Abstract
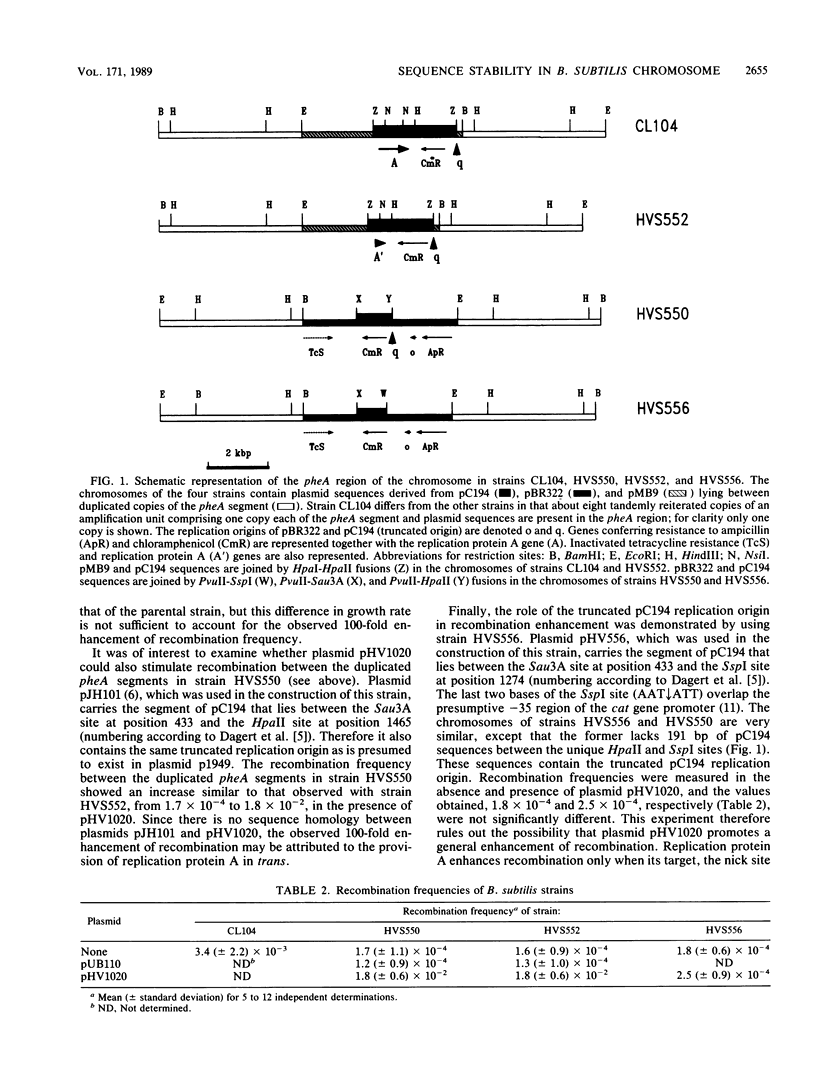
The instability of reiterated sequences in the Bacillus subtilis chromosome that was previously reported (M. Young, J. Gen. Microbiol. 130:1613-1621, 1984) results from the presence of a truncated pC194 replication origin together with an intact replication protein A gene in the amplified DNA. Removal of the truncated pC194 replication origin or inactivation of replication protein A stabilizes reiterated sequences, whereas provision of replication protein A in trans destabilizes them. We suggest that residual activity of protein A at the truncated replication origin generates single-stranded DNA, which stimulates recombination between repeated sequences and thus destabilizes amplified structures.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Galizzi A. Amplification of a chromosomal region in Bacillus subtilis. J Bacteriol. 1985 Jun;162(3):1203–1211. doi: 10.1128/jb.162.3.1203-1211.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Betlach M. C., Boyer H. W. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene. 1977;2(2):75–93. doi: 10.1016/0378-1119(77)90074-9. [DOI] [PubMed] [Google Scholar]
- Brunier D., Michel B., Ehrlich S. D. Copy choice illegitimate DNA recombination. Cell. 1988 Mar 25;52(6):883–892. doi: 10.1016/0092-8674(88)90430-8. [DOI] [PubMed] [Google Scholar]
- Dagert M., Jones I., Goze A., Romac S., Niaudet B., Ehrlich S. D. Replication functions of pC194 are necessary for efficient plasmid transduction by M13 phage. EMBO J. 1984 Jan;3(1):81–86. doi: 10.1002/j.1460-2075.1984.tb01764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Nguyen A., Lang D., Hoch J. A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldenwang W. G., Banner C. D., Ollington J. F., Losick R., Hoch J. A., O'Connor M. B., Sonenshein A. L. Mapping a cloned gene under sporulation control by inserttion of a drug resistance marker into the Bacillus subtilis chromosome. J Bacteriol. 1980 Apr;142(1):90–98. doi: 10.1128/jb.142.1.90-98.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi K., Tanimoto A., Nomura S., Yamane K., Yoda K., Harada S., Mori M., Furusato T., Takatsuki A., Yamasaki M. Amplification of the amyE-tmrB region on the chromosome in tunicamycin-resistant cells of Bacillus subtilis. Mol Gen Genet. 1986 Jul;204(1):36–43. doi: 10.1007/BF00330184. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hranueli D., Piggot P. J., Mandelstam J. Statistical estimate of the total number of operons specific for Bacillus subtilis sporulation. J Bacteriol. 1974 Sep;119(3):684–690. doi: 10.1128/jb.119.3.684-690.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannière L., Ehrlich S. D. Recombination between short repeated sequences is more frequent in plasmids than in the chromosome of Bacillus subtilis. Mol Gen Genet. 1987 Nov;210(1):116–121. doi: 10.1007/BF00337766. [DOI] [PubMed] [Google Scholar]
