Abstract
Imprinted genes are essential in embryonic development, and imprinting dysregulation contributes to human disease. We report two new human imprinted genes: KCNK9 is predominantly expressed in the brain, is a known oncogene, and may be involved in bipolar disorder and epilepsy, while DLGAP2 is a candidate bladder cancer tumor suppressor. Both genes lie on chromosome 8, not previously suspected to contain imprinted genes. We identified these genes, along with 154 others, based on the predictions of multiple classification algorithms using DNA sequence characteristics as features. Our findings demonstrate that DNA sequence characteristics, including recombination hot spots, are sufficient to accurately predict the imprinting status of individual genes in the human genome.
A gene is imprinted if the expression of one of its alleles is silenced depending on the parent from which that allele was inherited (Reik and Walter 2001). This functionally haploid state eliminates the protection that diploidy normally confers against the deleterious effects of recessive mutations. Moreover, the expression of imprinted genes can be deregulated epigenetically. Thus, imprinted genes represent susceptibility loci that can be functionally altered by both genetic and epigenetic events (Jirtle and Skinner 2007). Identifying genes that are imprinted in the human genome and determining the factors responsible for epigenetic establishment and maintenance of imprinting control are critical goals.
Identifying imprinted genes experimentally is challenging because the monoallelic expression of an imprinted gene may occur only in one of possibly several isoforms, only in particular tissues, or only at particular stages of development. Consequently, failure to confirm imprinting in a specific tissue at a specific stage of development for a specific splice variant does not eliminate the possibility that a different isoform may be imprinted in some other tissue at some other stage of development. Although estimates of imprinted gene prevalence in the human genome vary, they hover around 1%. Consequently, in the absence of any method for prioritizing genes, an average of 100 genes must be examined (perhaps in a broad range of tissues and at many stages of development) before a new imprinted gene can be identified. Indeed, experimental identification of human imprinted genes to date has been slow: The untranslated mRNA H19 was the first gene shown to be imprinted in human (Zhang and Tycko 1992), and since its discovery in 1992, only ∼40 additional human imprinted genes have been identified (Morison et al. 2005).
We set out to develop a computational method for predicting the genome-wide imprint status of human genes, the output of which could be used to prioritize genes for experimental identification. Since the concentration of certain types of repeated elements and other DNA sequence characteristics has been shown to differ between monoallelically and biallelically expressed genes (Greally 2002; Ke et al. 2002; Allen et al. 2003), we predicted each gene’s imprint status based on nearby sequence features. As no simple sequence patterns are known to accurately distinguish imprinted genes from nonimprinted ones, we adopted a sophisticated machine learning strategy, taking care to guard against the possibility of overfitting.
Although we previously demonstrated the feasibility of this kind of approach by identifying novel imprinted genes in the mouse genome (Luedi et al. 2005), simply mapping our murine predictions onto the human genome by homology would result in significant error. While some genes are imprinted in both mouse and human, others including Igf2r, Ascl2, Tspan32, Cd81, Tssc4, Nap1l4, Gatm, Dcn, and Impact are imprinted in mouse but not human (Morison et al. 2005; Monk et al. 2006). Conversely, the homeobox gene DLX5 is imprinted in human (Okita et al. 2003) but not mouse (although a subtle maternal preference was reported in the mouse brain) (Kimura et al. 2004; Horike et al. 2005). This discordance means our earlier mouse predictions are not ideal for predicting imprinted genes in human.
Here, we describe a new algorithm for predicting the genome-wide imprint status of human genes directly from sequence features in the human genome. Compared to our previous approach, we have included additional features and introduced additional learning algorithms to significantly reduce the possibility of methodological bias. Further, we focused our attention primarily on high-confidence predictions and demonstrated by cross-validation (CV) and independent testing that our new predictions are at the same time both more sensitive and more selective than before. Finally, we used our predictions to prioritize the experimental identification of two new human imprinted genes on chromosome 8, a chromosome not previously suspected to contain imprinted genes.
Results
Conceptual approach
We adopted a more conservative approach in identifying human imprinted genes because of their important role in the etiology of human health conditions. Specifically, we applied two separate classifier learning strategies—one based on support vector machines and the other on sparse logistic regression—each with a different feature selection process. With each strategy, we trained classifiers with two different similarity kernels: linear and radial basis function (RBF). Only genes predicted to be imprinted by all four classifiers were considered “high-confidence” predictions. Although all four classifiers use the same initial training set of known imprinted genes, the combined classifier approach helps to control for biases that might arise from different choices for feature selection, classifier learning, or similarity kernel.
All four classifiers were trained on DNA sequence features collected from 40 genes known to be imprinted in human and from 52 genes known not to be imprinted in human, plus 500 randomly selected genes suspected not to be imprinted in human. We assessed the generalization accuracy of the combined classifier by both CV and an independent negative test set. In a 40-fold CV, we obtained a sensitivity of 100% (40/40 imprinted genes correctly identified) and a specificity of 99% (545/552 presumably nonimprinted genes correctly identified). The independent negative test set consisted of 13 genes with random monoallelic expression and 88 genes with biallelic expression or synchronous replication, including four genes imprinted in mouse but not human. We correctly predicted all 101 genes not to be imprinted (see Supplemental Fig. 4 for a schematic depiction of the workflow).
Genome-wide prediction of candidate imprinted genes
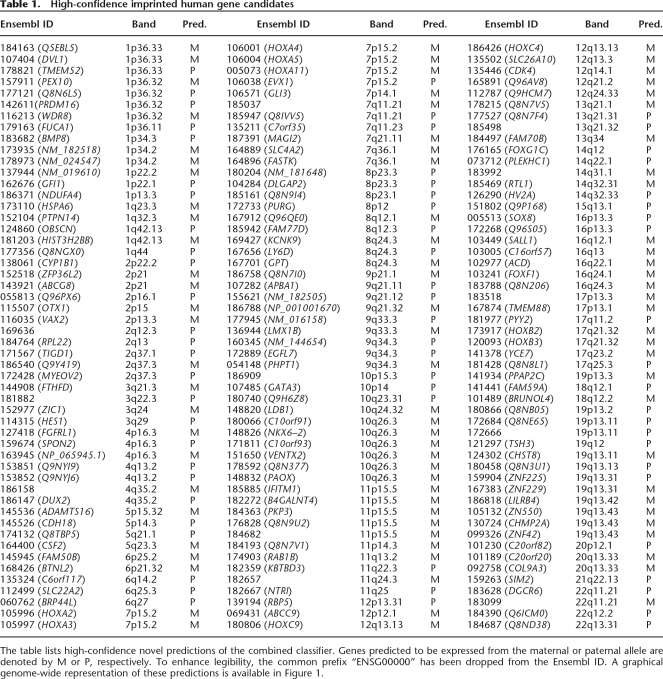
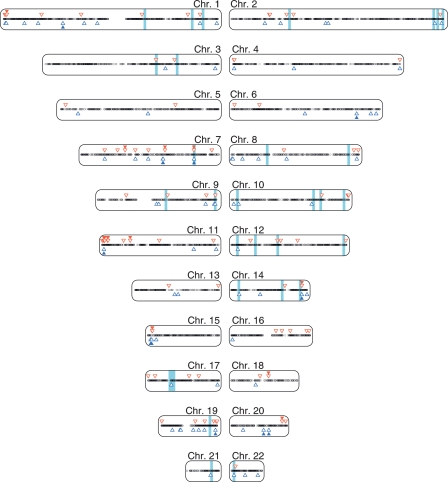
Applying the combined classifier to the entire human genome, we predicted 156 of 20,770 (0.75%) annotated autosomal genes (Ensembl v20) not previously known to be imprinted with high confidence (Table 1; Supplemental Table 1). Only chromosomes 7 and 11 showed a higher density of predicted and known imprinted genes compared to the rest of the autosome (P = 0.0014 and P = 0.0026, respectively, χ2-test with 1 df; Fig. 1). Seven chromosomal bands, however, contained a significantly higher density of imprinted gene candidates, including novel candidates related to various cancers (P < 2 × 10−8, χ2-test with 1 df; Supplemental Table 2).
