Abstract
A rare class of introns with AT–AC at their termini recently has been identified in metazoan genes. Splicing of these introns requires a different set of small nuclear ribonucleoprotein particles (snRNPs) (U11, U12, U5, and U4atac/U6atac) compared with the snRNPs (U1, U2, U5, and U4/U6) required for splicing the majority of pre-mRNA introns, but otherwise little is known regarding the excision of AT–AC introns. Here we use site-specific 4-thiouridine (4SU) crosslinking analysis to dissect the mechanism of 5′ splice site recognition during in vitro splicing of the AT–AC intron from the P120 pre-mRNA. Upon irradiation with 365-nm UV light, three P120 substrates, each with a single 4SU substitution near the 5′ splice site (at position +2, +4, or +7), produce two early ATP-independent crosslinks with similar kinetics. For one of the substrates, P120-4SU+2, a third ATP-requiring crosslink forms as the two early crosslinks diminish. RNase H digestion coupled with Northern blotting indicates that the two early crosslinks generated with P120-4SU+2 contain the U11 small nuclear RNA. Reverse transcription–PCR followed by cloning and sequencing demonstrates that the third crosslink involves U6atac. The dynamic appearance of the three crosslinks correlates with the kinetics of the splicing reaction and suggests that the 5′ splice site is recognized first by U11 and then by U6atac. Our results argue that the splicing of AT–AC introns is mechanistically similar to the splicing of the major class of introns and that the U11 and U6atac snRNPs in the AT–AC spliceosome fulfill analogous roles to U1 and U6, respectively, in the major spliceosome.
Keywords: 4-thiouridine, pre-mRNA, RNA processing, small nuclear ribonucleoprotein, spliceosome
Most pre-mRNAs contain introns starting with the dinucleotide GU and ending with the dinucleotide AG (the major class of introns). To produce functional messages, the introns are accurately removed through a process called pre-mRNA splicing. Before splicing, each intron associates with five small nuclear ribonucleoprotein particles (snRNP; U1, U2, U4, U5, and U6) and a large, as yet undetermined, number of protein factors (1–4) to form a multicomponent complex termed the spliceosome. Both genetic and biochemical studies have indicated that the assembly of the major class spliceosome is a step-wise event. At early times, the U1 snRNP interacts via base-pairing with intron sequences at the 5′ splice site (5–8). The U2 snRNP then recognizes (again via base-pairing) the branch site of the pre-mRNA (8–11) to form a presplicing complex. This complex is later joined by the U4/U5/U6 tri-snRNP, in which U4 and U6 are extensively base-paired to each other. Subsequently, new RNA–RNA interactions are created within the assembled spliceosome. U5 associates, largely via non-Watson–Crick interactions, with exon sequences immediately upstream of the 5′ splice site and downstream of the 3′ splice site (12–17). U6, which is released from pairing with U4 soon after joining the spliceosome, forms new base-pairing interactions with U2 (8, 18–21) and replaces U1 in interacting with the 5′ splice site (8, 22–26). At this stage, the catalytic core forms and the first transesterification takes place, creating the lariat 2/3 intermediate and the free 5′ exon. Using 4-thiouridine (4SU) crosslinking, Sontheimer and Steitz (13) demonstrated that the +2 position of the lariat 2/3 intermediate can be crosslinked to a conserved sequence (ACAGAG) of U6. This crosslink likely represents a functional interaction required for the second step of splicing that leads to intron removal and exon ligation.
Recently, a new class of introns that contain the dinucleotides AT and AC at their 5′ and 3′ termini, respectively, has been found in metazoan genes (27–29). Removal of AT–AC introns is catalyzed by a spliceosome that contains only one common (U5) and four different (U11, U12, U4atac, and U6atac) snRNPs compared with the major spliceosome (29–31). Despite very limited sequence conservation, U11, U12, and U4atac/U6atac can fold into secondary structures that are remarkably similar to U1, U2, and U4/U6, respectively, suggesting parallel roles for each pair of snRNPs in the two different spliceosomes (31–33). In particular, U11 and U6atac exhibit complementarity to the 5′ splice site, whereas U12 can potentially base-pair with the branch site of AT–AC introns. Both in vitro (30, 34) and in vivo (35) studies have demonstrated that U12 is indeed a U2 analog that recognizes the branch site of AT–AC introns. Using psoralen crosslinking, Tarn and Steitz (31) identified base-pairing interactions between U4atac and U6atac and between U12 and U6atac. These interactions exhibit striking resemblance to the interactions documented for U4 and U6 and for U2 and U6 (Helix I), respectively, in the major spliceosome. Most recently, Kolossova and Padgett (36) showed that compensatory changes in the 5′ end sequence of U11 could suppress certain mutations introduced into the 5′ splice site of an AT–AC intron, suggesting that U11 may play an analogous role to U1 at the 5′ splice site. However, a detailed picture of the dynamic RNA–RNA interactions that accompany assembly of the AT–AC spliceosome is lacking. In particular, how the 5′ splice site is recognized during splicing and which snRNPs are involved in the process have remained unclear.
