Tyr-43, which is one of the few absolutely conserved D12 residues, was a component of the activation domain (Saha and Shuman, 2001). Finally, in yeast and metazoan cap MTs sequence alignments (Figure 2A) show that there are significant insertions in the C-terminal helical region that packs against helix αZ, compared to Ecm1, which could play a similar stabilising role as D12 in vaccinia virus.
Figure 7.
Proposed model for the allosteric effect of the D12 subunit on MT activity. (A) Structural stabilisation of helix αZ of D1CTD through interactions with residues from αb′ of D12 subunit (region L37–I47) and αi from D1CTD (region F815–A821). Key residues involved in the stabilisation are labelled, as well as the two residues involved in interactions with the substrates (N570 with GTP, in magenta, and K573 with AdoHcy, in yellow). (B) Equivalent view for the same region in Ecm1, where the helix αZ is stabilised through residues located in region F253–L273. Comparison of both panels highlights the spatial equivalence of residues F253 and L264 in Ecm1 with L37 and F44 in D12, respectively.
Using a yeast complementation system, mutations in D1 that bypass the requirement for the stimulatory D12 subunit (Schwer et al, 2006) or suppress D12 mutations, have been selected (Schwer and Shuman, 2006). In vitro studies showed that the single mutations Tyr-752 to serine or alanine and Asn-753 to isoleucine, restore methylation activity of the catalytic domain alone to comparable or better levels than for the wild-type heterodimer, but do not correspondingly reduce much the Km for AdoMet, GTP or GpppA substrates. The mutant proteins are however still susceptible to enhancement of activity by hetero-dimerisation with D12, which by significantly decreasing Km values, leads to a gain-of-function over wild type by a factor of 10 in kcat for example for the N753I/D12 heterodimer. These results suggest that the mechanism of rate enhancement by the bypass mutants is distinct and additive to that of D12. Our structure shows that 752-YN aligns with 215-TL of Ecm1, not 214-FT as presumed previously (Schwer et al, 2006). These two residues are part of the 752–760 loop between strands β8 and β9 (Figure 1A), which forms one side of the guanine substrate-binding pocket and is not in contact with D12. Asn-753 is pointing into the pocket and changing it to isoleucine would make it functionally equivalent to Leu-216, with which it structurally aligns, as in Ecm1 this hydrophobic residues stacks over the base (on the opposite side to Phe-141). Tyr-752 points away from the pocket with its side chain stacking between prolines 754 and 760, thus, presumably stabilising the conformation of this loop region. It was found that the gain-of-function is only sustained by a reduction in the size of the side chain of position 752, notably to alanine, serine or cysteine (but not phenylalanine) (Schwer and Shuman, 2006). We hypothesise that the absence of an aromatic residue capable of stacking would destabilise the loop conformation, favouring a conformation closer to that in the transition state for cap methylation. This further destabilisation might explain the high temperature sensitivity of the mutant catalytic domains compared to wild-type, which, however, could be overcome again by hetero-dimerisation with D12 (Schwer et al, 2006).
Discussion
The vaccinia virus capping enzyme has been studied for more than 30 years yielding a vast amount of biochemical data and giving rise to important insights into the capping process and its coupling to transcription both in vaccinia virus and more generally all eukaryotic systems.
Our crystal structure of the MT subcomplex not only enables many of the previous results to be understood but also reveals completely unique and unexpected features of the vaccinia virus capping enzyme. Superficially, the structure of the vaccinia virus cap MT is rather similar to the previously determined structure of the equivalent enzyme from E. cuniculi and more generally, to all SAM-dependent MTs. However, our structure reveals the role of the critical N-terminal peptide in both contributing to the buried binding site of the methyl-donor AdoMet and providing essential residues for catalysis, notably Tyr-555, which packs against the ribose of AdoHcy and whose hydroxyl group is very close to the site of methylation. In fact, our mutagenesis experiments show that Tyr-555 is required for optimal catalysis but mainly due to its aromatic interactions, which help correctly orientate the methyl-donor, rather than any role of the hydroxyl group, as Phe-555 is still significantly active, whereas Ser-555 and Ala-555 are inactive.