Table 1.
High-confidence imprinted human gene candidates
The table lists high-confidence novel predictions of the combined classifier. Genes predicted to be expressed from the maternal or paternal allele are denoted by M or P, respectively. To enhance legibility, the common prefix “ENSG00000” has been dropped from the Ensembl ID. A graphical genome-wide representation of these predictions is available in Figure 1.
Figure 1.
Genome-wide distribution of genes proved (filled triangles) or predicted with high confidence (unfilled triangles) to be imprinted. Red downward triangles, blue upward triangles, and black dots indicate genes predicted to be maternally, paternally, or biallelically expressed, respectively. Light blue bars highlight a 3-Mb region centered on the linkage regions presented in Supplemental Table 6.
The high-density bands 15q12 and 7q21.3 contain exclusively known imprinted genes. Included in the high-density 11p15.5 band are well-known imprinted genes, such as H19 and IGF2, and five novel candidates located further distal, including PKP3, an oncogene involved in lung cancer (Furukawa et al. 2005). The high-density band 1p36.32 includes the known imprinted gene TP73 along with the novel candidate PRDM16, associated with leukemia (Du et al. 2005). The ortholog of this gene was also predicted to be imprinted in mouse (Luedi et al. 2005). The high-density 14q32.31 band contains the known imprinted gene MEG3 along with the novel candidate RTL1, which is imprinted in the mouse (Seitz et al. 2003) and sheep (Charlier et al. 2001). Finally, the high-density 10q26.3 band includes the novel candidate NKX6-2, which is preferentially expressed in the brain (Lee et al. 2001) and was predicted to be imprinted in the mouse (Luedi et al. 2005). NKX6-2 was predicted to be maternally expressed, along with four neighboring candidate genes. These candidates are 4.7–5.7 Mb from the marker D10S217, which is maternally linked to male sexual orientation (Mustanski et al. 2005). A germline differentially methylated region has been found within this interval (coordinate 135.1 Mb) (Strichman-Almashanu et al. 2002), lending further support to the prediction of imprinted genes within the immediate vicinity of this region.
Previous efforts to determine the sequence characteristics that discriminate imprinted from nonimprinted genes have demonstrated that imprinted loci are deficient in short interspersed transposable elements (SINEs), particularly in the more ancient MIRs (Greally 2002; Ke et al. 2002). We similarly find that imprinted genes contain a low concentration of SINEs in their flanking regions, especially Alu and MIR elements; however, we determined that the orientation of these repetitive elements is of greater discriminatory value than their concentration alone (Supplemental Table 4; Supplemental Fig. 1B). We observed the same result when classifying imprinted genes in mouse (Luedi et al. 2005). The relative orientation of repetitive elements in flanking sequences may contribute to physical chromosomal interactions that are important in controlling genomic imprinting. Physical chromosomal pairing has been observed in the vicinity of the imprinted Prader-Willi and Angelman syndrome loci (LaSalle and Lalande 1996), and it has been hypothesized that epigenetic differences between maternal and paternal chromosomes serve the evolutionary purpose of homologous pairing at meiosis (Pardo-Manuel de Villena et al. 2000). We further found that the family of endogenous retrovirus (ERV) elements was of greatest average importance among repetitive elements (Supplemental Fig. 1D), again just as in our murine classifier (Luedi et al. 2005). This time, however, repetitive elements within 1 kb downstream were least important (Supplemental Fig. 1C).
Among transcription factor binding sites, those of greatest importance in both feature selection strategies were CEBP, E2F, ICP4, IgPE2, NFuE1, NFuE3, PEA1, PEA2, Sp1, and SRF (Supplemental Fig. 1E). E2F family transcription factors are involved with cell proliferation, Sp1 elements have been shown to protect CpG islands from de novo methylation in the embryo (Brandeis et al. 1994), and SRF (serum response factor) is involved in the activation of “immediate early” genes (Schratt et al. 2001), in muscle differentiation (Vandromme et al. 1992; Soulez et al. 1996), and in mesoderm formation (Arsenian et al. 1998).
Prediction of parental preference
We trained a separate classifier to determine if the maternal or paternal allele of an imprinted gene is expressed. The training set included 19 maternally expressed genes and 20 paternally expressed genes (GRB10 was omitted due to its complex expression patterns) (Blagitko et al. 2000). In a 19-fold CV, we achieved a sensitivity of 85% (17/20 paternally expressed genes correctly identified) and a specificity of 79% (15/19 maternally expressed genes correctly identified). Our ability to accurately predict the expressed parental allele of known imprinted genes in both human and mouse (Luedi et al. 2005) lends support to the suggestion that different mechanisms may be responsible for regulating paternal versus maternal imprinting (Mancini-Dinardo et al. 2006).
We predicted maternal expression for 56% (88/156) of the candidate imprinted genes, comparable to the 64% frequency found for mouse imprinted genes (Luedi et al. 2005). Among the features of greatest significance for the prediction of parental expression preference were the ratios of the relative orientation of AluJ and ERVL elements downstream (Supplemental Table 5). E4F1 transcription factor binding sites were also significantly more prevalent in the 3–4-kb upstream region of maternally expressed genes than in paternally expressed genes.
Experimental identification of new imprinted genes
Guided by the high-confidence predictions of the combined classifier, we were able to experimentally identify two new imprinted human genes, each one monoallelically expressed from the predicted parental allele (Table 1): DLGAP2 (Disks Large-Associated Protein 2) and KCNK9 (potassium channel, subfamily K, member 9). These two genes were chosen based on a number of criteria (see Supplemental Methods), including their high probabilities of being imprinted and their locations at opposite telomeric regions of chromosome 8, a human chromosome not previously known to contain imprinted genes (Morison et al. 2005). These were the only two genes selected for detailed experimental validation.
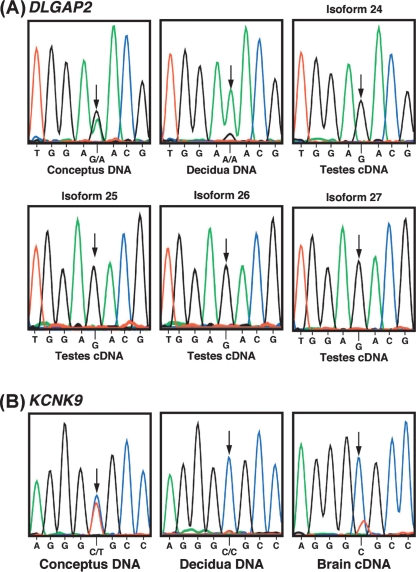
DLGAP2 is highly expressed and alternatively spliced in brain and testis (Ranta et al. 2000). It is contained within a 1.1-Mb interval on chromosome 8p23.3 that is frequently deleted in bladder cancer (Muscheck et al. 2000), making it a candidate tumor suppressor. cDNA containing polymorphic sites was generated by reverse transcription of total RNA isolated from brain and testis in heterozygous human conceptuses (N = 8; gestational age, 63–105 d). The four isoforms of DLGAP2 (splice variants 24, 25, 26, and 27) (Karolchik et al. 2003) were paternally expressed in the testis of all samples (Fig. 2A) with some evidence of imprinting relaxation in isoforms 24 (one of six individuals) and 26 (two of eight individuals), potentially resulting from the varying developmental ages of the samples, which can alter imprinted gene expression (Murphy and Jirtle 2003). In contrast, expression from both alleles was observed for all four isoforms of DLGAP2 in whole brain. MEST-AS (antisense) is another imprinted gene predominantly expressed in the testis, and like DLGAP2 is expressed only from the paternal allele (Li et al. 2002).
