Abstract
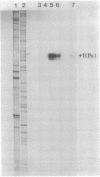
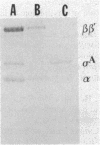
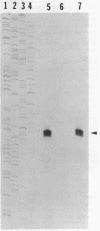
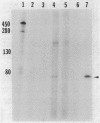
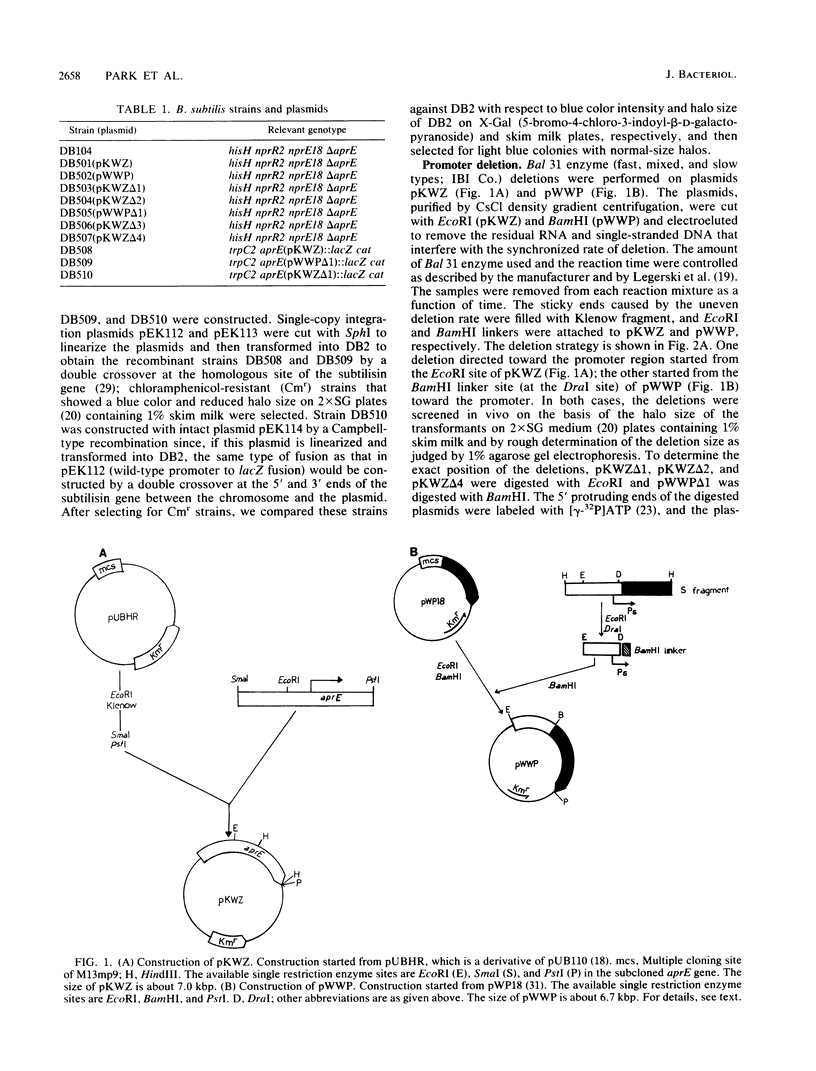
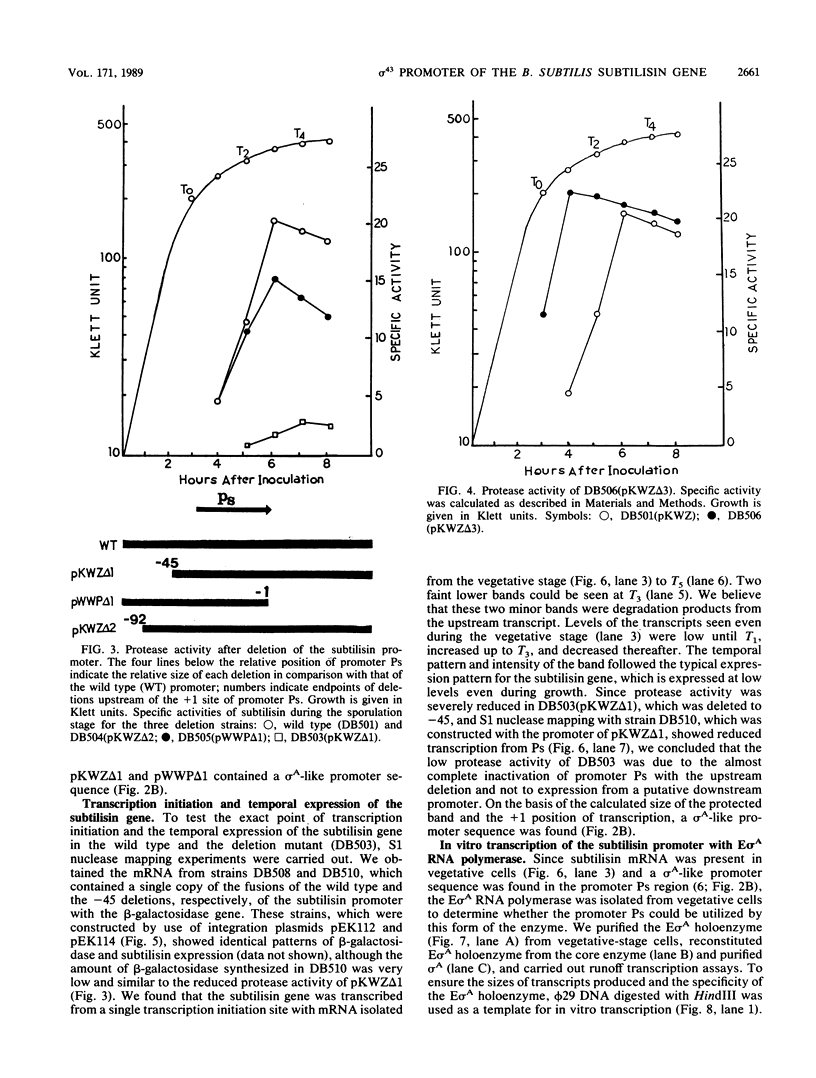
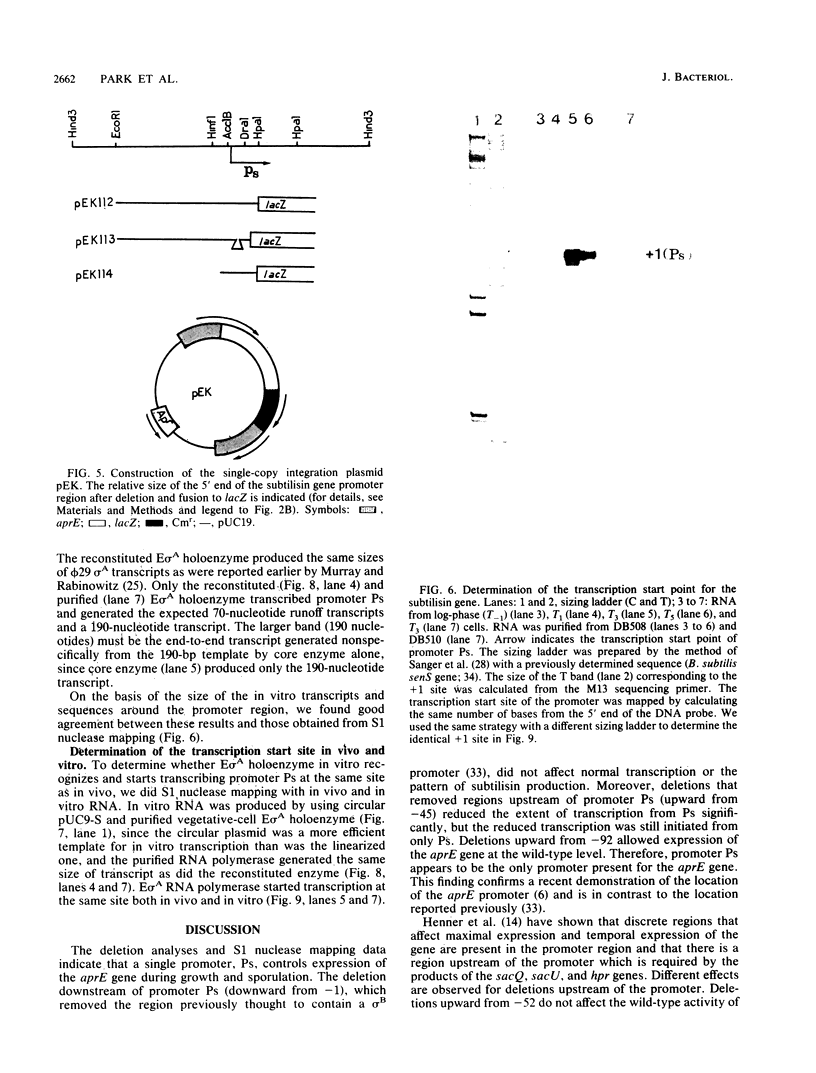
In vitro studies demonstrated that the Bacillus subtilis subtilisin gene (aprE) could be transcribed by RNA polymerase holoenzyme reconstituted from core and sigma A factor obtained from vegetative cells. Upstream deletions (from -45) reduced the amount of transcription from the promoter. A deletion downstream of the promoter that overlapped a putative downstream minor promoter did not affect transcription from the sigma A promoter, which indicated that the putative downstream promoter is not utilized in vivo. S1 nuclease mapping studies showed that there was a low level of transcription from the subtilisin promoter during the growth phase and that the site of transcription initiation was the same during log and stationary phases. We conclude from these findings that there is only one promoter for the subtilisin gene and that it can be transcribed by the sigma A form of RNA polymerase in vitro.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amory A., Kunst F., Aubert E., Klier A., Rapoport G. Characterization of the sacQ genes from Bacillus licheniformis and Bacillus subtilis. J Bacteriol. 1987 Jan;169(1):324–333. doi: 10.1128/jb.169.1.324-333.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Carter H. L., 3rd, Wang L. F., Doi R. H., Moran C. P., Jr rpoD operon promoter used by sigma H-RNA polymerase in Bacillus subtilis. J Bacteriol. 1988 Apr;170(4):1617–1621. doi: 10.1128/jb.170.4.1617-1621.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison B. L., Leighton T., Rabinowitz J. C. Purification of Bacillus subtilis RNA polymerase with heparin-agarose. In vitro transcription of phi 29 DNA. J Biol Chem. 1979 Sep 25;254(18):9220–9226. [PubMed] [Google Scholar]