As the N-terminal extension of the vaccinia MT domain carries residues essential for catalysis, we looked for conserved aromatic residues, in the N-terminal regions of other cap MTs. In all D1 homologues (including from the distantly related ASFV and mimiviruses), there exists the same conserved YF pair (YY in mimiviruses) in the peptide N-terminal to the first helix (αZ) of the MT fold (Supplementary Figure S7). Similarly, in the sequences of all known eukaryotic cellular cap MTs, one of the rather few strictly conserved residues is a tyrosine (typically in an HYN motif), preceding helix αZ (Figure 2A; Supplementary Figure S4), which could perhaps play a role similar to that of Tyr-555 in vaccinia virus. However, whereas the aromatic nature of this residue is essential for activity in vaccinia (see above), in the case of Ecm1, Tyr-27 of the HYN motif is within the first 30 residues, which if deleted, still allow yeast to grow in an in vivo complementation assay, albeit at a reduced rate (Fabrega et al, 2004), thus highlighting a difference between the two systems. Based on our results on the vaccinia cap MT and the fact that a 40 residue truncation in Ecm1 is lethal (Fabrega et al, 2004), we think it likely that in all cellular N7 cap MTs, the N-terminal extension to the MT domain folds back over the AdoMet-binding site and contributes residues important for catalysis. This implies that there must be additional residues critical for the catalytic mechanism, not visible in the Ecm1 crystal structure (in which residue 41 is the first ordered residue) that remains to be identified. Our structural results also suggests that the N-terminal peptide would have to open to allow AdoMet binding or AdoHcy product release consistent with the fact that the region is protease resistant in the presence of these compounds but not in their absence. An intriguing consequence of this is that as the N-terminal peptide forms the link to the preceding triphosphatase and guanylyl-transferase domains of D1, there may be significant quaternary structure re-arrangements of the whole enzyme during the capping process.
Another peculiarity of the vaccinia virus cap MT is its requirement for the D12 subunit for optimal activity. The fold of D12, unpredictable from its primary sequence, turns out to have a MT-like core, but with the AdoMet-binding domain truncated. Its closest structural homologues are the ribose 2′-O-MTs from reovirus and flavivirus (Supplementary Figure S2), strongly suggesting that D12 may have had an ancestral role in the final step of cap 1 synthesis. In this scenario, the subsequent acquisition by poxviruses of the VP39 protein to fulfil this enzymatic activity may have allowed the catalytic function of D12 to degenerate while preserving an essential allosteric role in enhancing the rate of the MT and perhaps other transcription related activities. Our structure suggests that the mechanism of this rate enhancement is likely to be structural stabilisation of the N-terminal helix αZ of the MT, which carries residues important for binding AdoMet and the cap substrate. Another point related to the evolution of capping enzymes is that the N7 cap MT of the DNA poxviruses has clear sequence as well as structural homology to eukaryotic cellular N7 cap MTs (Figure 2A). This is not the case of the N7 cap MTs from RNA viruses such as reovirus (Reinisch et al, 2000) and blue-tongue virus (Sutton et al, 2007), which have diverged considerable more in structure and have essentially no sequence homology to cellular and vaccinia cap N7 MTs, suggesting even a different evolutionary origin. Reovirus and blue-tongue virus capping enzymes both have distinct N7 and 2′-O MT domains, whereas the flavivirus cap MT, part of the NS5 protein, has recently been shown to be bifunctional, with both 2′-O and N7 cap MT activities (Ray et al, 2006). Several structures of flavivirus bifunctional cap MTs are now known (Dengue (Egloff et al, 2007), West Nile (Malet et al, 2007; Zhou et al, 2007), Meaban (Mastrangelo et al, 2007) and Murray Valley encephalitis virus (Assenberg et al, 2007)) and these are most closely related both in structure and sequence to other cap 2′-O MTs, including vaccinia VP39.