Figure 2.
Sequence comparisons of conceptus and maternal genomic DNA versus conceptus cDNA; arrow denotes polymorphic site. (A) The conceptus is polymorphic (G/A, dbSNP accession rs2235112) in DLGAP2, whereas the mother (maternal decidua) is homozygous (A/A). DLGAP2 isoforms 24, 25, 26, and 27 are expressed monoallelically in the testis from the paternal allele (representative DNA sequences from one individual out of eight informative conceptuses demonstrated to be imprinted at the DLGAP2 locus). (B) The conceptus is polymorphic (C/T, dbSNP accession rs2615374) in KCNK9, whereas the mother (maternal decidua) is homozygous (C/C). KCNK9 is expressed monoallelically in the brain from the maternal allele (representative DNA sequences from one individual out of nine informative conceptuses demonstrated to be imprinted at the KCNK9 locus).
KCNK9 resides at chromosomal location 8q24.3. It encodes for the TASK3 (Twik-like acid-sensitive K+) channel and is associated with a variety of human cancers (Patel and Lazdunski 2004). We tested for monoallelic expression in brain tissue because KCNK9 is predominantly expressed in the cerebellum (Medhurst et al. 2001), resides within 6.2 Mb of the marker D8S256 linked with bipolar disorder (McInnis et al. 2003) (Supplemental Table 6), and has been suggested to be a candidate for idiopathic absence epilepsies (Zara et al. 1995; Kananura et al. 2002) since it encodes for a potassium ion channel that mediates neuronal excitability. When RNA was isolated from the brains of conceptuses that were polymorphic at this locus (N = 9; gestational age, 63–98 d), KCNK9 was found to be exclusively expressed from the maternal allele in all samples (Fig. 2B).
Discussion
Parent-of-origin effects were first observed >3000 yr ago by mule breeders (Savory 1970), and the term genomic imprinting can be traced back to an article by Helen Crouse published over 45 yr ago (Crouse 1960). Nevertheless, it was not until 1991 that imprinted genes were first discovered (Barlow et al. 1991; Bartolomei et al. 1991; DeChiara et al. 1991), and the field of genomic imprinting is still considered a relatively young scientific field. Consequently, many unconfirmed—and in some cases competing—hypotheses remain regarding (1) the degree of conservation across species versus the impact that imprinting may have had on the process of speciation, (2) its origins and the mechanisms responsible for its spread throughout mammalian genomes during evolution, (3) the apparent preponderance of imprinted genes playing a role in development or regulation of growth, and (4) the influence of imprinting on conditions related to development, environment, and growth dysregulation (like cancer) (Hunter 2007). In the following sections, we consider some of these topics and indicate whether our predictions lend support to the various proposed hypotheses.
On the conservation of imprinting across species
When making predictions with a classifier, one must weigh the trade-off between sensitivity and specificity, or analogously, between false-positive rate and false-negative rate. In our previous mouse study (Luedi et al. 2005), we were more concerned with keeping the false-negative rate low. In the present human study, however, we sought to keep the false-positive rate low, defining the set of high-confidence imprinted gene candidates as the intersection of four different classifiers. Because of these different methodological choices, the number of imprinted genes predicted in the mouse and the number of high-confidence imprinted genes predicted in the human are not directly comparable. If a similar statistical methodology is adopted in the human as was used in the mouse, the number of human imprinted gene candidates increases but is still only a little more than half as large as the mouse set. While these numbers are still not directly comparable since the sequence features in the human data are slightly richer than those in mouse (see Methods), they are suggestive that the overall prevalence of imprinted genes is lower in human than in mouse.
We wished to examine the concordance between the high-confidence human imprinted candidates and the predictions for their orthologs in mouse. We identified a murine ortholog to 119 of the genes proved or predicted with high confidence to be imprinted in human. Only 38 (32%) of these genes are known or predicted to be imprinted in both species (Supplemental Table 3), and over half of these genes have subtelomeric chromosomal locations. This fraction does not change significantly if the same prediction method we used for the mouse is also applied to the human data. Hence, the lack of greater overlap is not solely due to differences in the statistical methodologies.
High levels of discordance of imprinting status between mouse and human have previously been reported (Morison et al. 2005; Monk et al. 2006). Indeed, the relative paucity of imprinted genes in human compared to mouse accords well with one of the most widely accepted theories for the origin of genomic imprinting, the “parental conflict hypothesis” (Haig and Westoby 1989). It has been speculated that mice may have expanded genomic imprinting in order for the placenta to accommodate a larger litter size and shorter gestational period, perhaps necessitating increased conservation of maternal resources (Monk et al. 2006). In contrast, human pregnancies tend to be singletons and of longer gestational time, alleviating evolutionary pressure on imprinted genes to conserve maternal resources. Hence, it seems plausible that relatively fewer genes be imprinted and maternally expressed in human (predicted proportion of 56%, versus 64% in mouse); this is also consistent with the lower predicted prevalence overall.
Although this disparity in the imprint status of genes in mouse and human may be a consequence of our computational approach, it also raises the possibility that despite their immense popularity as models of human disease, mice may not be a suitable choice for studying diseases resulting principally from the epigenetic deregulation of imprinted genes, or for assessing human risk from environmental factors that alter the epigenome.
On the evolution of imprinting
Interestingly, we found recombination data to be of considerable importance. For example, an 8-bp motif within THE1B elements that is overrepresented near recombination hotspots (Myers et al. 2005) is positively correlated with the presence of imprinted genes. In addition, the average distance between recombination hotspots and known imprinted genes is found to be about one third of that for all annotated genes. These observations lend support to the hypothesis that imprinted genes were originally linked in a few chromosomal regions and were dispersed throughout the genome by recombination events during mammalian evolution (Walter and Paulsen 2003).
In a cross-species comparison of imprinted regions between mouse and human, it has also been hypothesized that genomic imprinting may have evolved on the basis of dosage compensation following large-scale duplication events (Walter and Paulsen 2003). To investigate this, we asked if the imprinted gene candidates were more likely to have been duplicated than the rest of the autosome. When using FASTA (Pearson and Lipman 1988) to query each protein sequence against all other human proteins in our set, the distribution of the significance value for the second best hit was not different among imprinted gene candidates compared to the rest of the autosomal genes. Also, we found that the proportion of paralogs that are located on the same chromosome did not differ between the two classes of genes, nor was there a significant difference in distance to that paralog. In conclusion, these findings fail to corroborate the hypothesis of large-scale gene duplication as the driving force of imprinting evolution.
