Abstract
Prostate cancer is a common complex disease that disproportionately affects men of African descent. Recently, several different common variants on chromosome 8q24 have been shown to be associated with prostate cancer in multiple studies and ethnic groups. The objective of this study was to confirm the association of 8q24 markers with prostate cancer in African Americans. We genotyped 24 markers along 8q24 and 80 unlinked ancestry informative markers in a hospital-based case-control sample of 1057 African American men (490 prostate cancer cases and 567 controls). Association analyses of 8q24 markers with prostate cancer risk were adjusted for both global and local 8q24 admixture stratification using estimates from ancestry informative markers. We report that rs7008482, which maps to the 8q24.13 region, is an additional independent prostate cancer risk variant (P = 5 × 10−4), and we also replicate the association of rs16901979 with prostate cancer (P = 0.002). Other published risk variants in the region such as rs1447295 and rs6983267 showed a similar direction and magnitude of effect, but were not significant in our population. Both rs7008482 and rs16901979 independently predicted risk and remained significant (P < 0.001) after controlling for each other. Our data combined with additional replications of 8q24 markers provide compelling support for multiple regions of risk for prostate cancer on 8q24.
Prostate cancer (PCa) still remains the most common male-specific malignancy diagnosed in the United States. In 2007 alone, about 218,890 new cases of prostate cancer and 27,050 deaths will be attributed to this disease (Jemal et al. 2007). This slight decrease when compared with previous annual estimates is due to a new method being utilized in estimating new cancer cases by the American Cancer Society. The new estimate covers about 86% versus the previous 10% of the US population (Jemal et al. 2007). Despite the slight decrease in the estimates for 2007, African American (AA) men still have an even higher incidence and mortality rate when compared with other ethnic groups (Jemal et al. 2007).
Genome-wide linkage and association studies have been used to identify genomic loci contributing to prostate cancer susceptibility. Recently, the results of a genome-wide scan in Icelandic families suggested strong evidence for association between a microsatellite marker at 8q24 (DG8S737) and prostate cancer risk (Amundadottir et al. 2006). Initially, a maximum LOD score of 2.11 was achieved for microsatellite marker DG8S529 in a genome-wide scan of 323 Icelandic extended families. Additional genotyping in 869 unrelated Icelandic prostate cancer cases and 596 controls using 358 microsatellites within the region of interest confirmed the original linkage results, with allele −8 of marker DG8S373 showing the most significant evidence of association (OR = 1.79; P = 3.0 × 10−6) (Amundadottir et al. 2006). Additional analyses were performed in Icelandic cases and controls using a set of ∼60 single nucletide polymorphisms (SNPs) and additional microsatellites encompassing DG8S737. Among the 37 SNPs that were associated, rs1447295 was most significant (OR = 1.72; P = 1.7 × 10−9) (Amundadottir et al. 2006). Using an expanded cohort, the association with prostate cancer was confirmed among Swedish, European American, and African American men (Amundadottir et al. 2006). Interestingly, microsatellite DG8S737 was significant among the African American prostate cancer cases; however, the association with rs1447295 was not statistically significant. Nonetheless, the authors concluded that for DG8S737, the population attributable risk (PAR) of 16% in African Americans was considerably higher than the PAR for the European populations studied (5%–11%) and suggest that this allele may partially account for the significant difference in incidence seen among African American men (Amundadottir et al. 2006). The recent results of a whole-genome SNP scan from this same research group revealed a strong genome-wide association to markers mapping within 8q24 (Gudmundsson et al. 2007).
In an independent study, a multiethnic cohort of prostate cancer cases and controls was evaluated with ∼1500 genome-wide ancestry informative markers (AIMs). Analysis of the admixture data from ∼1600 African Americans showed a statistically significant association to markers mapping to 8q24, with the signal primarily associated with diagnosis <72 yr of age (Freedman et al. 2006). Specific analysis of the two originally associated markers, DG8S737 and rs1447295, revealed a positive association with prostate cancer among Japanese Americans, Native Hawaiians, Latino Americans, and European Americans. However, these analyses failed to yield statistically significant results in African Americans after correcting for a local rise in genetic admixture within the 8q24 region, suggesting that the true association to 8q24 in African Americans could not be explained by these alleles (Freedman et al. 2006). The association to 8q AIMs were statistically significant and a confirmation report from this same group, using fine mapping markers, suggests at least three independent loci within 8q24, with one SNP rs6983561 achieving a P-value of 7.9 × 10−19 (Haiman et al. 2007).