Here, we use 4SU site-specific crosslinking to dissect the mechanism of 5′ splice site recognition during the excision of an AT–AC intron. We show that U11 assembles, in an ATP-independent manner, onto the 5′ splice site at very early times. Later during spliceosome assembly, U6atac achieves close contact with the 5′ splice site. Our data suggest that the mechanism of 5′ splice site recognition in the splicing of AT–AC introns is analogous to that described for major class introns.
MATERIALS AND METHODS
Preparation of 4SU-Substituted P120 Splicing Substrates.
Preparation of three different 4SU-substituted P120 splicing substrates is described in (37). For each substrate, P120-4SU+2, P120-4SU+4, or P120-4SU+7, a single uridine residue downstream of the 5′ splice site (at position +2, +4, or +7) was replaced by 4SU.
P120 Pre-mRNA Splicing and 4SU Crosslinking.
In vitro splicing of the three P120 substrates was performed in HeLa nuclear extract according to Tarn and Steitz (30). To examine the time course of 4SU crosslinking, a 7-fold splicing reaction containing 1 × 106 cpm (≈10 ng) of a [5′, 3′-32P]p4SUp-labeled P120 substrate (or for an unlabeled 4SU-P120, 1 μl of splicing reaction containing ≈1 ng of the substrate) was prepared. At the indicated times, fractions (1/7) of the reaction mixture were removed, irradiated with 365-nm UV light for 10 min (13), treated with 100 μg proteinase K, extracted with phenol/chloroform/isoamyl alcohol and precipitated with ethanol. Recovered RNAs were resolved on 5% polyacrylamide/8 M urea gels and visualized by autoradiography.
Anti-Trimethyl Guanosine (TMG) Immunoprecipitation.
Immunoprecipitation was performed essentially as previously described (38). Crosslinked RNAs were excised from gels and incubated with anti-TMG antibodies (Oncogene Science). RNAs recovered from pellets and supernatants were analyzed in parallel on 5% polyacrylamide/8 M urea gels.
RNase H Digestion and Northern Blot Analysis.
To identify small nuclear RNA (snRNA) partners, crosslinked RNAs were subjected to oligodeoxynucleotide-directed RNase H digestion (39). The oligonucleotides used were: U1-L2, complementary to nucleotides 64–75 of U1; U2-b, complementary to nucleotides 27–49 of U2; U4-b, complementary to nucleotides 30–44 of U4; U5Ae, complementary to nucleotides 69–88 of U5A; U6-a, complementary to nucleotides 46–60 of U6; U11–10D and U11–10L, complementary to nucleotides 11–27 and 52–70 of U11, respectively; U12-b, complementary to nucleotides 2–23 of U12; U4atac-10a, complementary to nucleotides 63–82 of U4atac; and U6atac-9a, complementary to nucleotides 87–103 of U6atac. After RNase H digestion, RNAs were resolved on 5% polyacrylamide gels and electrophoretically transferred to Zeta-Probe GT membranes (Bio-Rad). Northern blotting was performed essentially as described (30). Anti-sense RNA probes for detecting U11 and U2 were produced by in vitro transcription as previously described (30). To detect the P120 substrate, the bridging oligonucleotide complementary to nucleotides −17 to +31 of the P120 RNA (used for two-piece ligation, see ref. 37) was 5′-end labeled and used as a probe.
Reverse Transcription (RT)–PCR Sequencing Analysis.
Crosslink 3 and control regions (see text) were excised and used as templates for RT, as described (39). The primer was complementary to the last 20 nucleotides of human U6atac (31). The cDNAs were recovered to serve as templates for PCR amplification (30 cycles). The 5′ primer corresponded to nucleotides 22–38 of U6atac and the 3′ primer was the same as for RT. The annealing and extending temperatures for PCR were 55°C and 72°C, respectively. After PCR, the product was visualized on a 2% agarose gel, excised, and recovered. The sequence of the PCR product was determined after cloning.
RESULTS
Three Species Crosslinked at the 5′ Splice Site Are Generated During P120 Splicing.