Although WHO officially declared smallpox eradicated in 1979, other animal poxviruses cause sporadic but significant epidemics in human populations, and poxviruses are still considered an emergent epidemiological (Lewis-Jones, 2004), as well as a bioterrorism threat (Smith and McFadden, 2002; Esposito et al, 2006). Owing to the complications and morbidity of vaccination against poxviruses (Belongia and Naleway, 2003) and its lack of effectiveness once infection has taken place (Stittelaar et al, 2006), the search for new antiviral agents is still being intensively pursued. The Shuman laboratory has frequently suggested that unusual features of the poxvirus capping enzyme could make it a good target for antiviral drug design and the unique conformation of the bound AdoMet/AdoHcy cofactor revealed by this work is a good example. For instance, a compound constrained to mimic the syn-conformation of AdoMet, could be a good starting point for design of inhibitors that should not cross-react with any other MT.
Materials and methods
See Supplementary data for additional information.
Protein expression and purification
His-tagged full-length vaccinia capping enzyme or the MT subcomplex were obtained by coexpression in Escherichia coli of the D1 (or D1CTD i.e. residues 545–844) and D12 subunits cloned in tandem. Proteins were sequentially purified with Ni-NTA resin followed by His-tag cleavage with His-tagged TEV protease, application to a heparin column, a second round of Ni-NTA resin, and finally Superdex200 chromatography. SeMet-substituted subcomplex was produced in a defined minimal medium containing 60 mg l−1 of SeMet and purified as for the native protein.
Limited proteolysis assays
Time course proteolysis of full-length D1–D12 capping enzyme was performed at room temperature in a 100 μl of reaction buffer containing 200 μg of full-length vaccinia virus capping enzyme, 0.4 μg of trypsin (ratio 1/500 w/w) and 3 mM of either AdoMet, AdoHcy or GTP. Aliquots were taken at different time points, denatured and loaded onto a 12.5% SDS-polyacrylamide gel.
Crystallisation and data collection
Protein solution at 10–20 mg ml−1 was mixed with an equal volume of a reservoir solution containing either 0.1 M sodium citrate (pH 5.6), 1.0 M ammonium sulphate and traces of trypsin (1/20 000 w/w) for hexagonal and monoclinic crystals, or 10% PEG 4000, 0.1 M HEPES pH 6.5, 0.2 M lithium sulphate, 0.3 M ammonium sulphate for triclinic crystals. Data from cryoprotected crystals were collected at the European Synchrotron Radiation Facility on beamlines ID14-4 and ID29. Images were processed using XDS (Kabsch, 1993; Table I). Crystals belonging to space group P6522 and C2 contain one subcomplex in the asymmetric unit, whereas P1 crystals contain four subcomplexes in the asymmetric unit. In all cases, 5 mM AdoMet or AdoHcy was required to obtain crystals, and 5 mM of m7GDP for the case of triclinic crystals.
Phase determination and structure refinement
The subcomplex structure was solved by a SeMet SAD experiment using the P6522 crystal form, 17 selenium sites being found by SHELXD (Uson and Sheldrick, 1999). Phasing with SOLVE and density modification with RESOLVE (Terwilliger, 2004), yielded an interpretable electron density map. A partial model was used to solve the monoclinic and triclinic crystal forms by molecular replacement. The four subcomplexes in the P1 unit cell were refined with REFMAC (Murshudov et al, 1997) to a final R-factor of 22.3% and Rfree of 26.5% at 2.7 Å using strict NCS restraints and separate TLS parameters for each of the eight chains. A simulated omit map for the N-terminal region of chain C and the AdoHcy molecule is shown in Supplementary Figure S1. According to MOLPROBITY (Lovell et al, 2003), 95.4% of 2238 residues are in favoured regions of the Ramachandran plot and 99.9% in allowed regions. Loops 639–642 and 730–738 in D1 and 120–123 in D12 are poorly ordered in most copies of the complex. Structural figures were drawn with PyMOL(Delano, 2002). Structure based sequence alignments were displayed with ESPRIPT (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
Site-directed mutagenesis and assays of methyl-transfer activity
D1CTD mutants were obtained by PCR amplification of the whole D1CTD–D12 and full-length D1–D12 coexpression vector. Protein expression and purification by Ni-NTA chromatography was performed as described above. For the MT assays, reaction mixtures containing 50 mM Tris–HCl pH 8, 5 mM DTT, 0.1 μM [α-32P]GTP (400 Ci/mmol, Amersham), 5 mM AdoMet (Sigma) and 10 μg of enzyme in a total volume of 20 μl were incubated for 30 min at 37°C. Aliquots of 2 μl at 10 and 30 min were spotted on TLC plates (Merck), developed with 0.25 M ammonium sulphate for 5 h and scanned with a bioimage analyzer (Fuji BAS1500).
