Abstract
The contribution to genetic diversity of genomic segmental copy number variations (CNVs) is less well understood than that of single-nucleotide polymorphisms (SNPs). While less frequent than SNPs, CNVs have greater potential to affect phenotype. In this study, we have performed the most comprehensive survey to date of CNVs in mice, analyzing the genomes of 42 Mouse Phenome Consortium priority strains. This microarray comparative genomic hybridization (CGH)-based analysis has identified 2094 putative CNVs, with an average of 10 Mb of DNA in 51 CNVs when individual mouse strains were compared to the reference strain C57BL/6J. This amount of variation results in gene content that can differ by hundreds of genes between strains. These genes include members of large families such as the major histocompatibility and pheromone receptor genes, but there are also many singleton genes including genes with expected phenotypic consequences from their deletion or amplification. Using a whole-genome association analysis, we demonstrate that complex multigenic phenotypes, such as food intake, can be associated with specific copy number changes.
In this post-genomic era, many researchers have begun focusing on between-individual genetic differences as sources of both benign and malignant phenotypic differences. For example, the International HapMap Consortium was founded in 2002 “to determine the common patterns of DNA sequence variation in the human genome” (The International HapMap Consortium 2003). So far, the bulk of this effort has focused on identifying single-nucleotide polymorphisms (SNPs) (Hinds et al. 2005; The International HapMap Consortium 2005). The large number of such variations, approximately one every 300 nucleotides in humans (Kruglyak and Nickerson 2001), clearly supports this focus on SNPs. Long before SNPs came to the forefront, large-scale genomic polymorphisms due to chromosomal deletions, duplications, and rearrangements were identified through microscopic chromosomal observation (Feuk et al. 2006). The infrequency and size of these karyotype alterations underscored their roles in major genomic diseases (Emanuel and Shaikh 2001; Shaw and Lupski 2004), but made it seem unlikely that they were in-volved in normal individual variation or milder forms of disease.
This view has been changing with the advent of newer technologies, primarily microarray-based comparative genomic hybridization (CGH) (Pinkel et al. 1998; Barrett et al. 2004), that have allowed for genome-wide submicroscopic surveys of segmental copy number variations (CNVs) (Feuk et al. 2006). These scans have identified the previously unappreciated scope of heterogeneity in genomic content in both humans (Sebat et al. 2004; Sharp et al. 2005; Redon et al. 2006) and mice (Li et al. 2004; Snijders et al. 2005; Graubert et al. 2007) due to CNVs. Although less frequent than SNPs, CNVs’ relatively large sizes lead to the involvement of a significant fraction of the genome, 12% in one study of CNVs between 270 individuals (Redon et al. 2006). Furthermore, while most SNPs would be expected to have no or only mild phenotypic effects, the effects of CNVs, ranging from increased gene dosage to full gene knockouts, would be expected to lead to greater and more frequent impacts on phenotype.
Human studies have, by necessity, looked at CNVs between moderate-sized groups of individuals (Sebat et al. 2004; Sharp et al. 2005; Redon et al. 2006). In contrast, murine studies (Li et al. 2004; Snijders et al. 2005; Graubert et al. 2007) have looked at CNV differences between inbred mouse strains. Since inbred mouse genomes have stabilized following generations of inbreeding, the prospect of making an exhaustive survey of CNVs in these strains is feasible. Inbred mouse strains have long served as important disease models and display a wide range of phenotypic variation (Bogue et al. 2007; Svenson et al. 2007). Cataloging their complement of CNVs would further our understanding of these models and the genetic differences that make individual strains relevant to specific human diseases. Furthermore, it would provide a better understanding of the processes underlying variation, evolution, and speciation.
This study is comprised of a CGH analysis of the genomes of 41 inbred mouse strains compared to the reference strain C57BL/6J. These strains represent the Mouse Phenome Database priority strains list (Bogue et al. 2007), designed to cover a wide range of genetic diversity while simultaneously including the most commonly used research strains. We show that over a hundred regions of a strain’s genome may be amplified or deleted in relation to the C57BL/6J reference. This intra-species variability results in many genes being completely or partially deleted, while many other genes are present at increased copy number in a given mouse strain. Some gene functions, including chemosensation and immune response, are preferentially found in these CNVs, but these multiparalog gene families are only a portion of the genes found in CNVs. In fact, copy number changes affect unique genes and genes which, when deleted, are known to have deleterious phenotypic effects. This surprising variability in the gene content of inbred mouse strains provides both challenges and opportunities in the use of these strains as models for understanding disease and gene function.
Results
CGH analysis
We performed CGH analysis on genomic DNA from 42 inbred mouse strains using Agilent 244K Mouse Genome Arrays. These arrays tile the mouse genome at an average density of one 60-mer probe per 6.4 kb. Duplicate male samples from each of 41 mouse strains were compared to the samples from the reference strain, C57BL/6J. The CGH data was then analyzed for the presence of CNVs using the Gain and Loss Analysis of DNA (GLAD) algorithm (Hupe et al. 2004) coupled with a t-test for significance. A final filtered, high-confidence set of CNVs was generated containing a total of 793 amplifications and 1303 deletions across the 41 strains (Fig. 1; Table 1; Supplemental Table S1). The amplifications have a mean length of 183 kb and the deletions a mean length of 207 kb (Fig. 2A). Given that C57BL/6J is used as the reference strain for all comparisons, all of these CNVs are defined in relation to that genome.