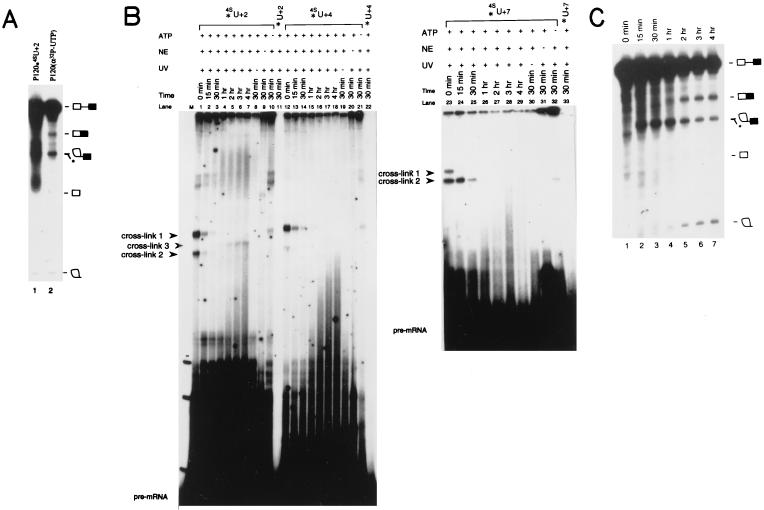
Three P120 substrates site specifically substituted with [5′, 3′-32P]p4SUp at position +2, +4, or +7 relative to the 5′ splice site were prepared (37). Before crosslinking analyses, the 4SU-substituted P120 substrates were tested for splicing efficiency and accuracy. Fig. 1A compares the in vitro splicing profile of P120-4SU+2 with that of unsubstituted P120 substrate after 4 hr of incubation. The 2/3 lariat intermediate and the lariat intron product appear identical in lanes 1 and 2. Because the 4SU-substituted P120 RNA was labeled only at the +2 position, the 5′ exon intermediate and the ligated mRNA are invisible in lane 1. To check the accuracy of splicing, a gel slice predicted to contain the spliced product was excised and the RNA recovered; using RT–PCR followed by cloning and sequencing, we confirmed that the mRNA was generated using the correct 5′ and 3′ splice sites (ref. 30, and data not shown). The other two 4SU-substituted P120 substrates (P120-4SU+4 and P120-4SU+7) were also efficiently spliced (data not shown).
Figure 1.
Splicing and crosslinking. (A) P120-4SU+2 with both the 5′ and 3′ phosphates of the 4SU+2 radioactive (lane 1) or uniformly labeled P120 (lane 2) substrate was used for splicing (30). The positions of the pre-mRNA, the spliced mRNA, the 2/3 intermediate, the free 5′ exon, and the lariat intron on an 8% polyacrylamide/8 M urea gel are indicated. ∗ indicates a degradation product that comigrates with the 2/3 intermediate (30). (B) A time course of crosslinking using site-specifically labeled P120-4SU+2 (lanes 1–10), P120-U+2 (lane 11), P120-4SU+4 (lanes 12–21), P120-U+4 (lane 22), P120-4SU+7 (lanes 23–32), or P120-U+7 (lane 33) substrate. A large splicing reaction containing each 4SU-substituted P120 was incubated and portions removed and irradiated with 365-nm UV light at the indicated times. Recovered RNAs were resolved on 5% polyacrylamide/8 M urea gels. Crosslinks 1, 2, and 3 are indicated by arrow heads. Lanes 8–11, 19–22, and 30–33 are 30-min controls where either UV irradiation (lanes 8, 19, 30), HeLa nuclear extract (lanes 9, 20, 31), or ATP (lanes 10, 21, 32) was omitted. (C) A parallel time course of splicing of uniformly labeled P120 pre-mRNA. At time points (lanes 1–7) identical to those chosen for irradiation (B), portions of the splicing reaction were removed. Recovered RNAs were resolved on an 8% polyacrylamide/8 M urea gel. The positions of the pre-mRNA, the spliced mRNA, the 2/3 intermediate, the free 5′ exon, and the lariat intron are indicated. A degradation product that comigrates with the 2/3 intermediate is denoted by ∗ (30).
The three 4SU-substituted P120 substrates then were subjected to crosslinking by irradiation with 365-nm UV light. The results of a time-course experiment for each of the substrates (P120-4SU+2, P120-4SU+4, and P120-4SU+7) are shown in Fig. 1B. At an early time in the splicing reaction, two crosslinked species (crosslink 1 and crosslink 2) are observed for each substrate. These two crosslinked species, even though they were generated using different substrates, exhibit similar kinetics: they are observed only during the first hour of the 4-hr splicing reaction. For the P120-4SU+2 substrate, as the two early crosslinks diminish, a new crosslinked species (Fig. 1B, crosslink 3) appears at the 1-hr time point and reaches a plateau by 2 hr. Importantly, generation of all three crosslinks is dependent on 365-nm UV irradiation (lanes 8, 19, 30), the presence of 4SU in the substrate (lanes 11, 22, 33, 34), and the presence of HeLa nuclear extract in the reaction mixture (lanes 9, 20, 31). Crosslinks 1 and 2 appear in the absence of ATP (lanes 10, 21, 32; longer exposures are required to visualize clearly both crosslinks, data not shown), whereas crosslink 3 does require ATP.