Other hypotheses for the evolution of genomic imprinting include the proposition that imprinting is a by-product of a host defense against foreign DNA (Barlow 1993; Yoder et al. 1997) or that during retrotransposition of a gene some regulatory elements may have been carried along with it that confer imprinted expression (Walter and Paulsen 2003). To investigate this, we asked whether our set of imprinted gene candidates was enriched for single-exon genes that may have been derived from multiexonic precursor paralogs. We observed no significant difference in the rate of imprinted gene candidates consisting of only a single exon, compared to the autosomal genes not predicted to be imprinted (18% vs. ∼16%). Contrary to the observation that almost all known imprinted genes derived from retrotransposition are paternally expressed (Walter and Paulsen 2003; Morison et al. 2005), we also found no statistically significant difference in the rate of intron-less genes among imprinted gene candidates with predicted maternal versus paternal expression.
On imprinting and development
A substantial number of known imprinted genes are involved in embryonic development (Reik and Walter 2001). Our predictions are consistent with this trend. Of the 146 genes with a systematic name that are proved or predicted with high confidence to be imprinted, 38% are associated with embryonic development (based on PubMed abstracts); this compares to 18% among a random set of 5000 autosomal genes predicted not to be imprinted (P < 1.7 × 10−9, χ2-test with 1 df). As one interesting example, the homeobox HOX genes play a key role in pre- and post-implantation development (Eun Kwon and Taylor 2004; Moens and Selleri 2006), and we predicted 23% of them to be imprinted (nine out of 39; P < 2 × 10−16, χ2-test with 1 df). Five of the high-confidence candidates are located in the HOXA cluster, two in each of the HOXB and HOXC clusters, and none in the HOXD cluster. Several imprinted genes are known to be regulated in mouse by the same Polycomb group proteins (Mager et al. 2003; Umlauf et al. 2004) that regulate HOX expression (Bantignies and Cavalli 2006). Thus, there may exist sequence characteristics shared in common between these two groups of genes; however, no Hox genes were predicted to be imprinted in the mouse (Luedi et al. 2005). This indicates that the high prevalence of HOX imprinted gene candidates in human does not result simply from shared sequence characteristics. Instead, it raises the possibility that monoallelic expression of HOX genes may have arisen during human evolution.
Conclusion
Genes involved in various human conditions are commonly identified by condition-oriented experimental approaches. Here, we show that potential susceptibility genes for a wide range of conditions can also be identified by defining the subset of genes that are functionally haploid because of imprinting. Mapping these imprinted genes to susceptibility loci that exhibit parent-of-origin inheritance (Supplemental Table 6) provides novel hypotheses about how complex human conditions can arise from environmental alteration of the epigenome.
Methods
Human genome data
DNA sequence and annotation data were obtained from Ensembl (http://www.ensembl.org, Version 20). We used a positive training set of 40 imprinted genes compiled from the Imprinted Gene Catalog (http://igc.otago.ac.nz/) and recent literature, as well as a negative training set of 52 genes, for which experimental evidence suggests biallelic expression. Additionally, we used random sets of 500 control genes presumed to be nonimprinted for a number of tasks. These random control genes were sampled from autosomal chromosomal bands known or not suspected to contain imprinted genes and were intended to represent the overall characteristics of biallelically expressed genes. Random control genes were used to compute top pairwise interaction terms, to carry out feature selection with the Equbits classifier, and finally to supplement the final training set that was used to learn our classifiers. To minimize bias, we resampled the set of 500 random control genes for each of these three tasks.
Feature measurements
We acquired DNA sequence feature measurements in an analogous fashion to our previous study in mouse (Luedi et al. 2005), including an additional set of data derived from repeat phase changes, recombination hotspots, and nucleosome formation potential, as explained below.
We introduced another statistic regarding the repetitive elements flanking a gene, which we will term “phase change.” We define a phase change as an instance of a repeated element changing its orientation compared to a neighboring element of the same family. We counted the number of such phase changes among retrotransposon classes such as Alus, MIRs, and LTRs within 100 kb upstream and downstream. In doing this, we noticed that within the downstream region of imprinted genes, compared to a random sample, a phase change occurred more frequently in one of the following LTRs: MLT1A0, MLT1B, MSTA, MSTB1, MLT1D, MLT2B4, or MLT1G1. Conversely, phase changes in an MLT1C LTR were underrepresented in the flanking regions of imprinted genes.
We also wanted to study whether data on recombination could be used to discern imprinted genes. We downloaded coordinates of recombination hotspots (Myers et al. 2005) from the International HapMap Project Web site (http://www.hapmap.org). Next, we mapped the recombination hotspots to our data set and for each gene computed the number of hotspots within 350 kb upstream and downstream, as well as the minimum distance to the closest recombination hotspot upstream and downstream. Interestingly, the retrovirus-like retrotransposon THE1B was found to be among certain sequence features that are overrepresented in hotspots (Myers et al. 2005). In particular, these authors found the 8-nucleotide motif CCACGTGG to be significantly more frequent in hotspot THE1Bs compared to THE1Bs elsewhere in the genome. The same oligonucleotide motif is also involved in serum-induced transcription at the G1/S-phase boundary in the hamster (Miltenberger et al. 1995) and is known as the G-box binding motif for plant basic leucine zipper (bZIP) proteins (Niu et al. 1999). We proceeded by counting the occurrence of this oligomer in all THE1B elements within the 100 kb regions flanking each gene.
The last additional class of feature measurements involved nucleosome formation potential profiles. Such in silico estimates of nucleosome packaging density in the promoter region has previously been used to distinguish tissue-specific genes from housekeeping genes and widely expressed genes (Levitsky et al. 2001). Using the Web interface at http://wwwmgs.bionet.nsc.ru/mgs/programs/recon, we acquired nucleosome formation potential estimates and summarized them as follows. We computed the sum within the region 0.82–0.61 kb upstream, the standard deviation 5.86–5.81 kb upstream, the mean 0–1 and 0.31–0.49 kb within the concatenated exons, and the standard deviation 6.7–6.75 and 7.02–7.07 kb downstream. These particular windows were picked following visual inspection of plotted potentials.
Statistical methods
To be more robust in our imprinted gene predictions, we used two strategies for feature selection and classifier learning: Equbits Foresight (which employs support vector machines and is documented at http://www.equbits.com) and SMLR (which adopts a Bayesian approach to sparse multinomial logistic regression and is documented in the literature and at http://www.cs.duke.edu/~amink/software/) (Krishnapuram et al. 2005). In each case, we learned two separate classifiers, one with a linear kernel and one with an RBF kernel. The operating point on the ROC for each classifier was chosen so as to minimize the number of false positives while retaining all true positives (Supplemental Fig. 2). To be more conservative in our final predictions, we required joint agreement among all four classifiers before predicting a gene to be imprinted. We refer to these as our high-confidence predictions. We chose to report the prediction set sorted by chromosome because the confidence estimates provided by the different classifiers could not be unified in a direct manner. The procedure of CV, training, testing, and prediction is conceptually depicted in Supplemental Figure 3.
When using Equbits to predict imprinted genes, a 40-fold CV procedure was used; to prevent overfitting or overestimation of prediction accuracy, feature selection using a linear kernel was performed afresh for each fold of the CV (without inclusion of the genes being held out during that fold). Once features were selected in each fold, classifiers for imprint status with linear and RBF kernels were learned for that fold. The number of retained features ranged from 613–638 during CV, whereas 626 features were retained in the final classifier.