Accession numbers
Accession codes at the Macromolecular Structure Database are 2vdw for the coordinates and r2vdwsf for the structure factors.
Supplementary Material
Supplementary Information
Supplementary Figures
Acknowledgments
We thank the EMBL-ESRF Joint Structural Biology group for access to ESRF beamlines. We are indebted to Raimond Ravelli for assistance in data collection and structure solution, to Thibaut Crepin for help with initial model building and to Catherine Mazza, Jan Kadlec and Carlo Petosa for helpful discussions. MDP received a long-term fellowship from the Human Frontier Science Program Organization and a Ramón y Cajal contract from the Spanish MEC. The original plasmid coexpressing both vaccinia virus capping enzyme subunits was kindly provided by Professor Shuman.
References
- Assenberg R, Ren J, Verma A, Walter TS, Alderton D, Hurrelbrink RJ, Fuller SD, Bressanelli S, Owens RJ, Stuart DI, Grimes JM (2007) Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues. J Gen Virol 88: 2228–2236 [DOI] [PubMed] [Google Scholar]
- Belongia EA, Naleway AL (2003) Smallpox vaccine: the good, the bad, and the ugly. Clin Med Res 1: 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MS, DeLange AM (1991) A temperature-sensitive lesion in the small subunit of the vaccinia virus-encoded mRNA capping enzyme causes a defect in viral telomere resolution. J Virol 65: 4042–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S (1997) mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev 11: 3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong P, Shuman S (1992) Methyltransferase and subunit association domains of vaccinia virus mRNA capping enzyme. J Biol Chem 267: 16424–16429 [PubMed] [Google Scholar]
- Cougot N, van Dijk E, Babajko S, Seraphin B (2004) ‘Cap-tabolism'. Trends Biochem Sci 29: 436–444 [DOI] [PubMed] [Google Scholar]
- Delano WL (2002) The PyMOL Molecular Graphics System, http://pymol.sourceforge.net [Google Scholar]
- Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B (2002) An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J 21: 2757–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff MP, Decroly E, Malet H, Selisko B, Benarroch D, Ferron F, Canard B (2007) Structural and functional analysis of methylation and 5′-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J Mol Biol 372: 723–736 [DOI] [PubMed] [Google Scholar]
- Esposito JJ, Sammons SA, Frace AM, Osborne JD, Olsen-Rasmussen M, Zhang M, Govil D, Damon IK, Kline R, Laker M, Li Y, Smith GL, Meyer H, Leduc JW, Wohlhueter RM (2006) Genome sequence diversity and clues to the evolution of variola (smallpox) virus. Science 313: 807–812 [DOI] [PubMed] [Google Scholar]
- Fabrega C, Hausmann S, Shen V, Shuman S, Lima CD (2004) Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol Cell 13: 77–89 [DOI] [PubMed] [Google Scholar]
- Fabrega C, Shen V, Shuman S, Lima CD (2003) Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell 11: 1549–1561 [DOI] [PubMed] [Google Scholar]
- Higman MA, Bourgeois N, Niles EG (1992) The vaccinia virus mRNA (guanine-N7-)-methyltransferase requires both subunits of the mRNA capping enzyme for activity. J Biol Chem 267: 16430–16437 [PubMed] [Google Scholar]
- Higman MA, Christen LA, Niles EG (1994) The mRNA (guanine-7-)methyltransferase domain of the vaccinia virus mRNA capping enzyme. Expression in Escherichia coli and structural and kinetic comparison to the intact capping enzyme. J Biol Chem 269: 14974–14981 [PubMed] [Google Scholar]