To correlate these crosslinks with the progress of the splicing reaction, we examined the time course of splicing (Fig. 1C). The disappearance of crosslinks 1 and 2 and the appearance of crosslink 3 occur at the same time (≈1 hr) that splicing intermediates and products become visible. This suggests sequential functional interactions between the 5′ splice site of the P120 intron and yet-to-be identified RNA species.
Evidence for U11 and U6atac as Crosslinking Partners with the P120 RNA.
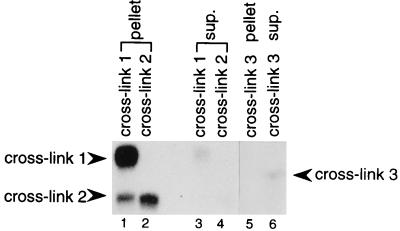
Because the P120-4SU+2 could generate all three crosslinked species quite efficiently, we used this substrate exclusively for the following characterizations. To ask whether the crosslinking partners might be U snRNAs, we first performed immunoprecipitation with anti-TMG antibody. The results for gel-purified crosslinks 1, 2, and 3 are shown in Fig. 2. While crosslinks 1 and 2 were nearly quantitatively precipitated by the anti-TMG antibody (compare lanes 1 and 2 with lanes 3 and 4), crosslink 3 remained in the supernatant (compare lane 5 with lane 6). These results suggest that crosslinked species 1 and 2, but not 3, contain a TMG-capped RNA.
Figure 2.
Immunoprecipitation of the crosslinked species with anti-TMG antibody. Crosslink 1 (lanes 1 and 3), crosslink 2 (lanes 2 and 4), and crosslink 3 (lanes 5 and 6) were excised from gels (Fig. 1B) and incubated with antibody. RNAs recovered from the pellets (lanes 1, 2, 5) and supernatants (lanes 3, 4, 6) were visualized on 5% polyacrylamide gels. The positions of the three crosslinks are indicated. The excised crosslink 1 band was contaminated with a small amount of crosslink 2 (lane 1).
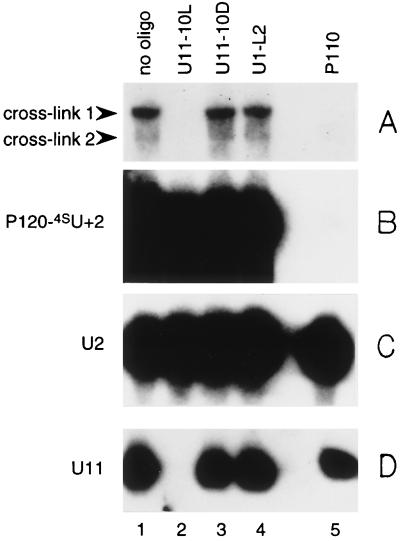
To determine the identity of U snRNA(s) in crosslinks 1 and 2, we performed a series of RNase H digestion analyses. Oligodeoxynucleotides complementary to known TMG-containing snRNAs U1, U2, U4, U5, U12, or U4atac all failed to direct the digestion of these two crosslinked RNAs (data not shown). In contrast, a U11 antisense oligodeoxynucleotide (U11–10L) directed complete digestion of crosslinks 1 and 2 (data not shown). Puzzlingly, however, another U11 antisense oligodeoxynucleotide (U11–10D) yielded negative results (data not shown). Oligonucleotide U11–10L is complementary to nucleotides 52–70 in U11, whereas U11–10D is predicted to base-pair with positions 11–27 (see Materials and Methods). Because the U11 sequence targeted by U11–10D is very close to the region proposed to interact with the 5′ splice site (27), it is possible that crosslinking might have created interference with U11–10D-directed RNase H digestion. Alternatively, the highly structured character of U11 RNA itself might render the U11–10D targeted region only marginally accessible even under “denaturing” conditions (unpublished observation).