When using SMLR to predict imprinted genes, a similar scheme was adopted. At each step of a 40-fold CV, feature selection was performed afresh (without inclusion of the genes being held out during that fold) by applying a sparsity-promoting prior directly on the weights of the features (no kernel). Once features were selected in this manner in each fold, classifiers for imprint status with linear and RBF kernels were learned. During CV, the number of retained features averaged 875, while 820 features were retained in the final classifier.
SMLR is written in portable Java, with a GUI, and is available with complete source code under a noncommercial use license from http://www.cs.duke.edu/~amink. In addition, all data, and all scripts used to produce the SMLR results, are also available.
To ensure that no straightforward relationships within the training data were obscured by sophisticated learning methods, we also performed CV using three simple classifiers (as implemented in Weka 3.4) (Witten and Frank 2005). A naïve Bayes classifier showed a sensitivity of 40% (16 out of 40 imprinted genes correctly recognized) and a specificity of 97% (535 out of 552 nonimprinted genes correctly classified). A decision stump simply classified all genes as nonimprinted. A random forest classifier showed a sensitivity of 20% (eight out of 40 correct) and a specificity of 95% (522 out of 552 correct). These experiments suggest that simple alternative classification approaches are not likely to result in comparable classification accuracy.
To simplify the prediction of parental expression preference, we used Equbits only with a linear kernel and the top 30 features. This procedure is exactly analogous to that used to predict parental preference in the mouse (Luedi et al. 2005).
We used χ2-tests to compare proportions and two-sided Student’s t-tests to compare means. To be conservative, Bonferroni’s method was used when correcting for multiple testing (α = 0.05).
Experimental procedures
From human conceptuses and matched maternal decidua, DNA was isolated in Qiagen buffer ATL and proteinase K (Qiagen Sciences, Inc.) followed by phenol-chloroformisoamyl alcohol extraction and ethanol precipitation. Each individual was screened for polymorphisms in KCNK9 (C/T, dbSNP accession rs2615374) and DLGAP2 (G/A, dbSNP accession rs2235112) by genomic DNA PCR with Qiagen Hotstart Taq polymerase (Qiagen Sciences, Inc.) as per manufacturer’s instructions. Following identification of heterozygous polymorphic individuals, total RNA was isolated from brain and testis by homogenization in RNA-Stat 60 (Tel-Test, Inc.); subsequent processing was performed as recommended by the manufacturer.
First-strand cDNA was primed with gene-specific primers, and synthesized from DNaseI-treated RNA using Superscript II as recommended by the manufacturer (Invitrogen). We used Qiagen Hotstart Taq polymerase (Qiagen Sciences, Inc.) in a 25 μL RT-PCR reaction volume, as per manufacturer’s instructions. RT-PCR products were separated by electrophoresis on a 1.5% agarose gel, and appropriately-sized fragments of cDNA were excised and gel-extracted (GenElute, Sigma Chemical Co.). Products were sequenced (ABI 377 sequencer, PE Biosystems), and analyzed for expression using FinchTV (Geospiza, Inc.).
In order to rule out any stochastic effects, we repeated the PCR and the sequencing reactions at least three times in all cases where exclusive monoallelic expression was observed. All sequencing reactions were performed in both directions.
This study was performed in accordance with current regulations and standards of the United States Department of Health and Human Services and National Institutes of Health.
Acknowledgments
We thank the Birth Defects Research Laboratory at the University of Washington for tissue samples. This work was supported in part by grants from NIH to F.S.D., to R.L.J. (ES13053, T32ES07031), and to R.L.J. and A.J.H. (ES015165); from DOE to R.L.J. (DE-FG02-05ER64101); and from NSF and the Sloan Foundation to A.J.H.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6584707
References
- Allen E., Horvath S., Tong F., Kraft P., Spiteri E., Riggs A.D., Marahrens Y., Horvath S., Tong F., Kraft P., Spiteri E., Riggs A.D., Marahrens Y., Tong F., Kraft P., Spiteri E., Riggs A.D., Marahrens Y., Kraft P., Spiteri E., Riggs A.D., Marahrens Y., Spiteri E., Riggs A.D., Marahrens Y., Riggs A.D., Marahrens Y., Marahrens Y. High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes. Proc. Natl. Acad. Sci. 2003;100:9940–9945. doi: 10.1073/pnas.1737401100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenian S., Weinhold B., Oelgeschlager M., Ruther U., Nordheim A., Weinhold B., Oelgeschlager M., Ruther U., Nordheim A., Oelgeschlager M., Ruther U., Nordheim A., Ruther U., Nordheim A., Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F., Cavalli G., Cavalli G. Cellular memory and dynamic regulation of polycomb group proteins. Curr. Opin. Cell Biol. 2006;18:275–283. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Barlow D.P. Methylation and imprinting: from host defense to gene regulation? Science. 1993;260:309–310. doi: 10.1126/science.8469984. [DOI] [PubMed] [Google Scholar]
- Barlow D.P., Stoger R., Herrmann B.G., Saito K., Schweifer N., Stoger R., Herrmann B.G., Saito K., Schweifer N., Herrmann B.G., Saito K., Schweifer N., Saito K., Schweifer N., Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- Bartolomei M.S., Zemel S., Tilghman S.M., Zemel S., Tilghman S.M., Tilghman S.M. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- Blagitko N., Mergenthaler S., Schulz U., Wollmann H.A., Craigen W., Eggermann T., Ropers H.H., Kalscheuer V.M., Mergenthaler S., Schulz U., Wollmann H.A., Craigen W., Eggermann T., Ropers H.H., Kalscheuer V.M., Schulz U., Wollmann H.A., Craigen W., Eggermann T., Ropers H.H., Kalscheuer V.M., Wollmann H.A., Craigen W., Eggermann T., Ropers H.H., Kalscheuer V.M., Craigen W., Eggermann T., Ropers H.H., Kalscheuer V.M., Eggermann T., Ropers H.H., Kalscheuer V.M., Ropers H.H., Kalscheuer V.M., Kalscheuer V.M. Human GRB10 is imprinted and expressed from the paternal and maternal allele in a highly tissue- and isoform-specific fashion. Hum. Mol. Genet. 2000;9:1587–1595. doi: 10.1093/hmg/9.11.1587. [DOI] [PubMed] [Google Scholar]