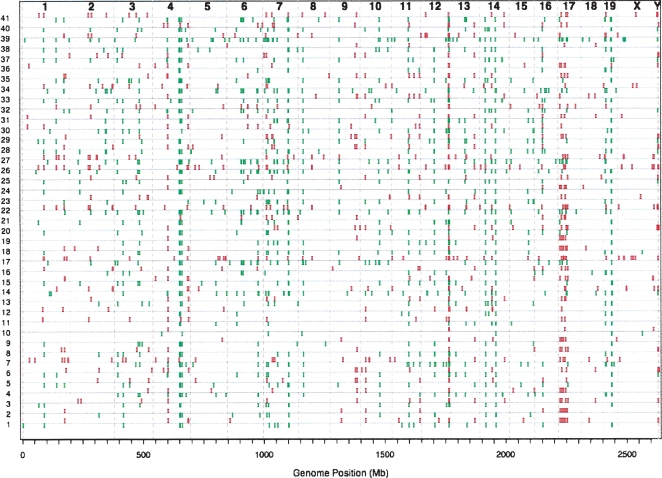
Figure 1.
All mouse CNVs. Predicted CNVs for each inbred mouse strain are displayed based on their genomic position. Amplifications are shown in red above the baseline for each strain, deletions are shown in green below. The strains are: (1) 129X1/SvJ; (2) 129S1/SvImJ; (3) AKR/J; (4) A/J; (5) BTBR T+ tf/J; (6) BUB/BnJ; (7) BALB/cJ; (8) C3H/HeJ; (9) C57BLKS/J; (10) C57BL/10J; (11) C57BR/cdJ; (12) C57L/J; (13) C58/J; (14) CAST/EiJ; (15) CBA/J; (16) CE/J; (17) CZECHII/EiJ; (18) DBA/1J; (19) DBA/2J; (20) FVB/Ntac; (21) I/LnJ; (22) JF1/Ms; (23) KK/HIJ; (24) LP/J; (25) MA/MyJ; (26) MOLF/EiJ; (27) MSM/Ms; (28) NOD/LtJ; (29) NON/LtJ; (30) NZB/BINJ; (31) NZW/LacJ; (32) PERA/EiJ; (33) PL/J; (34) PWK/PhJ; (35) RIIIS/J; (36) SEA/GnJ; (37) SJL/J; (38) SM/J; (39) SPRET/EiJ; (40) SWR/J; (41) WSB/EiJ.
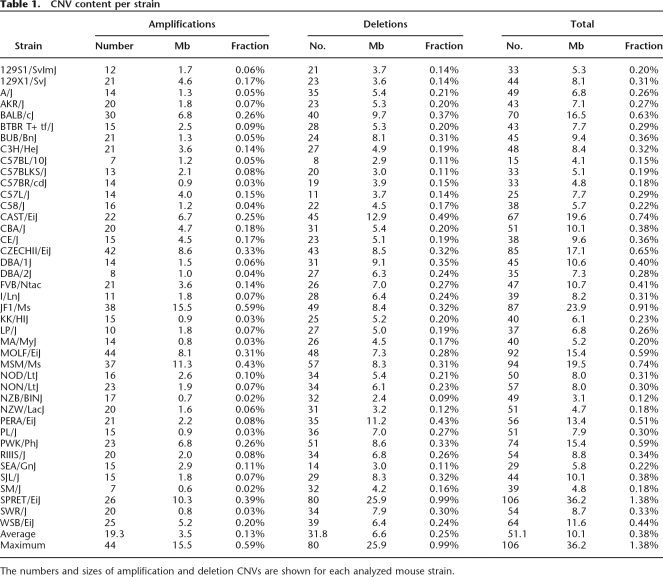
Table 1.
CNV content per strain
The numbers and sizes of amplification and deletion CNVs are shown for each analyzed mouse strain.
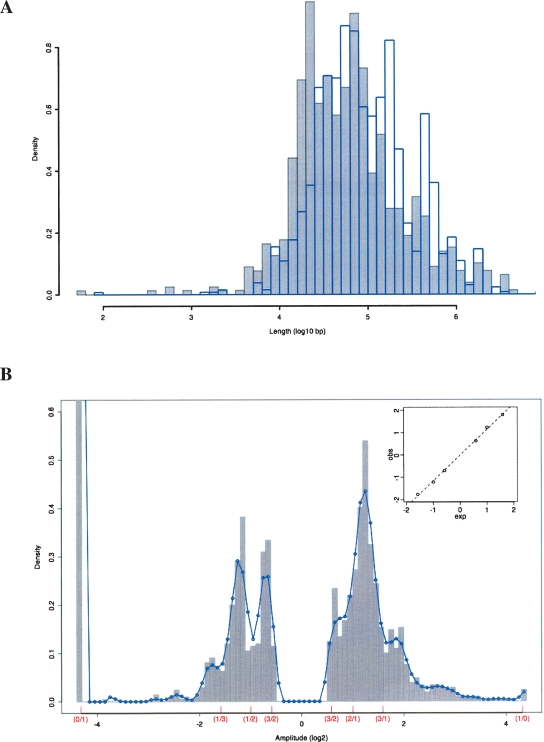
Figure 2.