Furthermore, results of an independent genome-wide scan in ∼1200 prostate cancer cases and ∼1200 matched controls were reported, with markers within 8q24 showing the strongest evidence of association (Yeager et al. 2007). Recently, there have been four independent confirmation reports replicating the association of prostate cancer with variants mapping within the 8q24 region of the genome (Schumacher et al. 2007; Severi et al. 2007; Suuriniemi et al. 2007; Wang et al. 2007). These associations along with the fact that the chromosome region 8q24 is amplified in most prostate tumors and has been associated with aggressive disease and poor prognosis (Jenkins et al. 1997; El Gedaily et al. 2001; Tan et al. 2003; Liu et al. 2006; Sun et al. 2007; Tan and Chow 2007) make it an interesting region to explore prostate cancer risk.
Here, we report on a structured association analysis of the original 8q24 markers (DG8S737 and rs1447295) reported by Amundadottiir et al. (2006) performed in 490 African American prostate cancer cases and 567 matched controls. In addition, we genotyped 23 additional SNPs mapping to chromosome 8 in all subjects. All of our cases and controls were also genotyped with a set of 80 genome-wide AIMs to correct for individual ancestry. As a number of the SNPs mapping to 8q were also AIMs, we were also able to correct for any local changes in admixture along 8q24.
Results
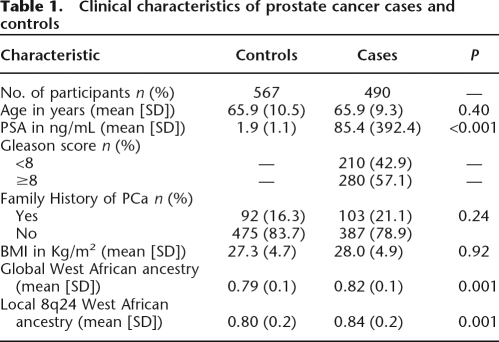
The clinical characteristics of our prostate cancer cases and controls are shown in Table 1. Cases and controls were not significantly different except for mean PSA and mean West African genetic ancestry values. Mean West African ancestry was significantly higher among the PCa cases than controls for both local 8q individual ancestry (LIA) and global individual ancestry (GIA) (P = 0.001) (Table 1).
Table 1.
Clinical characteristics of prostate cancer cases and controls
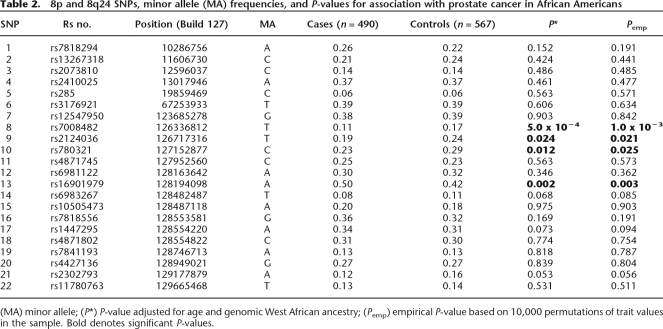
Twenty-four of the SNPs genotyped map to human chromosome 8. Three SNPs map to 8p and the other 21 SNPs map to 8q24. We tested genotype frequencies for significant departure from Hardy Weinberg (HW) proportions independently in the cases and controls. Two SNPs were excluded; rs10086908 due to strong departure from Hardy Weinberg equilibrium (P = 1 × 10−24) and rs1668875, because it was monomorphic in our population (Supplemental Table 1). Allele frequencies for the remaining 22 SNPs are detailed in Table 2. Minor allele frequencies ranged from 6% to 42% in the African American controls.
Table 2.
8p and 8q24 SNPs, minor allele (MA) frequencies, and P-values for association with prostate cancer in African Americans
(MA) minor allele; (P*) P-value adjusted for age and genomic West African ancestry; (Pemp) empirical P-value based on 10,000 permutations of trait values in the sample. Bold denotes significant P-values.
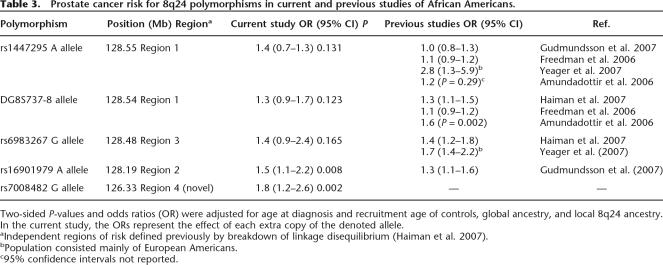
Three SNPs mapping to 8q24.13, rs7008482, rs2124036, and rs780321, and one SNP mapping to 8q24.21, rs16901979, appeared to influence prostate cancer risk in our African American cohort after controlling for age and GIA (Table 2). To be more confident in our findings we again performed our regression analyses; however, this time we controlled for differences in locus-specific ancestry along 8q24 using the LIA estimates. We determined that the slight associations for rs2124036 and rs780321 were likely due to local admixture. Two SNPs remained significantly associated with prostate cancer even after including LIA as a covariate, rs7008482 (OR = 1.8; CI = 1.2–2.6; P = 0.002) and rs16901979 (OR = 1.5, CI = 1.1–2.2; P = 0.008) (Table 3). Table 3 details prostate cancer risk associated with polymorphisms across several regions along 8q24 in this and previous studies of African Americans. Surprisingly, there is very little linkage disequilibrium between markers in close proximity in regions 1–3 (Witte 2007). The two significant SNPs we observed in this study were found in region 2 (rs16901979) and a new region denoted region 4 at 126.33Mb (rs7008482).