- Higman MA, Niles EG (1994) Location of the S-adenosyl-L-methionine binding region of the vaccinia virus mRNA (guanine-7-) methyltransferase. J Biol Chem 269: 14982–14987 [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26: 795–800 [Google Scholar]
- Konishi K, Fujioka M (1988) Rat liver glycine methyltransferase. Cooperative binding of S-adenosylmethionine and loss of cooperativity by removal of a short NH 2-terminal segment. J Biol Chem 263: 13381–13385 [PubMed] [Google Scholar]
- Lewis-Jones S (2004) Zoonotic poxvirus infections in humans. Curr Opin Infect Dis 17: 81–89 [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 50: 437–450 [DOI] [PubMed] [Google Scholar]
- Malet H, Egloff MP, Selisko B, Butcher RE, Wright PJ, Roberts M, Gruez A, Sulzenbacher G, Vonrhein C, Bricogne G, Mackenzie JM, Khromykh AA, Davidson AD, Canard B (2007) Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J Biol Chem 282: 10678–10689 [DOI] [PubMed] [Google Scholar]
- Mao X, Shuman S (1994) Intrinsic RNA (guanine-7) methyltransferase activity of the vaccinia virus capping enzyme D1 subunit is stimulated by the D12 subunit. Identification of amino acid residues in the D1 protein required for subunit association and methyl group transfer. J Biol Chem 269: 24472–24479 [PubMed] [Google Scholar]
- Mao X, Shuman S (1996) Vaccinia virus mRNA (guanine-7-)methyltransferase: mutational effects on cap methylation and AdoHcy-dependent photo-cross-linking of the cap to the methyl acceptor site. Biochemistry 35: 6900–6910 [DOI] [PubMed] [Google Scholar]
- Martin JL, McMillan FM (2002) SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr Opin Struct Biol 12: 783–793 [DOI] [PubMed] [Google Scholar]
- Mastrangelo E, Bollati M, Milani M, Selisko B, Peyrane F, Canard B, Grard G, de Lamballerie X, Bolognesi M (2007) Structural bases for substrate recognition and activity in Meaban virus nucleoside-2′-O-methyltransferase. Protein Sci 16: 1133–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Myette JR, Niles EG (1996) Characterization of the vaccinia virus RNA 5′-triphosphatase and nucleotide triphosphate phosphohydrolase activities. Demonstrate that both activities are carried out at the same active site. J Biol Chem 271: 11945–11952 [DOI] [PubMed] [Google Scholar]
- Niles EG, Christen L (1993) Identification of the vaccinia virus mRNA guanyltransferase active site lysine. J Biol Chem 268: 24986–24989 [PubMed] [Google Scholar]
- Pakhomova S, Luka Z, Grohmann S, Wagner C, Newcomer ME (2004) Glycine N-methyltransferases: a comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes. Proteins 57: 331–337 [DOI] [PubMed] [Google Scholar]
- Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY (2006) West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol 80: 8362–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch KM, Nibert ML, Harrison SC (2000) Structure of the reovirus core at 3.6 Å resolution. Nature 404: 960–967 [DOI] [PubMed] [Google Scholar]
- Saha N, Shuman S (2001) Effects of alanine cluster mutations in the D12 subunit of vaccinia virus mRNA (guanine-N7) methyltransferase. Virology 287: 40–48 [DOI] [PubMed] [Google Scholar]
- Saha N, Shuman S, Schwer B (2003) Yeast-based genetic system for functional analysis of poxvirus mRNA cap methyltransferase. J Virol 77: 7300–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann TH, Keeler C, Hodsdon ME (2004) Consequences of binding an S-adenosylmethionine analogue on the structure and dynamics of the thiopurine methyltransferase protein backbone. Biochemistry 43: 12198–12209 [DOI] [PubMed] [Google Scholar]