Distribution of CNV lengths and amplitudes. (A) A histogram of the density of the log10 lengths in base pairs for all amplification (gray) and deletion (blue) CNVs is shown. (B) A histogram of the density of the log2 amplitudes of all CNVs in the data set is plotted along with a smoothed curve fit to the histogram. Parenthesized fractions on the X-axis show the positions of expected ratios of test to reference strain copy numbers. Inset is a plot of the observed peaks of the smoothed histogram curve versus the expected copy number ratio positions and a fitted linear regression line. The truncated bar at log2(amplitude) = −4.32 has a height of 3.7.
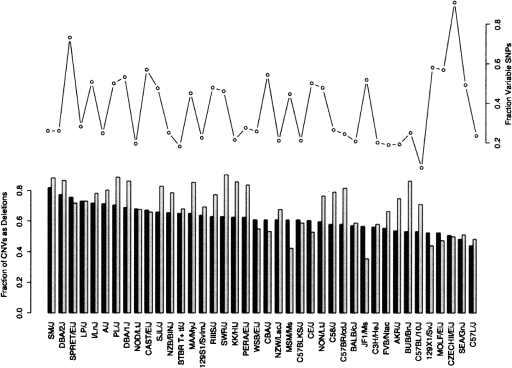
Since the observed CNVs are believed to be derived from discrete copy number changes occurring in both the test and the reference mouse genomes, we should primarily see CNV amplitudes that correspond to the ratios of these copy numbers. Indeed, an examination of the observed distribution of CNV amplitudes (Fig. 2B) reveals that there are peaks in CNV abundance at very close to their expected amplitudes (adjusted r2 = 0.998, P-value = 1.09 × 10−6). The slope of the fitted line is 1.15, indicating that our calculated CNV amplitudes are close to, but slightly higher than, they should be. The excess of deletions over amplifications, as can be seen in Figure 2B, is expected as the microarray probes are derived from the reference C57BL/6J sequence (Waterston et al. 2002) so there should be no probes for sequences deleted in C57BL/6J, leading us to miss this subset of CNVs that would otherwise appear as amplifications. Additionally, SNPs that fall within the probed regions may reduce hybridization signals and lead to false positive CNV deletion calls. To gauge whether this potential source of false-positive deletion CNVs plays an important confounding role, we compared the excess of deletions, as measured by the fraction of total CNVs comprised of deletions (by length or number) to the fraction of known SNP positions which vary between each strain and C57BL/6J (Fig. 3). No relationship between SNP content and CNV distribution can be seen, discounting SNPs as a significant source of error in this CNV data set.
Figure 3.
Deletion excess does not correlate with SNP content. A bar plot shows the fraction of all CNVs per strain which are deletions by number (black bars) or total length (white bars), sorted by decreasing deletion number fraction. The line plot shows the fraction of known SNPs between each strain and the reference, which vary.
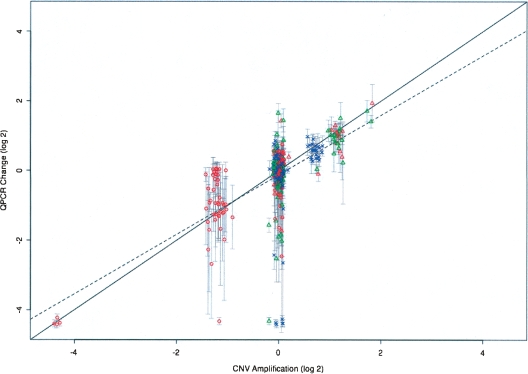
To further test the validity of this set of CNVs, they were compared to CNVs derived from randomized data. All probe ratio values in each CGH data set were scrambled 10 times, with CNV predictions performed each round. Using the same filtering criteria as for the real data sets, an average of 0.25 CNVs per randomized data set was predicted (data not shown). This compares to the average of 51.1 CNVs per strain actually identified. To further validate our results, quantitative PCR (QPCR) was performed on three genomic regions selected to represent a range of amplifications and deletions. Plotting the QPCR signal change, determined by comparing each strain to C57BL/6J, versus the amplification score for the corresponding CNV shows a strong relationship between the two data sets (slope = 0.86, P < 2 × 10−16; Fig. 4). Six additional CNV regions have been confirmed with similar or better correlations (data not shown). Within the overall good concordance, however, there are some disagreements between the QPCR and the CNV data sets. A few data points score as absent by QPCR (truncated to a log2 change of −4.2), but not by CGH. These may represent missed small deletions in the CNV data or instances of SNPs which abrogate the QPCR reaction but not CGH hybridization. The latter can occur since QPCR oligonucleotides are shorter, and thus less robust, than CGH probes, and all three QPCR oligos in each set must work in order to see any signal. Sequencing of genomic regions targeted for QPCR has, in fact, revealed previously unidentified SNPs (data not shown). A second type of discrepancy consists of a number of putative 1/3 or 1/2 amplitude deletions which do not appear changed when measured by QPCR. The source of this discrepancy awaits further analysis.