Table 3.
Prostate cancer risk for 8q24 polymorphisms in current and previous studies of African Americans.
Two-sided P-values and odds ratios (OR) were adjusted for age at diagnosis and recruitment age of controls, global ancestry, and local 8q24 ancestry. In the current study, the ORs represent the effect of each extra copy of the denoted allele.
aIndependent regions of risk defined previously by breakdown of linkage disequilibrium (Haiman et al. 2007).
bPopulation consisted mainly of European Americans.
c95% confidence intervals not reported.
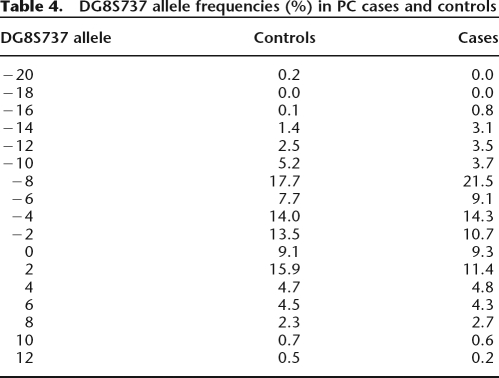
We included two markers (rs1447295 and DG8S737) in our study to directly compare the results previously observed in African Americans by others. The microsatellite marker DG8S737 was genotyped for all 490 African American cases and 567 age- and ethnicity-matched controls. Table 4 reveals the frequencies of the 17 alleles for DG8S737 observed in the sample. While the frequencies were consistent with what was observed in the literature for African Americans, we did not observe association between any of the DG8S373 alleles and prostate cancer (P = 0.065). Also, the previously reported risk allele for SNP rs1447295 revealed no association with prostate cancer in our population (Tables 2, 3). Additionally, two SNPs within a 1.2-kb region flanking and in LD with rs1447295 were typed, rs7818556 and rs4871802, neither of which revealed an association with prostate cancer (Table 2; Fig. 1). We did observe a slight association at rs1447295 when we stratified our populations by age of diagnosis (≤60; P = 0.03), similar to Schumacher et al. (2007) and Freedman et al. (2006), but when we controlled for locus-specific ancestry, the association became nonsignificant. Additionally, further analyses were carried out in which samples were stratified by family history and also pathological grade (Gleason score ≤7 vs. ≥8). We did not observe any SNP association with family history and Gleason score.
Table 4.
DG8S737 allele frequencies (%) in PC cases and controls
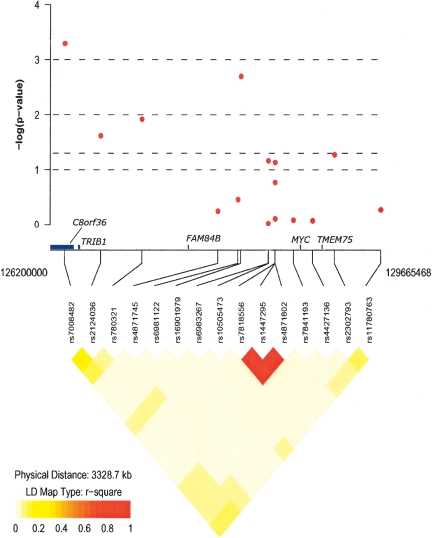
Figure 1.
Plot of empirical P-values from tests of association between SNPs across 8q24 region and prostate cancer risk in African Americans. Plot includes a display of linkage disequilibrium (R2) plot and positions of SNPs along chromosome 8q24 (1262000000–129665468). Association analysis P-values are adjusted for age and global West African ancestry.
To help determine the effect size of the rs7008482 and rs16901979 SNP associations in relation to admixture effects we performed three logistic regression analyses for both markers. First we examined the SNP association unadjusted for individual ancestry, then adjusting for genome-wide ancestry, and finally controlling for genome-wide and local 8q24 individual ancestry. Assuming an additive model of effect per allele copy, the odds ratios for the three analyses were 1.89, 1.86, and 1.83, respectively, for rs7008482 and 1.52, 1.51, and 1.45, respectively, for rs16901979. Thus, ancestry effects at the genome-wide and local level contributed little if any to the rs7008482 and rs16901979 associations with prostate cancer risk.