- Brandeis M., Frank D., Keshet I., Siegfried Z., Mendelsohn M., Nemes A., Temper V., Razin A., Cedar H., Frank D., Keshet I., Siegfried Z., Mendelsohn M., Nemes A., Temper V., Razin A., Cedar H., Keshet I., Siegfried Z., Mendelsohn M., Nemes A., Temper V., Razin A., Cedar H., Siegfried Z., Mendelsohn M., Nemes A., Temper V., Razin A., Cedar H., Mendelsohn M., Nemes A., Temper V., Razin A., Cedar H., Nemes A., Temper V., Razin A., Cedar H., Temper V., Razin A., Cedar H., Razin A., Cedar H., Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- Charlier C., Segers K., Wagenaar D., Karim L., Berghmans S., Jaillon O., Shay T., Weis-Senbach J., Cockett N., Gyapay G., Segers K., Wagenaar D., Karim L., Berghmans S., Jaillon O., Shay T., Weis-Senbach J., Cockett N., Gyapay G., Wagenaar D., Karim L., Berghmans S., Jaillon O., Shay T., Weis-Senbach J., Cockett N., Gyapay G., Karim L., Berghmans S., Jaillon O., Shay T., Weis-Senbach J., Cockett N., Gyapay G., Berghmans S., Jaillon O., Shay T., Weis-Senbach J., Cockett N., Gyapay G., Jaillon O., Shay T., Weis-Senbach J., Cockett N., Gyapay G., Shay T., Weis-Senbach J., Cockett N., Gyapay G., Weis-Senbach J., Cockett N., Gyapay G., Cockett N., Gyapay G., Gyapay G., et al. Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 2001;11:850–862. doi: 10.1101/gr.172701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse H.V. The controlling element in sex chromosome behavior. Genetics. 1960;45:1429–1443. doi: 10.1093/genetics/45.10.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara T.M., Robertson E.J., Efstratiadis A., Robertson E.J., Efstratiadis A., Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- Du Y., Jenkins N.A., Copeland N.G., Jenkins N.A., Copeland N.G., Copeland N.G. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun Kwon H., Taylor H.S., Taylor H.S. The role of HOX genes in human implantation. Ann. N. Y. Acad. Sci. 2004;1034:1–18. doi: 10.1196/annals.1335.001. [DOI] [PubMed] [Google Scholar]
- Furukawa C., Daigo Y., Ishikawa N., Kato T., Ito T., Tsuchiya E., Sone S., Nakamura Y., Daigo Y., Ishikawa N., Kato T., Ito T., Tsuchiya E., Sone S., Nakamura Y., Ishikawa N., Kato T., Ito T., Tsuchiya E., Sone S., Nakamura Y., Kato T., Ito T., Tsuchiya E., Sone S., Nakamura Y., Ito T., Tsuchiya E., Sone S., Nakamura Y., Tsuchiya E., Sone S., Nakamura Y., Sone S., Nakamura Y., Nakamura Y. Plakophilin 3 oncogene as prognostic marker and therapeutic target for lung cancer. Cancer Res. 2005;65:7102–7110. doi: 10.1158/0008-5472.CAN-04-1877. [DOI] [PubMed] [Google Scholar]
- Greally J.M. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc. Natl. Acad. Sci. 2002;99:327–332. doi: 10.1073/pnas.012539199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D., Westoby M., Westoby M. Parent-specific gene expression and the triploid endosperm. Am. Nat. 1989;134:147–155. [Google Scholar]
- Horike S., Cai S., Miyano M., Cheng J.F., Kohwi-Shigematsu T., Cai S., Miyano M., Cheng J.F., Kohwi-Shigematsu T., Miyano M., Cheng J.F., Kohwi-Shigematsu T., Cheng J.F., Kohwi-Shigematsu T., Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Hunter P. The silence of genes. Is genomic imprinting the software of evolution or just a battleground for gender conflict? EMBO Rep. 2007;8:441–443. doi: 10.1038/sj.embor.7400965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirtle R.L., Skinner M.K., Skinner M.K. Environmental epigenomics and disease susceptibility. Nat. Rev. Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kananura C., Sander T., Rajan S., Preisig-Muller R., Grzeschik K.H., Daut J., Derst C., Steinlein O.K., Sander T., Rajan S., Preisig-Muller R., Grzeschik K.H., Daut J., Derst C., Steinlein O.K., Rajan S., Preisig-Muller R., Grzeschik K.H., Daut J., Derst C., Steinlein O.K., Preisig-Muller R., Grzeschik K.H., Daut J., Derst C., Steinlein O.K., Grzeschik K.H., Daut J., Derst C., Steinlein O.K., Daut J., Derst C., Steinlein O.K., Derst C., Steinlein O.K., Steinlein O.K. Tandem pore domain K+-channel TASK-3 (KCNK9) and idiopathic absence epilepsies. Am. J. Med. Genet. 2002;114:227–229. doi: 10.1002/ajmg.10201. [DOI] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Schwartz M., Sugnet C.W., Thomas D.J., Sugnet C.W., Thomas D.J., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X., Thomas S.N., Robinson D.O., Collins A., Thomas S.N., Robinson D.O., Collins A., Robinson D.O., Collins A., Collins A. The distinguishing sequence characteristics of mouse imprinted genes. Mamm. Genome. 2002;13:639–645. doi: 10.1007/s00335-002-3038-x. [DOI] [PubMed] [Google Scholar]
- Kimura M.I., Kazuki Y., Kashiwagi A., Kai Y., Abe S., Barbieri O., Levi G., Oshimura M., Kazuki Y., Kashiwagi A., Kai Y., Abe S., Barbieri O., Levi G., Oshimura M., Kashiwagi A., Kai Y., Abe S., Barbieri O., Levi G., Oshimura M., Kai Y., Abe S., Barbieri O., Levi G., Oshimura M., Abe S., Barbieri O., Levi G., Oshimura M., Barbieri O., Levi G., Oshimura M., Levi G., Oshimura M., Oshimura M. Dlx5, the mouse homologue of the human-imprinted DLX5 gene, is biallelically expressed in the mouse brain. J. Hum. Genet. 2004;49:273–277. doi: 10.1007/s10038-004-0139-2. [DOI] [PubMed] [Google Scholar]
- Krishnapuram B., Figueiredo M., Carin L., Hartemink A.J., Figueiredo M., Carin L., Hartemink A.J., Carin L., Hartemink A.J., Hartemink A.J. Sparse multinomial logistic regression: Fast algorithms and generalization bounds. IEEE Trans. Pattern Anal. Mach. Intell. 2005;27:957–968. doi: 10.1109/TPAMI.2005.127. [DOI] [PubMed] [Google Scholar]
- LaSalle J., Lalande M., Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science. 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Davison J.A., Vidal S.M., Belouchi A., Davison J.A., Vidal S.M., Belouchi A., Vidal S.M., Belouchi A., Belouchi A. Cloning, expression and chromosomal location of NKX6B to 10q26, a region frequently deleted in brain tumors. Mamm. Genome. 2001;12:157–162. doi: 10.1007/s003350010247. [DOI] [PubMed] [Google Scholar]
- Levitsky V.G., Podkolodnaya O.A., Kolchanov N.A., Podkolodny N.L., Podkolodnaya O.A., Kolchanov N.A., Podkolodny N.L., Kolchanov N.A., Podkolodny N.L., Podkolodny N.L. Nucleosome formation potential of eukaryotic DNA: Calculation and promoters analysis. Bioinformatics. 2001;17:998–1010. doi: 10.1093/bioinformatics/17.11.998. [DOI] [PubMed] [Google Scholar]