Figure 4.
QPCR analysis provides validation for CGH-derived CNV data. QPCR was performed on 42 strains, generating eight different PCR products spanning three genomic regions. The mean of the log2-transformed ratio between the QPCR signal for each strain and the C57BL/6J QPCR signal is plotted against the log2-transformed median fold-change for all the probes within the corresponding CNVs. QPCR was performed for loci on chromosomes 7, 9, and 17, plotted as red circles, blue crosses, and green triangles, respectively. Error bars show ±1 SD for the QPCR data. The expected correlation of the two data sets is indicated by the solid line with slope = 1.0; the observed correlation, slope = 0.86, is indicated by the dashed line.
Comparison with previous mouse CGH analyses
Several previous studies have used BAC arrays (Li et al. 2004; Snijders et al. 2005) or oligo arrays (Graubert et al. 2007) to perform similar CNV analyses on more limited subsets of mouse strains. Graubert and colleagues had performed the most comprehensive mouse strain CNV analysis to date. A comparison of the Graubert CNV loci amplitudes with our CGH probe signals shows that they correlate well, with a Pearson correlation of 0.80. Of the 72 Graubert CNV loci that could be compared, 67% are identified by this work, with a mean Spearman correlation for CNV direction (amplification or deletion) across that subset of 0.79. The 24 Graubert CNV loci not identified correspond to regions of low probe coverage on the Agilent arrays used in this study and point to a shortcoming in the array platform. In contrast, of the 2094 CNVs identified in this study only 26% of them fall into regions identified as CNV loci in Graubert et al. (2007). This high miss rate in Graubert et al. likely derives from their “conservative” CNV calling algorithm as well as the low amplitude fold-changes which arise from the Nimblegen microarray platform used (Graubert et al. 2007; data not shown). Unlike the generally good concordance between CNVs identified in Graubert et al. and those in our data set, there is virtually no overlap between the BAC array-derived CNVs in Li et al. (2004) and our CNVs, although we do identify the only one of the Li et al. CNVs that both had validation data and could be mapped to the current genome assembly.
Inbred mouse strain CNV content
A visual inspection of the distribution of CNVs (Fig. 1) shows a generally uniform distribution of CNVs across all the autosomes, contrasting with a low density of CNVs on the X chromosome. A very common deletion at the distal end of chromosome 12 overlaps the immunoglobulin heavy chain locus and likely represents B-cell contamination of the reference C57BL/6J material as this deletion is seen in independent C57BL/6J mouse samples (data not shown). The strains with the largest numbers of CNVs (SPRET/EiJ, CZECHII/EiJ, PWK/PhJ, MOLF/EiJ, JF1/Ms, CAST/EiJ), being wild-derived, are the most genetically distant from the reference C57BL/6J strain as one would expect. Likewise, the strains with the fewest CNVs are the C57BL/6J sibling strains, C57BL/10J, C57L/J, and C57BLKS/J. As expected, a phylogram based on similarities between the CNV content of the tested mouse strains approximates their breeding history (Supplemental Fig. S1).
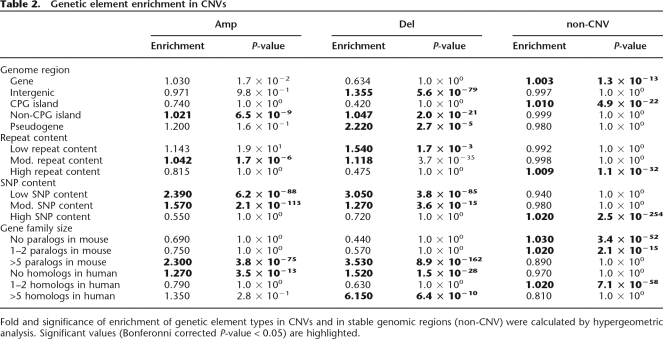
We performed enrichment analyses to understand the characteristics of the genomic regions within CNVs. Specifically, hypergeometric enrichment tests were performed on the overlap between gene and other genetic element positions, as annotated in the UCSC (Kent et al. 2002; Hsu et al. 2006) and Ensembl mouse genome databases (Birney et al. 2004), and a CNV data set formed by combining CNV predictions from all the analyzed strains. A miniscule but significant enrichment for genes in stable, non-CNV regions can be observed (Table 2), coupled with a more robust enrichment for intergenic DNA in deletions—36% more intergenic regions are found in regions of deletions than would be expected if the distributions of intergenic regions and deletions were unrelated. Likewise, CPG islands, regions of DNA rich in housekeeping genes (Gardiner-Garden and Frommer 1987), are very slightly enriched in stable genomic regions, and CPG- less DNA segments are 4.7% less abundant in deletions and 2.1% less abundant in amplifications than would be expected. Known pseudogenes, as annotated in the Ensembl database, are enriched by over twofold in deletions. These findings all point to the exclusion of functional DNA regions from areas of CNV instability and the concomitant enhancement of nonfunctional DNA in regions of CNVs. In contrast to these results, regions of both high repeat (simple and complex) and high SNP content appear to be enriched, albeit slightly, in stable genomic regions, while regions of low repeat and SNP content are enriched within CNVs (Table 2). The explanations for both of these may be ascertainment bias; in the case of the SNPs, they are more likely to have been genotyped in regions containing genes and, in the case of the repeats, they are less likely to be covered by probes on the arrays.