- Jannière L., Niaudet B., Pierre E., Ehrlich S. D. Stable gene amplification in the chromosome of Bacillus subtilis. Gene. 1985;40(1):47–55. doi: 10.1016/0378-1119(85)90023-x. [DOI] [PubMed] [Google Scholar]
- McKenzie T., Hoshino T., Tanaka T., Sueoka N. Correction. A revision of the nucleotide sequence and functional map of pUB110. Plasmid. 1987 Jan;17(1):83–85. doi: 10.1016/0147-619x(87)90015-1. [DOI] [PubMed] [Google Scholar]
- McKenzie T., Hoshino T., Tanaka T., Sueoka N. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid. 1986 Mar;15(2):93–103. doi: 10.1016/0147-619x(86)90046-6. [DOI] [PubMed] [Google Scholar]
- Mori M., Tanimoto A., Yoda K., Harada S., Koyama N., Hashiguchi K., Obinata M., Yamasaki M., Tamura G. Essential structure in the cloned transforming DNA that induces gene amplification of the Bacillus subtilis amyE-tmrB region. J Bacteriol. 1986 Jun;166(3):787–794. doi: 10.1128/jb.166.3.787-794.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet B., Jannière L., Ehrlich S. D. Recombination between repeated DNA sequences occurs more often in plasmids than in the chromosome of Bacillus subtilis. Mol Gen Genet. 1984;197(1):46–54. doi: 10.1007/BF00327921. [DOI] [PubMed] [Google Scholar]
- Noirot P., Petit M. A., Ehrlich S. D. Plasmid replication stimulates DNA recombination in Bacillus subtilis. J Mol Biol. 1987 Jul 5;196(1):39–48. doi: 10.1016/0022-2836(87)90509-2. [DOI] [PubMed] [Google Scholar]
- Primrose S. B., Ehrlich S. D. Isolation of plasmid deletion Mutants and study of their instability. Plasmid. 1981 Sep;6(2):193–201. doi: 10.1016/0147-619x(81)90066-4. [DOI] [PubMed] [Google Scholar]
- Sargent M. G., Bennett M. F. Amplification of a major membrane-bound DNA sequence of Bacillus subtilis. J Bacteriol. 1985 Feb;161(2):589–595. doi: 10.1128/jb.161.2.589-595.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner V., Ehrlich S. D. Efficiency of homologous DNA recombination varies along the Bacillus subtilis chromosome. J Bacteriol. 1988 Sep;170(9):3978–3982. doi: 10.1128/jb.170.9.3978-3982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. R., Morgan A. E. Chromosomal-DNA amplification in Bacillus subtilis. J Bacteriol. 1985 Aug;163(2):445–453. doi: 10.1128/jb.163.2.445-453.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M. Gene amplification in Bacillus subtilis. J Gen Microbiol. 1984 Jul;130(7):1613–1621. doi: 10.1099/00221287-130-7-1613. [DOI] [PubMed] [Google Scholar]
- Young M. The mechanism of insertion of a segment of heterologous DNA into the chromosome of Bacillus subtilis. J Gen Microbiol. 1983 May;129(5):1497–1512. doi: 10.1099/00221287-129-5-1497. [DOI] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Are single-stranded circles intermediates in plasmid DNA replication? EMBO J. 1986 Mar;5(3):631–637. doi: 10.1002/j.1460-2075.1986.tb04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]