- Schnierle BS, Gershon PD, Moss B (1992) Cap-specific mRNA (nucleoside-O2′-)-methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proc Natl Acad Sci USA 89: 2897–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Hausmann S, Schneider S, Shuman S (2006) Poxvirus mRNA cap methyltransferase. Bypass of the requirement for the stimulatory subunit by mutations in the catalytic subunit and evidence for intersubunit allostery. J Biol Chem 281: 18953–18960 [DOI] [PubMed] [Google Scholar]
- Schwer B, Shuman S (2006) Genetic analysis of poxvirus mRNA cap methyltransferase: suppression of conditional mutations in the stimulatory D12 subunit by second-site mutations in the catalytic D1 subunit. Virology 352: 145–156 [DOI] [PubMed] [Google Scholar]
- Shuman S (1989) Functional domains of vaccinia virus mRNA capping enzyme. Analysis by limited tryptic digestion. J Biol Chem 264: 9690–9695 [PubMed] [Google Scholar]
- Shuman S (2001) Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol 66: 1–40 [DOI] [PubMed] [Google Scholar]
- Shuman S (2002) What messenger RNA capping tells us about eukaryotic evolution. Nat Rev Mol Cell Biol 3: 619–625 [DOI] [PubMed] [Google Scholar]
- Shuman S, Moss B (1988) Factor-dependent transcription termination by vaccinia virus RNA polymerase. Evidence that the cis-acting termination signal is in nascent RNA. J Biol Chem 263: 6220–6225 [PubMed] [Google Scholar]
- Shuman S, Surks M, Furneaux H, Hurwitz J (1980) Purification and characterization of a GTP-pyrophosphate exchange activity from vaccinia virions. Association of the GTP-pyrophosphate exchange activity with vaccinia mRNA guanylyltransferase. RNA (guanine-7-)methyltransferase complex (capping enzyme). J Biol Chem 255: 11588–11598 [PubMed] [Google Scholar]
- Smith GL, McFadden G (2002) Smallpox: anything to declare? Nat Rev Immunol 2: 521–527 [DOI] [PubMed] [Google Scholar]
- Stittelaar KJ, Neyts J, Naesens L, van Amerongen G, van Lavieren RF, Holy A, De Clercq E, Niesters HG, Fries E, Maas C, Mulder PG, van der Zeijst BA, Osterhaus AD (2006) Antiviral treatment is more effective than smallpox vaccination upon lethal monkeypox virus infection. Nature 439: 745–748 [DOI] [PubMed] [Google Scholar]
- Sutton G, Grimes JM, Stuart DI, Roy P (2007) Bluetongue virus VP4 is an RNA-capping assembly line. Nat Struct Mol Biol 14: 449–451 [DOI] [PubMed] [Google Scholar]
- Takata Y, Huang Y, Komoto J, Yamada T, Konishi K, Ogawa H, Gomi T, Fujioka M, Takusagawa F (2003) Catalytic mechanism of glycine N-methyltransferase. Biochemistry 42: 8394–8402 [DOI] [PubMed] [Google Scholar]
- Terwilliger T (2004) SOLVE and RESOLVE: automated structure solution, density modification, and model building. J Synchrotron Radiat 11: 49–52 [DOI] [PubMed] [Google Scholar]
- Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J (2007) Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. EMBO J 26: 4391–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uson I, Sheldrick GM (1999) Advances in direct methods for protein crystallography. Curr Opin Struct Biol 9: 643–648 [DOI] [PubMed] [Google Scholar]
- Vos JC, Sasker M, Stunnenberg HG (1991) Vaccinia virus capping enzyme is a transcription initiation factor. EMBO J 10: 2553–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH (2007) Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. EMBO J 26: 4402–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H (2007) Structure and function of flavivirus NS5 methyltransferase. J Virol 81: 3891–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Figures