Table 2.
Genetic element enrichment in CNVs
Fold and significance of enrichment of genetic element types in CNVs and in stable genomic regions (non-CNV) were calculated by hypergeometric analysis. Significant values (Bonferonni corrected P-value < 0.05) are highlighted.
To understand what kinds of genes are found in CNVs, we performed enrichment analyses on gene types. Mouse genes were divided into three categories based on Ensembl paralogy annotation: those with no known paralogs in mice, those with few (1–2) paralogs, and those with many (>5) paralogs. Enrichment analysis reveals that genes with no or few paralogs are enriched in stable genomic regions while genes in large multigene families are strongly enriched in both amplifications and deletions (Table 2), suggesting that there is greater flexibility in copy number for more redundant genes. A similar analysis was performed based on the numbers of annotated human homologs for each mouse gene. In this case, mouse genes with no known human homologs were found enriched 52% in deletions and 27% in amplifications, mouse genes with few human homologs were enriched in stable genomic areas, and mouse genes with many human homologs were greatly enriched, by >615%, in regions of deletions. This makes sense as mouse genes without human homologs are less likely to be essential genes and mouse genes with many human homologs are most likely members of large gene families, both characteristics which make their strict copy number control less vital. To further understand the types of genes within and without CNVs, we performed an enrichment analysis based on annotation by the Gene Ontology consortium (Ashburner et al. 2000) in the form of GO terms (Supplemental Table S2). Genes involved in pheromone binding were strongly enriched in both amplifications (10.8-fold) and deletions (16.4-fold), as were genes involved in antigen binding (5.9-fold and 7.2-fold, respectively) and antigen presentation by MHC class I receptors (5.1-fold and 5.2-fold, respectively). In addition, other immune-related gene annotations (“defense response”) as well as steroid-processing gene annotations (“3-beta-hydroxy-delta5-steroid dehydrogenase activity,” “steroid delta-isomerase activity,” “C21-steroid hormone biosynthetic process”) were found enriched in deletions. In contrast to the pheromone- and immune-related genes, which are members of large, rapidly evolving families, gene types found enriched in stable genomic regions include those related to many basic cellular processes such as nucleotide binding, protein folding, and cell cycle regulation.
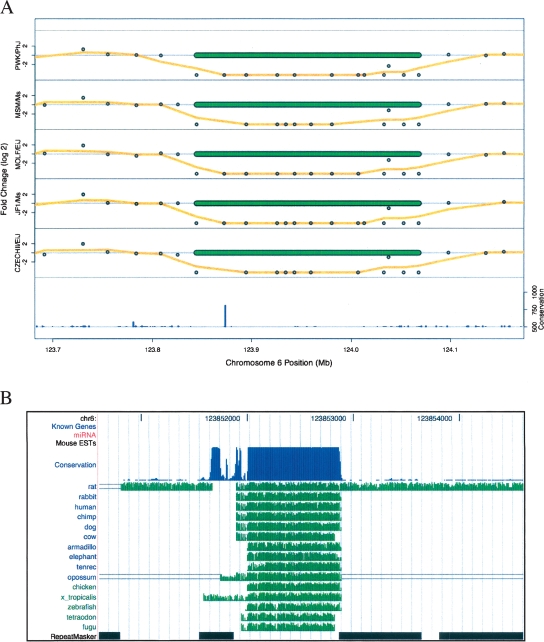
Alignment of multiple vertebrate genomes has led to the identification of highly conserved elements (HCEs) that cover ∼0.14% of the human genome (Siepel et al. 2005). The functional importance of these regions should be reflected in a paucity of these HCEs in deletion CNVs. Neither statistically significant enhancement nor exclusion of the top 1% scoring HCEs was observed in deletions (P = 0.25 and 0.77, respectively). An enrichment was only observed in amplifications and only when taking into account the top scoring 0.1% HCEs (enrichment = 38%, P = 0.0019). In fact, we did observe the deletion of HCEs. An example of an HCE found in a deletion CNV is shown in Figure 5A. This HCE, which has a conservation score in the 99.8th percentile for the mouse genome, is deleted in five mouse strains. A more detailed examination of this region (Fig. 5B) shows no overlap with any known genes, ESTs, or miRNAs (Kent et al. 2002; Blanchette et al. 2004), suggesting the presence of a novel genetic element.
Figure 5.
Deletions in five strains remove a highly conserved element on chromosome 6. (A) The log2 values of CGH probe fold-change values are shown as points along with their running mean (window size = 3) as an orange line for each of five strains. Extreme fold-changes are truncated at ±20 (log2 of ±4.2). The locations of the calculated deletions for each strain are shown as green bars. The bar graph at the bottom of the plot shows the conservation score for this genomic locus. (B) A detailed view of the chromosome 6 highly conserved element deleted in five mouse strains shows extensive cross-species homology as plotted by the UCSC genome browser (Kent et al. 2002). The conservation track, based on scores from the phastCons algorithm (Siepel et al. 2005), along with individual species alignments using the MultiZ algorithm (Blanchette et al. 2004) are shown. Known genes, miRNA, mouse EST, and RepeatMasker tracks are also displayed, but none are present in this region.