Finally, we tested the possibility of whether SNPs rs7008482 and rs16901979 could fully explain the association with prostate cancer, or whether the associations were dependent on each other. To do this we performed a stepwise regression using each SNP individually as causal while controlling for the others. No other SNPs in the region could explain the association when either rs7008482 or rs16901979 were considered causal in the analysis.
Discussion
Recently, multiple independent genetic variants on chromosome region 8q24 have been implicated in prostate cancer risk (Witte 2007). In this study, we report on a genetic association study in 1057 African Americans for 8q24 prostate cancer risk loci. We have combined genotyping of the originally associated markers at 8q24.21 (Amundadottir et al. 2006) with genotyping of additional chromosome 8 markers, including a series of admixture informative SNPs. Neither the −8 allele at microsatellite marker DG8S737 nor SNP rs1447295 was significantly associated with prostate cancer risk in our African American cohort (Table 4). In other studies of African Americans, no significant association of prostate cancer was observed with DG8S737 (Freedman et al. 2006); however, a positive association has been reported between prostate cancer and SNP rs1447295 in an African American cohort when the data were stratified according to age (≤65 yr) (Schumacher et al. 2007). These results are similar to those reported by others for African Americans (Freedman et al. 2006; Gudmundsson et al. 2007). Ignoring differences in local ancestry across African American subjects may account for the differences observed in previous studies. Yet, strong and replicable association with both DG8S737 and rs1447295 are seen among cohorts of European ancestry (Schumacher et al. 2007; Severi et al. 2007; Suuriniemi et al. 2007; Wang et al. 2007). In a study of 1121 European descent familial and sporadic PCa cases, Wang et al. (2007) reported significant association with both familial (P = 0.0004) and aggressive prostate cancer (P = 0.0005) and SNP rs1447295 (Wang et al. 2007). Furthermore, positive associations were also observed for the microsatellite DG8S737 and risk of familial prostate cancer (P = 0.031) and aggressive prostate cancer (P = 0.0004) (Wang et al. 2007). In an independent study, Suuriniemi et al. (2007) reported significant association of rs1447295 with prostate cancer risk in a cohort of 597 European American prostate cancer cases and 548 matched controls. In their study, there was no evidence of association for the microsatellite DG8S737 and prostate cancer risk. Finally, in a large expanded study of both breast and prostate cancer cohorts, Schumacher et al. (2007) reported strong association of increased prostate cancer risk with rs1447295 (P = 1.23 × 10−6), primarily among European Americans. However, among African Americans, significance was achieved (P = 0.011) only for early onset cases (age <65) (Schumacher et al. 2007).
Our data validate the overall results of the previous studies, suggesting the presence of multiple independent prostate cancer susceptibility loci at 8q24. Results from a genome-wide association study on 1453 Icelandic prostate cancer cases and 3064 controls using ∼300,000 SNPs revealed significant association to the original SNP rs1447295 at 8q24.1 (chromosome 8 position 128.6 Mb), and to a second SNP, rs16901979, which maps to proximal 8q24.21 (chromosome 8 position 128.2 Mb). In a separate independent genome-wide association study of 1172 European American prostate cancer cases and 1157 matched controls using 550,000 genome-wide SNPs revealed evidence of association with prostate cancer at SNP rs6983267, which maps ∼70 kb proximal to the originally associated 8q24 SNP, rs1447295 (Yeager et al. 2007). A highly significant association (P = 9.42 × 10−13) was observed at the proximal SNP, rs6983267, in an internal validation of 4296 prostate cancer cases and 4299 matched controls, further supporting multiple independent loci contributing to prostate cancer risk within the 8q24 region of the genome (Yeager et al. 2007). We note that rs6983267 was not significantly associated with prostate cancer in our study, although the magnitude of effect was similar to that found by others (Haiman et al. 2007; Yeager et al. 2007). Given that 95% confidence interval around the odds ratio estimate did not exclude previous findings (see Table 3), it may be that our inability to observe a significant association for rs6983267 is likely due to chance because of a smaller sample size than previous studies. For instance, rs6983267 was strongly associated with prostate cancer in a follow-up fine-mapping study of the 8q24 region (Haiman et al. 2007). In the Haiman et al. (2007) study, 2973 SNPs were genotyped in ∼7500 prostate cancer cases or controls from multiple ethnicities. In their study, up to five SNPs independently predicted risk of prostate cancer, with the most significant SNP being rs6983561 (P = 7.9 × 10−19). They reported three regions consisting of clusters of SNPs spanning from chromosome 8q24 position 128.1–128.6 Mb, each representing an independent predictor of prostate cancer risk (Haiman et al. 2007).