- Doi R. H. Role of ribonucleic acid polymerase in gene selection in procaryotes. Bacteriol Rev. 1977 Sep;41(3):568–594. doi: 10.1128/br.41.3.568-594.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E., Henner D. J., Perego M., Hoch J. A. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J Bacteriol. 1988 Jan;170(1):289–295. doi: 10.1128/jb.170.1.289-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E., Howard S. M., Hoch J. A. Effect of stage 0 sporulation mutations on subtilisin expression. J Bacteriol. 1986 Apr;166(1):173–179. doi: 10.1128/jb.166.1.173-179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R., Doi R. H. Two polypeptides associated with the ribonucleic acid polymerase core of Bacillus subtilis during sporulation. J Bacteriol. 1977 Jan;129(1):422–432. doi: 10.1128/jb.129.1.422-432.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur N. K., Cabane K., Smith I. Structure and expression of the Bacillus subtilis sin operon. J Bacteriol. 1988 Mar;170(3):1046–1053. doi: 10.1128/jb.170.3.1046-1053.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman M. Z., Chamberlin M. J. Developmental and genetic regulation of Bacillus subtilis genes transcribed by sigma 28-RNA polymerase. Cell. 1983 Nov;35(1):285–293. doi: 10.1016/0092-8674(83)90231-3. [DOI] [PubMed] [Google Scholar]
- Goldfarb D. S., Wong S. L., Kudo T., Doi R. H. A temporally regulated promoter from Bacillus subtilis is transcribed only by an RNA polymerase with a 37,000 dalton sigma factor. Mol Gen Genet. 1983;191(2):319–325. doi: 10.1007/BF00334833. [DOI] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Halling S. M., Sanchez-Anzaldo F. J., Fukuda R., Doi R. H., Meares C. F. Zinc is associated with the beta subunit of DNA-dependent RNA polymerase of Bacillus subtilis. Biochemistry. 1977 Jun 28;16(13):2880–2884. doi: 10.1021/bi00632a012. [DOI] [PubMed] [Google Scholar]