CNV/phenotype whole-genome association
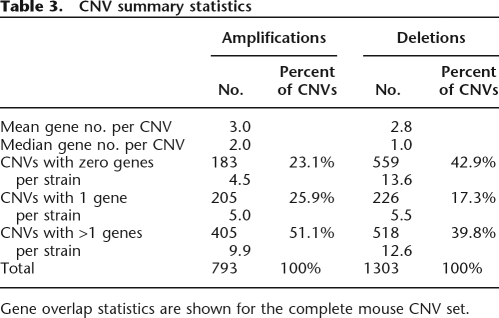
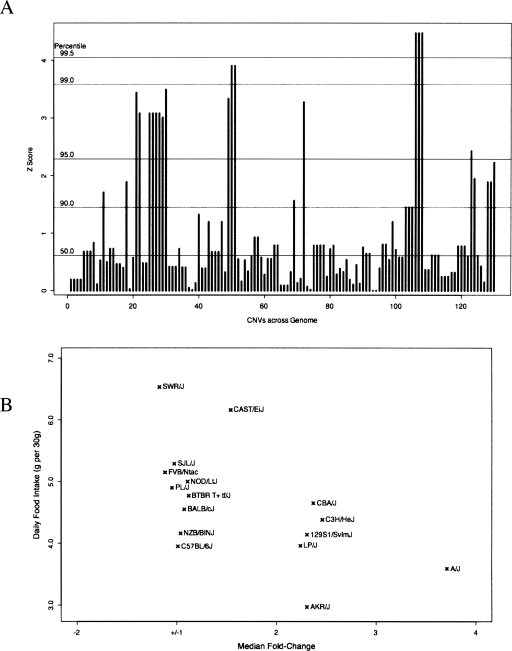
The recent availability of large amounts of mouse SNP genotyping data has allowed researchers to perform SNP/phenotype association studies in mice (Grupe et al. 2001; Wang et al. 2005; Liu et al. 2006). Individual CNVs may be expected to have a greater likelihood of exerting phenotypic effects than individual SNPs, offsetting their lower frequency. Furthermore, in contrast to the somatic CNVs found in human tumors (Hurst et al. 2004) and cell lines (Brookman-Amissah et al. 2005) which often span scores if not hundreds of genes, a large fraction of observed murine CNVs overlap only a single gene (Table 3). To assay the usefulness of performing CNV/phenotype association analysis, we tested the association between food intake—a complex multigenic trait known to vary greatly between mouse strains (Seburn 2001)—and CNVs in our data set. Association studies were performed by calculating the ratios of within- and between-group sums-of-squares when per-strain food-intake measurements were grouped by CNV allele at each CNV-containing locus throughout the genome. These values were compared to the values derived from 100 randomizations of allele memberships at each locus. Those loci which had z-scores greater than the 99.5th percentile of randomized scores were considered hits. From this genome-wide association analysis, only one set of three consecutive loci scored above the threshold (Fig. 6A). Mouse strains with the highest food intake, such as SWR/J and SJL/J, show no amplification across that region, strains with a duplication, such as C3H/J and LP/J, show intermediate food intake levels, and the A/J strain, with one of the lowest food intake levels, has an apparent quadruplication across the locus (Fig. 6B). Interestingly, this region overlaps with the glucagon-like peptide 1 receptor (Glp1r) gene, a gene which plays an important role in satiety and weight homeostasis (Navarro et al. 1996; Turton et al. 1996) and that has previously been shown to have a genetic linkage to energy intake in mice (Kumar et al. 2007).
Table 3.
CNV summary statistics
Gene overlap statistics are shown for the complete mouse CNV set.
Figure 6.
Whole genome CNV association with food intake. (A) Association scores are plotted for all tested CNV loci. CNV loci are plotted in genomic order along the X-axis with the height of each bar representing the z-score-transformed association score for that locus. Horizontal lines indicate z-score percentiles from randomized data. (B) Daily food intake shows a relationship to genomic amplification levels. Food intake from the Seburn1 MPD data set (Seburn 2001) is measured in gram of food per 30-g body weight. Genomic amplification levels are measured as median fold-change of probe intensities compared to C57BL/6J values for probes on chromosome 17 between positions 30,627,006 and 30,650,272.
Discussion
There is no doubt that SNPs play a major role in intra-species variation (The International HapMap Consortium 2005), while genomic segmental amplifications and deletions have long been understood to provide important raw material for evolution (Nei et al. 1997; Hancock 2005; Nei and Rooney 2005). More recently, researchers have started to understand the importance of CNVs as sources of variation within species (Li et al. 2004; Sharp et al. 2005; Conrad et al. 2006; Feuk et al. 2006; Redon et al. 2006; Graubert et al. 2007; Wong et al. 2007). This study reports the most comprehensive CNV analysis of the mouse genome yet performed. The generation of inbred mouse strains through brother-sister crosses has led to mice homozygous at all loci, capturing a snapshot of the genetic diversity present in the mouse population from which the strains were derived. Thus, in a way not possible with the CNV data on human individuals, this mouse strain CNV census provides us a static picture of CNV diversity in a species.