- Li T., Vu T.H., Lee K.O., Yang Y., Nguyen C.V., Bui H.Q., Zeng Z.L., Nguyen B.T., Hu J.F., Murphy S.K., Vu T.H., Lee K.O., Yang Y., Nguyen C.V., Bui H.Q., Zeng Z.L., Nguyen B.T., Hu J.F., Murphy S.K., Lee K.O., Yang Y., Nguyen C.V., Bui H.Q., Zeng Z.L., Nguyen B.T., Hu J.F., Murphy S.K., Yang Y., Nguyen C.V., Bui H.Q., Zeng Z.L., Nguyen B.T., Hu J.F., Murphy S.K., Nguyen C.V., Bui H.Q., Zeng Z.L., Nguyen B.T., Hu J.F., Murphy S.K., Bui H.Q., Zeng Z.L., Nguyen B.T., Hu J.F., Murphy S.K., Zeng Z.L., Nguyen B.T., Hu J.F., Murphy S.K., Nguyen B.T., Hu J.F., Murphy S.K., Hu J.F., Murphy S.K., Murphy S.K., et al. An imprinted PEG1/MEST antisense expressed predominantly in human testis and in mature spermatozoa. J. Biol. Chem. 2002;277:13518–13527. doi: 10.1074/jbc.M200458200. [DOI] [PubMed] [Google Scholar]
- Luedi P.P., Hartemink A.J., Jirtle R.L., Hartemink A.J., Jirtle R.L., Jirtle R.L. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager J., Montgomery N.D., de Villena F.P., Magnuson T., Montgomery N.D., de Villena F.P., Magnuson T., de Villena F.P., Magnuson T., Magnuson T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 2003;33:502–507. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- Mancini-Dinardo D., Steele S.J., Levorse J.M., Ingram R.S., Tilghman S.M., Steele S.J., Levorse J.M., Ingram R.S., Tilghman S.M., Levorse J.M., Ingram R.S., Tilghman S.M., Ingram R.S., Tilghman S.M., Tilghman S.M. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes & Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnis M.G., Lan T.H., Willour V.L., McMahon F.J., Simpson S.G., Addington A.M., MacKinnon D.F., Potash J.B., Mahoney A.T., Chellis J., Lan T.H., Willour V.L., McMahon F.J., Simpson S.G., Addington A.M., MacKinnon D.F., Potash J.B., Mahoney A.T., Chellis J., Willour V.L., McMahon F.J., Simpson S.G., Addington A.M., MacKinnon D.F., Potash J.B., Mahoney A.T., Chellis J., McMahon F.J., Simpson S.G., Addington A.M., MacKinnon D.F., Potash J.B., Mahoney A.T., Chellis J., Simpson S.G., Addington A.M., MacKinnon D.F., Potash J.B., Mahoney A.T., Chellis J., Addington A.M., MacKinnon D.F., Potash J.B., Mahoney A.T., Chellis J., MacKinnon D.F., Potash J.B., Mahoney A.T., Chellis J., Potash J.B., Mahoney A.T., Chellis J., Mahoney A.T., Chellis J., Chellis J., et al. Genome-wide scan of bipolar disorder in 65 pedigrees: Supportive evidence for linkage at 8q24, 18q22, 4q32, 2p12, and 13q12. Mol. Psychiatry. 2003;8:288–298. doi: 10.1038/sj.mp.4001277. [DOI] [PubMed] [Google Scholar]
- Medhurst A.D., Rennie G., Chapman C.G., Meadows H., Duckworth M.D., Kelsell R.E., Gloger I., Pangalos M.N., Rennie G., Chapman C.G., Meadows H., Duckworth M.D., Kelsell R.E., Gloger I., Pangalos M.N., Chapman C.G., Meadows H., Duckworth M.D., Kelsell R.E., Gloger I., Pangalos M.N., Meadows H., Duckworth M.D., Kelsell R.E., Gloger I., Pangalos M.N., Duckworth M.D., Kelsell R.E., Gloger I., Pangalos M.N., Kelsell R.E., Gloger I., Pangalos M.N., Gloger I., Pangalos M.N., Pangalos M.N. Distribution analysis of human two pore domain potassium channels in tissues of the central nervous system and periphery. Brain Res. Mol. Brain Res. 2001;86:101–114. doi: 10.1016/s0169-328x(00)00263-1. [DOI] [PubMed] [Google Scholar]
- Miltenberger R.J., Sukow K.A., Farnham P.J., Sukow K.A., Farnham P.J., Farnham P.J. An E-box-mediated increase in cad transcription at the G1/S-phase boundary is suppressed by inhibitory c-Myc mutants. Mol. Cell. Biol. 1995;15:2527–2535. doi: 10.1128/mcb.15.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens C.B., Selleri L., Selleri L. Hox cofactors in vertebrate development. Dev. Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Monk D., Arnaud P., Apostolidou S., Hills F.A., Kelsey G., Stanier P., Feil R., Moore G.E., Arnaud P., Apostolidou S., Hills F.A., Kelsey G., Stanier P., Feil R., Moore G.E., Apostolidou S., Hills F.A., Kelsey G., Stanier P., Feil R., Moore G.E., Hills F.A., Kelsey G., Stanier P., Feil R., Moore G.E., Kelsey G., Stanier P., Feil R., Moore G.E., Stanier P., Feil R., Moore G.E., Feil R., Moore G.E., Moore G.E. Limited evolutionary conservation of imprinting in the human placenta. Proc. Natl. Acad. Sci. 2006;103:6623–6628. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morison I.M., Ramsay J.P., Spencer H.G., Ramsay J.P., Spencer H.G., Spencer H.G. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Murphy S.K., Jirtle R.L., Jirtle R.L. Imprinting evolution and the price of silence. Bioessays. 2003;25:577–588. doi: 10.1002/bies.10277. [DOI] [PubMed] [Google Scholar]
- Muscheck M., Sukosd F., Pesti T., Kovacs G., Sukosd F., Pesti T., Kovacs G., Pesti T., Kovacs G., Kovacs G. High density deletion mapping of bladder cancer localizes the putative tumor suppressor gene between loci D8S504 and D8S264 at chromosome 8p23.3. Lab. Invest. 2000;80:1089–1093. doi: 10.1038/labinvest.3780114. [DOI] [PubMed] [Google Scholar]
- Mustanski B.S., Dupree M.G., Nievergelt C.M., Bocklandt S., Schork N.J., Hamer D.H., Dupree M.G., Nievergelt C.M., Bocklandt S., Schork N.J., Hamer D.H., Nievergelt C.M., Bocklandt S., Schork N.J., Hamer D.H., Bocklandt S., Schork N.J., Hamer D.H., Schork N.J., Hamer D.H., Hamer D.H. A genomewide scan of male sexual orientation. Hum. Genet. 2005;116:272–278. doi: 10.1007/s00439-004-1241-4. [DOI] [PubMed] [Google Scholar]
- Myers S., Bottolo L., Freeman C., McVean G., Donnelly P., Bottolo L., Freeman C., McVean G., Donnelly P., Freeman C., McVean G., Donnelly P., McVean G., Donnelly P., Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Niu X., Renshaw-Gegg L., Miller L., Guiltinan M.J., Renshaw-Gegg L., Miller L., Guiltinan M.J., Miller L., Guiltinan M.J., Guiltinan M.J. Bipartite determinants of DNA-binding specificity of plant basic leucine zipper proteins. Plant Mol. Biol. 1999;41:1–13. doi: 10.1023/a:1006206011502. [DOI] [PubMed] [Google Scholar]