To confirm the presence of U11 in crosslinks 1 and 2, we performed Northern analysis after RNase H digestion of crosslinked species generated using unlabeled P120-4SU+2. After irradiation of the splicing reaction mixture, total RNAs were recovered and portions subjected to RNase H digestion, each directed by a different oligonucleotide, including the two antisense U11 oligonucleotides. Gel-fractionated RNAs from digested samples then were transferred to nitrocellulose membrane and probed separately with antisense U11 RNA, antisense U2 RNA, and antisense P120 DNA oligonucleotides. The results shown in Fig. 3 reveal that oligonucleotide U11–10L (lane 2), which directs RNase H cleavage of crosslinks 1 and 2 (A) targets the degradation of free U11 (D). In contrast, oligonucleotide U11–10D (lane 3), which does not direct the cleavage of crosslinks 1 and 2 (A), does not target the degradation of free U11 RNA (D). As expected, the oligonucleotide complementary to P120 (lane 5) directed cleavage of both crosslinks 1 and 2 (A) and of free P120 RNA (B). U2 RNA remained unchanged in the presence of all oligonucleotides (C). None of these RNAs were digested in the presence of a control oligonucleotide (U1-L2, lane 4).
Figure 3.
Characterization of crosslinks 1 and 2 by RNase H digestion followed by Northern analysis. Unlabeled P120-4SU+2 was added to a splicing reaction and crosslinking performed at time 0. Recovered RNAs were subjected to RNase H digestion with no oligonucleotide (lane 1); with two different antisense U11 oligodeoxynucleotides, U11–10L (lane 2) or U11–10D (lane 3); with antisense U1 oligodeoxynucleotide U1-L2 (lane 4); or with antisense P120 oligodeoxynucleotide P110 (lane 5) (see Materials and Methods). The treated RNAs then were resolved on a 5% polyacrylamide gel and transferred to Zeta- Probe GT membrane (Bio-Rad) for Northern blotting. The top (A) and bottom (D) of the gel, which included crosslinks 1 and 2 and free U11, respectively, were probed with an antisense U11 RNA probe. In B, the part of the gel that included the P120 pre-mRNA was probed with an antisense P120 DNA probe (37). In C, the part of the gel that included U2 snRNA was probed with an antisense U2 RNA probe. Crosslinks 1 and 2, P120-4SU+2, U2, and U11 are indicated.
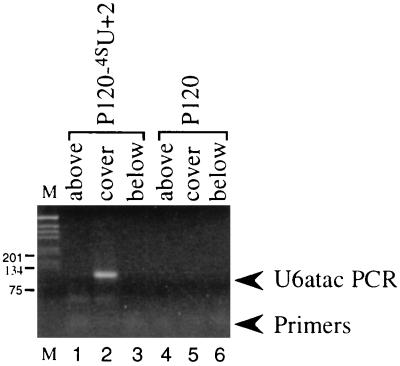
Analysis of crosslink 3 proved difficult for two reasons. First, its amount was always very low (much less than crosslinks 1 and 2). Second, most HeLa nuclear extracts produce high background in the region of the gel surrounding crosslink 3. Thus, we were unable to use direct assays, such as targeted RNase H digestion or Northern blotting, to characterize the small RNA partner in this crosslinked species. Nonetheless, the anti-TMG immunoprecipitation results (Fig. 2) clearly indicated that the partner in crosslink 3 does not contain a TMG cap. Because among all the snRNAs in the AT–AC spliceosome U6atac is the only one that possesses a 5′ non-TMG cap structure, we considered this snRNA to be the best candidate for a crosslink 3 component. To confirm the presence of U6atac, we used an indirect assay involving RT followed by PCR. P120-4SU+2 and unsubstituted P120 substrate were separately incubated with HeLa nuclear extract for 4 hr under splicing conditions and then irradiated with UV light. Total recovered RNAs were resolved in parallel lanes on a denaturing gel. The region that includes crosslink 3 and regions above and below crosslink 3 were excised from both gel lanes. Recovered RNAs served as templates for RT–PCR. The 5′ primer was designed to correspond to nucleotides 22–38, a sequence downstream of the region of U6atac proposed to interact with the intron 5′ splice site (31). The 3′ primer was complementary to the last 20 nucleotides of U6atac. The results are shown in Fig. 4: only the RNA sample from the gel slice that includes the crosslink 3 band gave a RT–PCR product (lane 2). The identity of the product as U6atac was confirmed by subsequent cloning and sequencing.
Figure 4.
Characterization of crosslink 3 by RT–PCR analysis. Unlabeled P120-4SU+2, wild-type P120, and labeled P120-4SU+2 (as a marker) were incubated individually in HeLa nuclear extract for 4 hr under splicing conditions. The reactions then were irradiated with 365-nm UV light for 10 min. RNAs were recovered and resolved on a 5% polyacrylamide/8 M urea gel. Regions above, covering, and below the crosslink 3 band in unlabeled lanes were excised to serve as templates for RT-PCR with U6atac primers (see Materials and Methods). The PCR products were resolved on a 2% agarose gel and stained with ethidium bromide. Lanes 1–3 show products from P120-4SU+2, and lanes 4–6 are from unsubstituted P120 substrate, with the regions above (lanes 1 and 4), covering (lanes 2 and 5), or below (lanes 3 and 6) the crosslink 3 band indicated. Lane M is a 1-kb marker (GIBCO/BRL).