CNVs and in particular segmental amplifications are an important force in evolution, providing the raw material for the birth of new genes (Nei et al. 1997; Hancock 2005; Nei and Rooney 2005). The C57 and C58 strains were derived from two female mice living at the Granby mouse farm in 1921 (Festing 1998; Behringer et al. 2003) and likely sharing most of their CNVs. The C57L, C57BR, and C57BL strains were all derived from offspring of the original C57 cross. Most of the CNVs found between C57BL/6J and C57BR/cdJ, C57L/J, and C58/J strains—33, 25, and 38, respectively—likely arose in the intervening 86 yr. C57BL/6J and C57BL/10J are sister strains separated prior to 1937 (Festing 1998; Beck et al. 2000), and their 15 CNVs likely arose in the roughly 70 yr since their split. This rapid appearance of CNVs shows how quickly structural genomic variation can develop in a species. The pheromone and MHC receptor genes are both members of large families which play roles in sexual selection (Nei et al. 1997; Singh 2001). We have observed the enrichment of both of these families in CNVs. This may be a glimpse of evolution in action, frozen by the isolation and subsequent breeding to homozygosity of inbred strains.
In this study, we have shown that tens of megabases of the genome can be altered by CNVs, leading to genomes that vary by hundreds of genes. This surprising amount of copy number polymorphism exceeds that previously reported in mice (Li et al. 2004; Graubert et al. 2007). The variation observed covers not only genes which are of decreased importance to laboratory-raised animals like pheromone receptors, MHC receptors, and antibacterial defensins, which we find amplified and deleted in large blocks, but also genes which in other contexts have been shown to have critical effects on phenotype. An example of this is deletion of most of the Abca4 gene in the JF1/Ms strain (data not shown). A laboratory knockout of this gene in 129S4/SvJae mice leads to abnormal rod morphology in the eye, mimicking details of Stargardt disease, which is caused by mutations in ABCA4 in humans (Weng et al. 1999). We would predict that JF1/Ms mice would have a similar eye defect. The occurrence of these natural gene knockouts underscores the importance of understanding the genetic backgrounds in mouse strains that are used as model organisms. We also demonstrate that we can identify associations between the copy number of specific genomic loci and a complex phenotype of multigenic origin, specifically between the metabolically important Glp1r locus (Navarro et al. 1996; Turton et al. 1996) and levels of food intake.
While we have been able to rediscover most of the CNVs described in the previously most complete mouse strain CNV analysis (Graubert et al. 2007), we have extended that set of CNVs by roughly sevenfold. Like the Graubert study, which was also oligo array-based, we were unable to replicate most of the CNVs in the previous two BAC array-based mouse CNV studies, Li et al. (2004) and Snijders et al. (2005). The design of the oligo probes used in this and the Graubert studies were guided by recent genome assemblies and attempted to avoid overlap with repetitive regions. In contrast, the older BAC-based arrays were vulnerable to issues including tracking and annotation problems, the presence of chimeric BACs, incorrect genome mapping, and the presence of repetitive elements. While care was surely taken to ensure that the spotted BAC arrays were of high quality, few labs could match the reproducibility and robustness of modern commercially produced oligo arrays, let alone their sensitivity and depth of coverage. Likely these factors are all responsible for the inability to reproduce these older studies. The danger of the oligo-based arrays is that SNPs present within genomic regions probed by the arrays could affect hybridization intensities, leading to false CNV calls. However, since the relative frequency of deletions, the most likely type of false-positive CNV, has no correlation with known SNP content nor are CNVs enhanced for the presence of SNPs, the confounding role played by SNPs is likely small.
Mice have long been an important model organism with numerous disease-related phenotypes displayed by different strains (Beck et al. 2000). The phenotypic variety between inbred mouse strains is derived from genomes that vary not only through SNPs but also through differences in gene content and dosage due to CNVs. Although this study does not explain the processes which generate CNVs nor the specific phenotypic consequences of most of these CNVs, we believe this comprehensive analysis of the CNV content of the mouse genome lays the groundwork for a better understanding of murine phenotypes and genotypes in the post-genomic era.
Methods
CGH
Genomic DNA samples for CGH were either isolated from tail snips from mice ordered from Jackson Laboratories (Bar Harbor, Maine) or directly ordered as DNA from Jackson Laboratories. DNA from two male mice from each of 42 strains along with the C57BL/6J reference were labeled and hybridized to 244K 60-mer Mouse Genome arrays (Agilent Technologies). Agilent’s protocols were modified (see Supplemental Methods for detailed protocol) to allow low volume hybridization in MAUI mixing chambers (BioMicro Systems). Hybridizations for each strain were performed in duplicate using samples from different mice for the dye-flipped replicates. Arrays were scanned with an Agilent scanner and analyzed with the Agilent Feature Extraction software.
Statistical analysis
Each pair of dye-flipped replicates was combined by averaging loess-normalized hybridization intensities. A zero-threshold, determined by chromosome Y hybridization intensities in separate female mouse hybridizations, was subtracted from all intensities. Probes were dropped when intensities varied by more than twofold between replicates or where both experimental and reference intensities were close to zero. Fold-change values were capped at 20× or 1/20× when experimental or reference intensities, respectively, were close to zero.