- Okita C., Meguro M., Hoshiya H., Haruta M., Sakamoto Y., Oshimura M., Meguro M., Hoshiya H., Haruta M., Sakamoto Y., Oshimura M., Hoshiya H., Haruta M., Sakamoto Y., Oshimura M., Haruta M., Sakamoto Y., Oshimura M., Sakamoto Y., Oshimura M., Oshimura M. A new imprinted cluster on the human chromosome 7q21-q31, identified by human-mouse monochromosomal hybrids. Genomics. 2003;81:556–559. doi: 10.1016/s0888-7543(03)00052-1. [DOI] [PubMed] [Google Scholar]
- de Pardo-Manuel Villena F., de la Casa-Esperon E., Sapienza C., de la Casa-Esperon E., Sapienza C., Sapienza C. Natural selection and the function of genome imprinting: Beyond the silenced minority. Trends Genet. 2000;16:573–579. doi: 10.1016/s0168-9525(00)02134-x. [DOI] [PubMed] [Google Scholar]
- Patel A.J., Lazdunski M., Lazdunski M. The 2P-domain K+ channels: role in apoptosis and tumorigenesis. Pflugers Arch. 2004;448:261–273. doi: 10.1007/s00424-004-1255-8. [DOI] [PubMed] [Google Scholar]
- Pearson W.R., Lipman D.J., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranta S., Zhang Y., Ross B., Takkunen E., Hirvasniemi A., de la Chapelle A., Gilliam T.C., Lehesjoki A.E., Zhang Y., Ross B., Takkunen E., Hirvasniemi A., de la Chapelle A., Gilliam T.C., Lehesjoki A.E., Ross B., Takkunen E., Hirvasniemi A., de la Chapelle A., Gilliam T.C., Lehesjoki A.E., Takkunen E., Hirvasniemi A., de la Chapelle A., Gilliam T.C., Lehesjoki A.E., Hirvasniemi A., de la Chapelle A., Gilliam T.C., Lehesjoki A.E., de la Chapelle A., Gilliam T.C., Lehesjoki A.E., Gilliam T.C., Lehesjoki A.E., Lehesjoki A.E. Positional cloning and characterisation of the human DLGAP2 gene and its exclusion in progressive epilepsy with mental retardation. Eur. J. Hum. Genet. 2000;8:381–384. doi: 10.1038/sj.ejhg.5200440. [DOI] [PubMed] [Google Scholar]
- Reik W., Walter J., Walter J. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Savory T. The mule. Sci. Am. 1970;223:102–109. [Google Scholar]
- Schratt G., Weinhold B., Lundberg A.S., Schuck S., Berger J., Schwarz H., Weinberg R.A., Ruther U., Nordheim A., Weinhold B., Lundberg A.S., Schuck S., Berger J., Schwarz H., Weinberg R.A., Ruther U., Nordheim A., Lundberg A.S., Schuck S., Berger J., Schwarz H., Weinberg R.A., Ruther U., Nordheim A., Schuck S., Berger J., Schwarz H., Weinberg R.A., Ruther U., Nordheim A., Berger J., Schwarz H., Weinberg R.A., Ruther U., Nordheim A., Schwarz H., Weinberg R.A., Ruther U., Nordheim A., Weinberg R.A., Ruther U., Nordheim A., Ruther U., Nordheim A., Nordheim A. Serum response factor is required for immediate-early gene activation yet is dispensable for proliferation of embryonic stem cells. Mol. Cell. Biol. 2001;21:2933–2943. doi: 10.1128/MCB.21.8.2933-2943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H., Youngson N., Lin S.P., Dalbert S., Paulsen M., Bachellerie J.P., Ferguson-Smith A.C., Cavaille J., Youngson N., Lin S.P., Dalbert S., Paulsen M., Bachellerie J.P., Ferguson-Smith A.C., Cavaille J., Lin S.P., Dalbert S., Paulsen M., Bachellerie J.P., Ferguson-Smith A.C., Cavaille J., Dalbert S., Paulsen M., Bachellerie J.P., Ferguson-Smith A.C., Cavaille J., Paulsen M., Bachellerie J.P., Ferguson-Smith A.C., Cavaille J., Bachellerie J.P., Ferguson-Smith A.C., Cavaille J., Ferguson-Smith A.C., Cavaille J., Cavaille J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- Soulez M., Rouviere C.G., Chafey P., Hentzen D., Vandromme M., Lautredou N., Lamb N., Kahn A., Tuil D., Rouviere C.G., Chafey P., Hentzen D., Vandromme M., Lautredou N., Lamb N., Kahn A., Tuil D., Chafey P., Hentzen D., Vandromme M., Lautredou N., Lamb N., Kahn A., Tuil D., Hentzen D., Vandromme M., Lautredou N., Lamb N., Kahn A., Tuil D., Vandromme M., Lautredou N., Lamb N., Kahn A., Tuil D., Lautredou N., Lamb N., Kahn A., Tuil D., Lamb N., Kahn A., Tuil D., Kahn A., Tuil D., Tuil D. Growth and differentiation of C2 myogenic cells are dependent on serum response factor. Mol. Cell. Biol. 1996;16:6065–6074. doi: 10.1128/mcb.16.11.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strichman-Almashanu L.Z., Lee R.S., Onyango P.O., Perlman E., Flam F., Frieman M.B., Feinberg A.P., Lee R.S., Onyango P.O., Perlman E., Flam F., Frieman M.B., Feinberg A.P., Onyango P.O., Perlman E., Flam F., Frieman M.B., Feinberg A.P., Perlman E., Flam F., Frieman M.B., Feinberg A.P., Flam F., Frieman M.B., Feinberg A.P., Frieman M.B., Feinberg A.P., Feinberg A.P. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 2002;12:543–554. doi: 10.1101/gr.224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D., Goto Y., Cao R., Cerqueira F., Wagschal A., Zhang Y., Feil R., Goto Y., Cao R., Cerqueira F., Wagschal A., Zhang Y., Feil R., Cao R., Cerqueira F., Wagschal A., Zhang Y., Feil R., Cerqueira F., Wagschal A., Zhang Y., Feil R., Wagschal A., Zhang Y., Feil R., Zhang Y., Feil R., Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Vandromme M., Gauthier-Rouviere C., Carnac G., Lamb N., Fernandez A., Gauthier-Rouviere C., Carnac G., Lamb N., Fernandez A., Carnac G., Lamb N., Fernandez A., Lamb N., Fernandez A., Fernandez A. Serum response factor p67SRF is expressed and required during myogenic differentiation of both mouse C2 and rat L6 muscle cell lines. J. Cell Biol. 1992;118:1489–1500. doi: 10.1083/jcb.118.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Paulsen M., Paulsen M. The potential role of gene duplications in the evolution of imprinting mechanisms. Hum. Mol. Genet. 2003;12:215–220. doi: 10.1093/hmg/ddg296. [DOI] [PubMed] [Google Scholar]
- Witten I.H., Frank E., Frank E. Data mining: Practical machine learning tools and techniques. Morgan Kaufmann; San Francisco: 2005. [Google Scholar]
- Yoder J.A., Walsh C.P., Bestor T.H., Walsh C.P., Bestor T.H., Bestor T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Zara F., Bianchi A., Avanzini G., Di Donato S., Castellotti B., Patel P.I., Pandolfo M., Bianchi A., Avanzini G., Di Donato S., Castellotti B., Patel P.I., Pandolfo M., Avanzini G., Di Donato S., Castellotti B., Patel P.I., Pandolfo M., Di Donato S., Castellotti B., Patel P.I., Pandolfo M., Castellotti B., Patel P.I., Pandolfo M., Patel P.I., Pandolfo M., Pandolfo M. Mapping of genes predisposing to idiopathic generalized epilepsy. Hum. Mol. Genet. 1995;4:1201–1207. doi: 10.1093/hmg/4.7.1201. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Tycko B., Tycko B. Monoallelic expression of the human H19 gene. Nat. Genet. 1992;1:40–44. doi: 10.1038/ng0492-40. [DOI] [PubMed] [Google Scholar]