In our study, genotyping of additional 8q24 SNPs that show ancestral allele frequency differences implicated two separate risk loci on 8q24. Two SNPs provide evidence for 126.8 Mb and 128.4 Mb regions influencing PCa risk. SNPs rs7008482 and rs1001979 revealed a highly significant association with disease even after correction for age, and local and global individual ancestry. Therefore, the association we observe is unlikely to be biased due to admixture. Our most significantly associated SNP, rs7008482, represents a new region of independent risk (region 4), which is mapped to 8q24.13, ∼2.2 Mb proximal to the DG8S737/rs1447295 region at 8q24.21. SNP rs7008482 lies within an intronic region of the NSMCE2 (also called MMS21) gene, which has been shown to be involved in DNA replication, recombination, and repair (Potts and Yu 2005). The SNPs we utilized in this study were chosen for their ancestry information rather than for direct association analyses. This being the case, the SNPs were not evenly spaced, and it is likely that finer-scale genotyping will lead to additional associations in this region. Further genotyping within this region and functional analyses are underway. We also note that among our African American subjects, we did not observe an association to the 128.6 Mb region, which maps to the distal 8q24.21 locus originally reported by Amundadottir et al. 2006; although the region has been shown by others to influence risk for prostate cancer (Gudmundsson et al. 2007; Schumacher et al. 2007; Severi et al. 2007; Suuriniemi et al. 2007; Wang et al. 2007; Yeager et al. 2007).
Taken together, multiple studies, including ours, strongly support the existence of several independent susceptibility loci within the 8q24 region of the genome. While other studies reported associations within the 128–129 Mb region of 8q24, our data show an even more proximal association near position 126.2 Mb of the 8q24 region of the genome. Our data add to the continually growing body of data supporting the presence of prostate cancer risk loci at 8q24 and provide justification for further genetic investigations in this region to facilitate the identification of causal alleles. As prostate cancer disproportionately affects African Americans, the discovery of true risk alleles could have important implications for early detection of prostate cancer in this high-risk population.
Methods
Subjects
Unrelated men (n = 1057) self-described as African American were recruited between the years 2001 and 2005 from the Division of Urology at Howard University Hospital (HUH) in Washington, DC. Incident prostate cancer cases (n = 490) were identified by urologists within the division or study coordinator and confirmed by review of medical records. Control subjects (n = 567) unrelated to the cases and matched for age (±5 yr) were also ascertained from the PCA screening population of the Division of Urology at HUH. Individuals who were ever diagnosed with benign prostatic hyperplasia (BPH) and/or had an elevated prostate-specific antigen test (>2.5 ng/mL), or have had an abnormal digital rectal examination (DRE) were not included as controls. The demographic characteristics of participants in the screening program were similar to the patient population seen in the Division of Urology clinics (Table 1). Recruitment of prostate cancer cases and controls occurred concurrently and were unselected with respect to family history. All participants were between 40 and 85 yr of age. Clinical characteristics including Gleason grade, PSA, age at diagnosis, and family history were obtained for all cases from medical records. Disease aggressiveness was defined as “Low” (Gleason grade <8) or “High” (Gleason grade ≥8). The Howard University IRB approved the study and written consent was obtained from all participants.
Genotyping
A total of 104 SNPs were genotyped using the Sequenom MassARRAY platform and IPLEXchemistry. Briefly, iPLE assays were designed utilizing the Sequenom Assay Design software, allowing for single base extension (SBE) designs used for multiplexing. PCR and SBE primer sequences are available upon request. Briefly, multiplex PCR were performed to amplify 5–10 ng of genomic DNA. PCR reactions were treated with shrimp alkaline phosphatase (SAP) to neutralize unincorporated dNTPs. Subsequently, a post-PCR single-base extension reaction was performed for each multiplex reaction using concentrations of 0.625 μM for low-mass primers and 1.25 μM for high-mass primers. Reactions were diluted with 16 μL of H2O and fragments were purified with resin, spotted onto Sequenom SpectroCHIP microarrays, and scanned by MALDI-TOF mass spectrometry. Individual SNP genotype calls were generated using Sequenom TYPE software, which automatically calls allele-specific peaks according to their expected masses. SNP cluster plots for the 24 chromosome 8 SNPs are available online (Supplemental Fig. 1). SNP information is available at dbSNP (www.ncbi.nlm.nih.gov/SNP). SNP data quality was high genotyping success rate was >98.9% for the SNPs after SNPs rs10086908 and rs1668875 were removed due to departure from HWE. The microsatellite DG8S737 was genotyped using published primer sequences (Freedman et al. 2006). Microsatellite genotyping was performed using fluorescently labeled primers and capillary electrophoresis using previously described methods (Gillanders et al. 2004). Genotypes were called using GeneMapper V3.5.1 software and reviewed by two experienced technicians. Allele sizes were determined in GeneMapper using the Local Southern algorithm. Genotyping quality control for SNPs and the microsatellite was accessed using duplicate DNA samples. Concordant rates >99% were observed for all markers.