DISCUSSION
Crosslinking experiments during in vitro splicing of an AT–AC intron containing site-specific 4SU substitutions near the 5′ splice site revealed three distinct crosslinked species. For P120-4SU+2, crosslinks 1 and 2 (which have different gel mobilities) both contain U11 snRNA, suggesting that two different sites in U11 may be involved. The other two 4SU substituted substrates (P120-4SU+4 or P120-4SU+7) appeared to yield the same two crosslinked species, which we did not characterize in such detail. Although it is possible that these species contain some snRNA other than U11, the fact that they appear with similar kinetics, are likewise ATP-independent, and have similar gel mobilities to crosslinks 1 and 2 generated with P120-4SU+2 (Fig. 1B) argues that the crosslinks generated with P120-4SU+4 and P120-4SU+7 also involve U11. We therefore deduce that these crosslinks all result from functional interactions between the 5′ splice site and the U11 snRNA at an early stage of AT–AC spliceosome assembly, in agreement with previous proposals placing nucleotides U+2, U+4, and U+7 in close contact with the 5′ end of U11 (refs. 27 and 36; see Fig. 5). Although the model predicts that nucleotides U+4 and U+7 participate in Watson–Crick base-pairing interactions, which should be unfavorable for 4SU crosslinking (12, 13), the sequence complementarity between U11 and the 5′ splice site of the P120 intron is quite limited (Fig. 5). Thus, the double-stranded region may not be as rigid as most long A-form RNA–RNA duplexes, allowing at least transient opportunity for capture by 4SU crosslinking. Primarily because of the low abundance of crosslinks 1 and 2, many attempts to identify the exact crosslinked residue(s) on U11 RNA were unsuccessful.
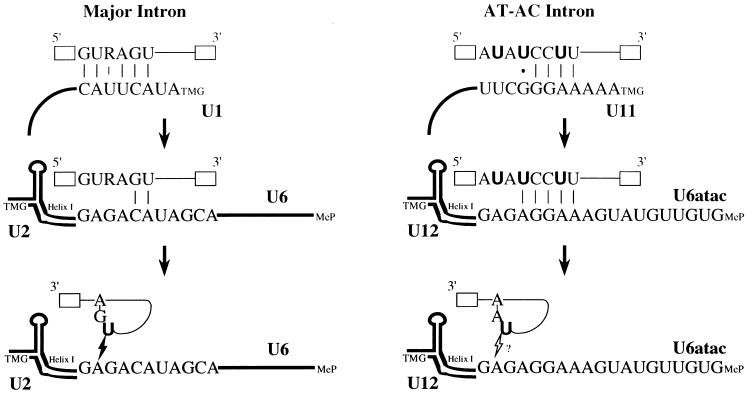
Figure 5.
Possible mechanisms of 5′ splice site recognition during splicing of major versus AT–AC introns. (Left) Sequential interactions of the 5′ splice site of a major class intron, as documented in previous studies (5–8, 13, 18–26, 40). (Right) Summary of our conclusions on how the 5′ splice site of an AT–AC intron is recognized during splicing. Crosslinks 1 and 2 are consistent with previous proposals (27, 31) that the 5′ splice site makes close contact with U11 at early times. After spliceosome assembly, U6atac replaces U11 in crosslinking to the 5′ splice site (crosslink 3). Whether crosslink 3 represents an interaction of U6atac with the 5′ splice site of the pre-mRNA before the first step of splicing or with the 2/3 intermediate after the first step is not known. Because mapping of the crosslinks was not possible, residues on U11 and U6atac interacting with 4SU (in bold) are not designated. In the pre-mRNAs, open boxes and thin lines represent exons and introns, respectively. In the snRNAs, the 5′ cap structures are indicated as TMG (U1, U2, U11 and U12) or MeP, γ-monomethyl phosphate (U6 and U6atac). Helix I indicates the conserved base-pairing interaction between U6 and U2 (21) and between U6atac and U12 (31). The lightning bolt between U6 and the 2/3 lariat intermediate represents the 4SU (in bold) crosslink detected in the major spliceosome (13). The open lightning bolt with a question mark describes a possible interaction (crosslink 3) between U6atac and the lariat 2/3 intermediate in the AT–AC spliceosome.