- Henner D. J., Ferrari E., Perego M., Hoch J. A. Location of the targets of the hpr-97, sacU32(Hy), and sacQ36(Hy) mutations in upstream regions of the subtilisin promoter. J Bacteriol. 1988 Jan;170(1):296–300. doi: 10.1128/jb.170.1.296-300.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higerd T. B., Hoch J. A., Spizizen J. Hyperprotease-producing mutants of Bacillus subtilis. J Bacteriol. 1972 Nov;112(2):1026–1028. doi: 10.1128/jb.112.2.1026-1028.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordănescu S. Relationships between cotransducible plasmids in Staphylococcus aureus. J Bacteriol. 1977 Jan;129(1):71–75. doi: 10.1128/jb.129.1.71-75.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs E., Clad A. Electroelution of fixed and stained membrane proteins from preparative sodium dodecyl sulfate-polyacrylamide gels into a membrane trap. Anal Biochem. 1986 May 1;154(2):583–589. doi: 10.1016/0003-2697(86)90033-3. [DOI] [PubMed] [Google Scholar]
- Kawamura F., Doi R. H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984 Oct;160(1):442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerski R. J., Hodnett J. L., Gray H. B., Jr Extracellular nucleases of pseudomonas BAL 31. III. Use of the double-strand deoxyriboexonuclease activity as the basis of a convenient method for the mapping of fragments of DNA produced by cleavage with restriction enzymes. Nucleic Acids Res. 1978 May;5(5):1445–1464. doi: 10.1093/nar/5.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton T. J., Doi R. H. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971 May 25;246(10):3189–3195. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Murray C. L., Rabinowitz J. C. Nucleotide sequences of transcription and translation initiation regions in Bacillus phage phi 29 early genes. J Biol Chem. 1982 Jan 25;257(2):1053–1062. [PubMed] [Google Scholar]