Statistical analyses
Estimating genetic ancestry
Both local 8q24 individual ancestry (LIA) and global individual ancestry (GIA) were determined for each individual using AIMs for West African and European genetic ancestry. Thirteen of the 22 SNPs typed along chromosome 8 were AIMs and were used to estimate local genetic ancestry along 8q24. An additional 80 AIMs distributed across the genome were used to estimate global ancestry and to correct for any admixture association (see Supplemental Table 2 for marker information and frequencies). Both LIA and GIA were estimated separately from the genotype data using the Bayesian Markov Chain-Monte Carlo (MCMC) method implemented in the program STRUCTURE 2.1 (Falush et al. 2003). STRUCTURE 2.1 was run under the admixture model using prior population information and independent allele frequencies for GIA estimates and the linkage model for the LIA estimates. We ran the MCMC method using K = 2 parental populations and a burn-in length of 30,000 for 70,000 repetitions.
Structured association analyses
We tested the chromosome 8 SNPs for association with prostate cancer by performing conditional logistic regression using the program PLINK (Purcell et al. 2007). In order to help determine whether our associations were valid, we performed permutation tests. Empirical P-values that corrected for multiple tests were generated by 10,000 permutations of the trait values in the sample using the Max (T) procedure. In addition, odds ratios and P-values were determined by logistic regression analyses using SAS version 6.91 (SAS Institute, Inc.). For all analyses, genetic effects were adjusted for age (at time of diagnosis for case subjects and at time of ascertainment for controls). Statistical control of admixture stratification was achieved by introducing individual ancestry estimates (LIA and GIA) also as covariates in the analyses. Associations by grade (Gleason score ≤7 vs. ≥8) were also examined by logistic regression in case-only analyses.
Acknowledgments
We thank all of the men who volunteered to participate in this genetic study. This research was funded in part by the National Institutes of Health (S06GM08016) and the Department of Defense (DAMD W81XWH-07-1-0203 and DAMD W81XWH-06-1-0066).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6782707
References
- Amundadottir L.T., Sulem P., Gudmundsson J., Helgason A., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., Sulem P., Gudmundsson J., Helgason A., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., Gudmundsson J., Helgason A., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., Helgason A., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., Benediktsdottir K.R., Cazier J.B., Sainz J., Cazier J.B., Sainz J., Sainz J., et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- El Gedaily A., Bubendorf L., Willi N., Fu W., Richter J., Moch H., Mihatsch M.J., Sauter G., Gasser T.C., Bubendorf L., Willi N., Fu W., Richter J., Moch H., Mihatsch M.J., Sauter G., Gasser T.C., Willi N., Fu W., Richter J., Moch H., Mihatsch M.J., Sauter G., Gasser T.C., Fu W., Richter J., Moch H., Mihatsch M.J., Sauter G., Gasser T.C., Richter J., Moch H., Mihatsch M.J., Sauter G., Gasser T.C., Moch H., Mihatsch M.J., Sauter G., Gasser T.C., Mihatsch M.J., Sauter G., Gasser T.C., Sauter G., Gasser T.C., Gasser T.C. Discovery of new DNA amplification loci in prostate cancer by comparative genomic hybridization. Prostate. 2001;46:184–190. doi: 10.1002/1097-0045(20010215)46:3<184::aid-pros1022>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J.K., Stephens M., Pritchard J.K., Pritchard J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman M.L., Haiman C.A., Patterson N., McDonald G.J., Tandon A., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M., Haiman C.A., Patterson N., McDonald G.J., Tandon A., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M., Patterson N., McDonald G.J., Tandon A., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M., McDonald G.J., Tandon A., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M., Tandon A., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M., Penney K., Steen R.G., Ardlie K., John E.M., Steen R.G., Ardlie K., John E.M., Ardlie K., John E.M., John E.M., et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl. Acad. Sci. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillanders E.M., Xu J., Chang B.L., Lange E.M., Wiklund F., Bailey-Wilson J.E., Baffoe-Bonnie A., Jones M., Gildea D., Riedesel E., Xu J., Chang B.L., Lange E.M., Wiklund F., Bailey-Wilson J.E., Baffoe-Bonnie A., Jones M., Gildea D., Riedesel E., Chang B.L., Lange E.M., Wiklund F., Bailey-Wilson J.E., Baffoe-Bonnie A., Jones M., Gildea D., Riedesel E., Lange E.M., Wiklund F., Bailey-Wilson J.E., Baffoe-Bonnie A., Jones M., Gildea D., Riedesel E., Wiklund F., Bailey-Wilson J.E., Baffoe-Bonnie A., Jones M., Gildea D., Riedesel E., Bailey-Wilson J.E., Baffoe-Bonnie A., Jones M., Gildea D., Riedesel E., Baffoe-Bonnie A., Jones M., Gildea D., Riedesel E., Jones M., Gildea D., Riedesel E., Gildea D., Riedesel E., Riedesel E., et al. Combined genome-wide scan for prostate cancer susceptibility genes. J. Natl. Cancer Inst. 2004;96:1240–1247. doi: 10.1093/jnci/djh228. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J., Sulem P., Manolescu A., Amundadottir L.T., Gudbjartsson D., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., Sulem P., Manolescu A., Amundadottir L.T., Gudbjartsson D., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., Manolescu A., Amundadottir L.T., Gudbjartsson D., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., Amundadottir L.T., Gudbjartsson D., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., Gudbjartsson D., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., Bergthorsson J.T., Agnarsson B.A., Baker A., Agnarsson B.A., Baker A., Baker A., et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- Haiman C.A., Patterson N., Freedman M.L., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., Patterson N., Freedman M.L., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., Freedman M.L., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., Neubauer J., Tandon A., Schirmer C., McDonald G.J., Tandon A., Schirmer C., McDonald G.J., Schirmer C., McDonald G.J., McDonald G.J., et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A., Siegel R., Ward E., Murray T., Xu J., Thun M.J., Siegel R., Ward E., Murray T., Xu J., Thun M.J., Ward E., Murray T., Xu J., Thun M.J., Murray T., Xu J., Thun M.J., Xu J., Thun M.J., Thun M.J. Cancer statistics, 2007. CA Cancer J. Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- Jenkins R.B., Qian J., Lieber M.M., Bostwick D.G., Qian J., Lieber M.M., Bostwick D.G., Lieber M.M., Bostwick D.G., Bostwick D.G. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 1997;57:524–531. [PubMed] [Google Scholar]
- Liu W., Chang B., Sauvageot J., Dimitrov L., Gielzak M., Li T., Yan G., Sun J., Sun J., Adams T.S., Chang B., Sauvageot J., Dimitrov L., Gielzak M., Li T., Yan G., Sun J., Sun J., Adams T.S., Sauvageot J., Dimitrov L., Gielzak M., Li T., Yan G., Sun J., Sun J., Adams T.S., Dimitrov L., Gielzak M., Li T., Yan G., Sun J., Sun J., Adams T.S., Gielzak M., Li T., Yan G., Sun J., Sun J., Adams T.S., Li T., Yan G., Sun J., Sun J., Adams T.S., Yan G., Sun J., Sun J., Adams T.S., Sun J., Sun J., Adams T.S., Sun J., Adams T.S., Adams T.S., et al. Comprehensive assessment of DNA copy number alterations in human prostate cancers using Affymetrix 100K SNP mapping array. Genes Chromosomes Cancer. 2006;45:1018–1032. doi: 10.1002/gcc.20369. [DOI] [PubMed] [Google Scholar]
- Potts P.R., Yu H., Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sklar P., de Bakker P.I., Daly M.J., de Bakker P.I., Daly M.J., Daly M.J., et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher F.R., Feigelson H.S., Cox D.G., Haiman C.A., Albanes D., Buring J., Calle E.E., Chanock S.J., Colditz G.A., Diver W.R., Feigelson H.S., Cox D.G., Haiman C.A., Albanes D., Buring J., Calle E.E., Chanock S.J., Colditz G.A., Diver W.R., Cox D.G., Haiman C.A., Albanes D., Buring J., Calle E.E., Chanock S.J., Colditz G.A., Diver W.R., Haiman C.A., Albanes D., Buring J., Calle E.E., Chanock S.J., Colditz G.A., Diver W.R., Albanes D., Buring J., Calle E.E., Chanock S.J., Colditz G.A., Diver W.R., Buring J., Calle E.E., Chanock S.J., Colditz G.A., Diver W.R., Calle E.E., Chanock S.J., Colditz G.A., Diver W.R., Chanock S.J., Colditz G.A., Diver W.R., Colditz G.A., Diver W.R., Diver W.R., et al. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–2956. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- Severi G., Hayes V.M., Padilla E.J., English D.R., Southey M.C., Sutherland R.L., Hopper J.L., Giles G.G., Hayes V.M., Padilla E.J., English D.R., Southey M.C., Sutherland R.L., Hopper J.L., Giles G.G., Padilla E.J., English D.R., Southey M.C., Sutherland R.L., Hopper J.L., Giles G.G., English D.R., Southey M.C., Sutherland R.L., Hopper J.L., Giles G.G., Southey M.C., Sutherland R.L., Hopper J.L., Giles G.G., Sutherland R.L., Hopper J.L., Giles G.G., Hopper J.L., Giles G.G., Giles G.G. The common variant rs1447295 on chromosome 8q24 and prostate cancer risk: Results from an Australian population-based case-control study. Cancer Epidemiol. Biomarkers Prev. 2007;16:610–612. doi: 10.1158/1055-9965.EPI-06-0872. [DOI] [PubMed] [Google Scholar]
- Sun J., Liu W., Adams T.S., Sun J., Li X., Turner A.R., Chang B., Kim J.W., Zheng S.L., Isaacs W.B., Liu W., Adams T.S., Sun J., Li X., Turner A.R., Chang B., Kim J.W., Zheng S.L., Isaacs W.B., Adams T.S., Sun J., Li X., Turner A.R., Chang B., Kim J.W., Zheng S.L., Isaacs W.B., Sun J., Li X., Turner A.R., Chang B., Kim J.W., Zheng S.L., Isaacs W.B., Li X., Turner A.R., Chang B., Kim J.W., Zheng S.L., Isaacs W.B., Turner A.R., Chang B., Kim J.W., Zheng S.L., Isaacs W.B., Chang B., Kim J.W., Zheng S.L., Isaacs W.B., Kim J.W., Zheng S.L., Isaacs W.B., Zheng S.L., Isaacs W.B., Isaacs W.B., et al. DNA copy number alterations in prostate cancers: A combined analysis of published CGH studies. Prostate. 2007;67:692–700. doi: 10.1002/pros.20543. [DOI] [PubMed] [Google Scholar]
- Suuriniemi M., Agalliu I., Schaid D.J., Johanneson B., McDonnell S.K., Iwasaki L., Stanford J.L., Ostrander E.A., Agalliu I., Schaid D.J., Johanneson B., McDonnell S.K., Iwasaki L., Stanford J.L., Ostrander E.A., Schaid D.J., Johanneson B., McDonnell S.K., Iwasaki L., Stanford J.L., Ostrander E.A., Johanneson B., McDonnell S.K., Iwasaki L., Stanford J.L., Ostrander E.A., McDonnell S.K., Iwasaki L., Stanford J.L., Ostrander E.A., Iwasaki L., Stanford J.L., Ostrander E.A., Stanford J.L., Ostrander E.A., Ostrander E.A. Confirmation of a positive association between prostate cancer risk and a locus at chromosome 8q24. Cancer Epidemiol. Biomarkers Prev. 2007;16:809–814. doi: 10.1158/1055-9965.EPI-06-1049. [DOI] [PubMed] [Google Scholar]
- Tan J.M., Chow V.T., Chow V.T. Cellular expression, localization and interactions of the product of the human MOST-1 gene associated with breast and prostate cancers. Int. J. Oncol. 2007;30:81–89. [PubMed] [Google Scholar]
- Tan J.M., Tock E.P., Chow V.T., Tock E.P., Chow V.T., Chow V.T. The novel human MOST-1 (C8orf17) gene exhibits tissue specific expression, maps to chromosome 8q24.2, and is overexpressed/amplified in high grade cancers of the breast and prostate. Mol. Pathol. 2003;56:109–115. doi: 10.1136/mp.56.2.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., McDonnell S.K., Slusser J.P., Hebbring S.J., Cunningham J.M., Jacobsen S.J., Cerhan J.R., Blute M.L., Schaid D.J., Thibodeau S.N., McDonnell S.K., Slusser J.P., Hebbring S.J., Cunningham J.M., Jacobsen S.J., Cerhan J.R., Blute M.L., Schaid D.J., Thibodeau S.N., Slusser J.P., Hebbring S.J., Cunningham J.M., Jacobsen S.J., Cerhan J.R., Blute M.L., Schaid D.J., Thibodeau S.N., Hebbring S.J., Cunningham J.M., Jacobsen S.J., Cerhan J.R., Blute M.L., Schaid D.J., Thibodeau S.N., Cunningham J.M., Jacobsen S.J., Cerhan J.R., Blute M.L., Schaid D.J., Thibodeau S.N., Jacobsen S.J., Cerhan J.R., Blute M.L., Schaid D.J., Thibodeau S.N., Cerhan J.R., Blute M.L., Schaid D.J., Thibodeau S.N., Blute M.L., Schaid D.J., Thibodeau S.N., Schaid D.J., Thibodeau S.N., Thibodeau S.N. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–2950. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- Witte J.S. Multiple prostate cancer risk variants on 8q24. Nat. Genet. 2007;39:579–580. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- Yeager M., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., Fearnhead P., Yu K., Chatterjee N., Yu K., Chatterjee N., Chatterjee N., et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]