Also for crosslink 3, generated with P120-4SU+2, extremely low abundance precluded mapping of the crosslinked residue on U6atac snRNA. On the other hand, our ability to obtain a U6atac RT-PCR product suggested that the site of crosslinking to the 5′ splice site is upstream of the sequence complementary to the 5′ primer (nucleotides 22–38 of U6atac). Within this 5′ region of U6atac lies a sequence (AAGGAGAG) that is predicted to function analogously to the conserved sequence ACAGAG of U6 snRNA, which interacts with the 5′ splice site during splicing of major class introns (ref. 31, and see Fig. 5). We therefore suggest that the sequence AAGGAGAG in U6atac has become crosslinked to the 5′ splice site of the AT–AC intron in our experiments.
Because of the low abundance of crosslink 3 and the high background in the region of the gel surrounding this crosslink, we were also unsuccessful in determining the nature of the substrate (pre-mRNA or splicing intermediate) involved in crosslink 3. If it is the pre-mRNA that becomes crosslinked to U6atac snRNA, our results would support the proposal that base pairing between U6atac and the 5′ splice site occurs before the first catalytic step (ref. 31, and see Fig. 5). Genetic suppression (22, 23), psoralen crosslinking (8), and direct UV crosslinking (25) experiments all have indicated an interaction between the 5′ splice site of major class introns and the conserved ACAGAG sequence in the U6 snRNA (Fig. 5). Interestingly, however, 4SU site-specific crosslinking analyses have failed to detect this interaction (13). Instead, a crosslink between the U+2 residue of the 2/3 intermediate and the 3′-most A residue of the conserved ACAGAG sequence in U6 has been characterized for the major spliceosome (refs. 13 and 26, and see Fig. 5). Because the 4SU was placed at the same position (+2) in both cases (this study and ref. 13) and the geometry of the two spliceosomal active sites (AT–AC spliceosome versus major spliceosome) are likely to be very similar (31), it is possible that crosslink 3 detected here may likewise be a crosslink between U+2 of the 2/3 intermediate and U6atac (Fig. 5). This crosslink therefore would represent a similar functional interaction to that identified for the second step of splicing in the major spliceosome (13, 31).
In summary, our results on AT–AC intron splicing nicely confirm and extend models of pre-mRNA splicing based on previous crosslinking (8, 12, 13, 24–26), biochemical (40), and genetic suppression (5–7, 22, 23) studies of the major spliceosome. Like the early interaction between U1 and the 5′ splice site of the majority of introns, U11 becomes crosslinked to the 5′ splice site of the AT–AC intron substrate at very early times (Fig. 5). These early interactions (or crosslinks) are not ATP-dependent and gradually diminish as spliceosome assembly proceeds. Subsequently, U6atac appears to replace U11 in interacting with the 5′ splice site in the catalytically active spliceosome, just as U6 replaces U1 (Fig. 5). Alternatively, conformational changes occurring after the first step may allow U6atac to interact with the 5′ splice site of the lariat 2/3 intermediate, similar to an interaction detected between U6 and the 5′ splice site of 2/3 intermediate in the major spliceosome (refs. 13 and 26, and see Fig. 5). The comparable kinetics of appearance of the dynamic interactions observed here and in previous studies (8, 12, 13) reinforce the notion that the splicing of AT–AC and major class introns is mechanistically highly conserved. Our data strongly support the idea that U11 and U6atac snRNAs in the AT–AC spliceosome are indeed analogs of U1 and U6, respectively, in the major class spliceosome.
Acknowledgments
We thank T. W. Nilsen, W.-Y. Tarn, T. McConnell, and J. Mermoud for critical reading of the manuscript. Y.-T.Y. was supported by the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation. This work was supported by Grant GM26154 from the U.S. Public Health Service.