- Nagami Y., Tanaka T. Molecular cloning and nucleotide sequence of a DNA fragment from Bacillus natto that enhances production of extracellular proteases and levansucrase in Bacillus subtilis. J Bacteriol. 1986 Apr;166(1):20–28. doi: 10.1128/jb.166.1.20-28.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinderknecht H., Geokas M. C., Silverman P., Haverback B. J. A new ultrasensitive method for the determination of proteolytic activity. Clin Chim Acta. 1968 Aug;21(2):197–203. doi: 10.1016/0009-8981(68)90127-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43(1-2):85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Kawata M., Nagami Y., Uchiyama H. prtR enhances the mRNA level of the Bacillus subtilis extracellular proteases. J Bacteriol. 1987 Jul;169(7):3044–3050. doi: 10.1128/jb.169.7.3044-3050.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. F., Doi R. H. Promoter switching during development and the termination site of the sigma 43 operon of Bacillus subtilis. Mol Gen Genet. 1987 Apr;207(1):114–119. doi: 10.1007/BF00331498. [DOI] [PubMed] [Google Scholar]
- Wong S. L., Doi R. H. Determination of the signal peptidase cleavage site in the preprosubtilisin of Bacillus subtilis. J Biol Chem. 1986 Aug 5;261(22):10176–10181. [PubMed] [Google Scholar]
- Wong S. L., Price C. W., Goldfarb D. S., Doi R. H. The subtilisin E gene of Bacillus subtilis is transcribed from a sigma 37 promoter in vivo. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1184–1188. doi: 10.1073/pnas.81.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. L., Wang L. F., Doi R. H. Cloning and nucleotide sequence of senN, a novel 'Bacillus natto' (B. subtilis) gene that regulates expression of extracellular protein genes. J Gen Microbiol. 1988 Dec;134(12):3269–3276. doi: 10.1099/00221287-134-12-3269. [DOI] [PubMed] [Google Scholar]
- Yang M., Ferrari E., Chen E., Henner D. J. Identification of the pleiotropic sacQ gene of Bacillus subtilis. J Bacteriol. 1986 Apr;166(1):113–119. doi: 10.1128/jb.166.1.113-119.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Shimotsu H., Ferrari E., Henner D. J. Characterization and mapping of the Bacillus subtilis prtR gene. J Bacteriol. 1987 Jan;169(1):434–437. doi: 10.1128/jb.169.1.434-437.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]