ABBREVIATIONS
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein particle
- 4SU
4-thiouridine
- RT
reverse transcription
- TMG
trimethyl guanosine
References
- 1.Moore M J, Query C C, Sharp P A. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 303–357. [Google Scholar]
- 2.Ruby S W, Abelson J. Trends Genet. 1991;7:79–85. doi: 10.1016/0168-9525(91)90276-V. [DOI] [PubMed] [Google Scholar]
- 3.Steitz J A, Black D L, Gerke V, Parker K A, Kramer A, Frendewey D, Keller W. In: Structure and Function of Major Small Nuclear Ribonucleoprotein Particles. Birnstiel M L, editor. Heidelberg: Springer; 1988. pp. 115–154. [Google Scholar]
- 4.Nilsen T W. Cell. 1994;78:1–4. doi: 10.1016/0092-8674(94)90563-0. [DOI] [PubMed] [Google Scholar]
- 5.Zhuang Y, Weiner A M. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 6.Seraphin B, Rosbash M. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siliciano P G, Guthrie C. Genes Dev. 1988;2:1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- 8.Wassarman D A, Steitz J A. Science. 1992;257:1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- 9.Zhuang Y, Weiner A M. Genes Dev. 1989;3:1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Manley J L. Genes Dev. 1989;3:1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- 11.Parker R, Siliciano P, Guthrie C. Cell. 1987;49:220–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 12.Wyatt J R, Sontheimer E J, Steitz J A. Genes Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- 13.Sontheimer E J, Steitz J A. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 14.Newman A, Norman C. Cell. 1991;65:115–123. doi: 10.1016/0092-8674(91)90413-s. [DOI] [PubMed] [Google Scholar]
- 15.Newman A, Norman C. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 16.Cortes J J, Sontheimer E J, Seiwert S D, Steitz J A. EMBO J. 1993;12:5181–5189. doi: 10.1002/j.1460-2075.1993.tb06213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman A J, Teigelkamp S, Beggs J D. RNA. 1995;1:968–980. [PMC free article] [PubMed] [Google Scholar]
- 18.Hausner T-P, Giglio L M, Weiner A M. Genes Dev. 1990;4:2146–2156. doi: 10.1101/gad.4.12a.2146. [DOI] [PubMed] [Google Scholar]
- 19.Datta B, Weiner A M. Nature (London) 1991;352:821–824. doi: 10.1038/352821a0. [DOI] [PubMed] [Google Scholar]
- 20.Wu J, Manley J L. Nature (London) 1991;352:818–821. doi: 10.1038/352818a0. [DOI] [PubMed] [Google Scholar]
- 21.Madhani H D, Guthrie C. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- 22.Lesser CF, Guthrie C. Science. 1993;262:1982–1988. doi: 10.1126/science.8266093. [DOI] [PubMed] [Google Scholar]
- 23.Kandels-Lewis S, Seraphin B. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- 24.Sawa H, Shimura Y. Genes Dev. 1992;6:244–254. doi: 10.1101/gad.6.2.244. [DOI] [PubMed] [Google Scholar]
- 25.Sawa H, Shimura Y. Proc Natl Acad Sci USA. 1992;89:11269–11273. doi: 10.1073/pnas.89.23.11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim C H, Abelson J. RNA. 1996;2:995–1010. [PMC free article] [PubMed] [Google Scholar]
- 27.Hall S L, Padgett R A. J Mol Biol. 1994;239:357–365. doi: 10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- 28.Mount S M. Science. 1996;271:1690–1692. doi: 10.1126/science.271.5256.1690. [DOI] [PubMed] [Google Scholar]
- 29.Tarn W-Y, Steitz J A. Trends Biochem Sci. 1997;22:132–137. doi: 10.1016/s0968-0004(97)01018-9. [DOI] [PubMed] [Google Scholar]
- 30.Tarn W-Y, Steitz J A. Cell. 1996;84:801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- 31.Tarn W-Y, Steitz J A. Science. 1996;273:1824–1832. doi: 10.1126/science.273.5283.1824. [DOI] [PubMed] [Google Scholar]
- 32.Yu Y-T, Tarn W-Y, Yario T A, Steitz J A. Exp Cell Res. 1996;229:276–281. doi: 10.1006/excr.1996.0372. [DOI] [PubMed] [Google Scholar]
- 33.Nilsen T W. Science. 1996;273:1813. doi: 10.1126/science.273.5283.1813. [DOI] [PubMed] [Google Scholar]
- 34.Wu Q, Krainer A R. Science. 1996;274:1005–1008. doi: 10.1126/science.274.5289.1005. [DOI] [PubMed] [Google Scholar]
- 35.Hall S L, Padgett R A. Science. 1996;271:1716–1718. doi: 10.1126/science.271.5256.1716. [DOI] [PubMed] [Google Scholar]
- 36.Kolossova I, Padgett R A. RNA. 1997;3:227–233. [PMC free article] [PubMed] [Google Scholar]
- 37.Yu, Y.-T. & Steitz, J. A. (1997) RNA, in press.
- 38.Yu Y-T, Maroney P A, Darzynkiewicz E, Nilsen T W. RNA. 1995;1:46–54. [PMC free article] [PubMed] [Google Scholar]
- 39.Hannon G J, Maroney P A, Denker J A, Nilsen T W. Cell. 1990;61:1247–1255. doi: 10.1016/0092-8674(90)90689-c. [DOI] [PubMed] [Google Scholar]
- 40.Konforti B B, Koziolkiewicz M J, Konarska M M. Cell. 1993;75:863–873. doi: 10.1016/0092-8674(93)90531-t. [DOI] [PubMed] [Google Scholar]