Segmental copy number changes were identified by the GLAD algorithm R implementation (default settings except nmax = 12) (Hupe et al. 2004). Segments were compared both to the rest of the genome and to just the local genomic region for that sample set by Student’s t-test and retained when the one-tailed P-value passed an FDR cutoff of 0.01. These CNVs were further filtered by these criteria: (P-value ≥ 3) and (probe length > 3 and ç.75| > 1.45 or probe length = 3 and fc.75 > 1.95), where P-value is from the global t-test, probe length is the number of probes which define a CNV, and fc.75 is the 75th percentile fold-change value for the CNV probes. Randomized data sets were generated by taking each data set being analyzed and scrambling the fold-change scores in relation to the genomic locations. Ten such randomized data sets were generated for each strain.
Enrichment analyses for genome regions were done by the standard hypergeometric test. The mouse genome was divided into contiguous 10-kb regions with regions of very low array probe coverage excluded. These genome bins were scored for overlap with each type of genetic element and with amplification and deletion CNVs. Hypergeometric P-values were generated based on these bins. Significance was determined by an FDR cutoff of 0.05 on one-tailed P-values. Enrichment amount was determined by dividing the observed true/true counts by the expected true/true counts in a 2 × 2 contingency table. Gene family size and GO-term enrichment analyses were performed similarly except that the bins were individual genes.
For whole-genome phenotype/CNV association analysis, all CNVs were combined into a nonoverlapping set of CNV loci. Those loci for which there were at least two alleles with at least four strains each were retained for further analysis. At each locus the ratio of within-group sums-of-squares (WGSS) to between-group sums-of-squares (BGSS) for the analyzed phenotypic data was calculated. Additionally, at each locus, one hundred randomizations of allele membership were performed and WGSS/BGSS ratios for the randomized data were calculated. All ratios were converted to z-scores based on the distribution of the randomized ratios and hits were selected based on a hit threshold of the 99.5th percentile of randomized ratios.
QPCR validation
Primers and probes were designed using the Universal Probe Library (UPL) system (Mouritzen et al. 2005) with online tools (https://www.roche-applied-science.com/sis/rtpcr/upl/adc.jsp). Genomic positions, sequences of primers, UPL probe numbers, and detailed reaction conditions are described in Supplemental Methods. Reactions were run in the ABI Prism 7900HT real-time thermocycler and analyzed using SDS2.1 software (Applied Biosystems). All reactions were performed in duplicate and repeated at least twice. Relative copy number values were obtained by comparison to standard curves of C57BL/6J genomic DNA.
Acknowledgments
We thank David Shuford for helping to obtain and prepare genomic DNA samples.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to NCBI GEO under accession no. GSE9186.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6754607
References
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Dolinski K., Dwight S.S., Eppig J.T., Dwight S.S., Eppig J.T., Eppig J.T., et al. Gene Ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett M.T., Scheffer A., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Scheffer A., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Ben-Dor A., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Sampas N., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Lipson D., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Kincaid R., Tsang P., Curry B., Baird K., Meltzer P.S., Tsang P., Curry B., Baird K., Meltzer P.S., Curry B., Baird K., Meltzer P.S., Baird K., Meltzer P.S., Meltzer P.S., et al. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc. Natl. Acad. Sci. 2004;101:17765–17770. doi: 10.1073/pnas.0407979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J.A., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M., Eppig J.T., Festing M.F., Fisher E.M., Festing M.F., Fisher E.M., Fisher E.M. Genealogies of mouse inbred strains. Nat. Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Behringer R.R., Nagy A., Gerstenstein M., Vintersten K., Nagy A., Gerstenstein M., Vintersten K., Gerstenstein M., Vintersten K., Vintersten K. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. [Google Scholar]
- Birney E., Andrews T.D., Bevan P., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Andrews T.D., Bevan P., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Bevan P., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Chen Y., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Clarke L., Coates G., Cuff J., Curwen V., Cutts T., Coates G., Cuff J., Curwen V., Cutts T., Cuff J., Curwen V., Cutts T., Curwen V., Cutts T., Cutts T., et al. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Rosenbloom K., Clawson H., Green E.D., Clawson H., Green E.D., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue M.A., Grubb S.C., Maddatu T.P., Bult C.J., Grubb S.C., Maddatu T.P., Bult C.J., Maddatu T.P., Bult C.J., Bult C.J. Mouse Phenome Database (MPD) Nucleic Acids Res. 2007;35:D643–D649. doi: 10.1093/nar/gkl1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookman-Amissah N., Duchesnes C., Williamson M.P., Wang Q., Ahmed A., Feneley M.R., Mackay A., Freeman A., Fenwick K., Iravani M., Duchesnes C., Williamson M.P., Wang Q., Ahmed A., Feneley M.R., Mackay A., Freeman A., Fenwick K., Iravani M., Williamson M.P., Wang Q., Ahmed A., Feneley M.R., Mackay A., Freeman A., Fenwick K., Iravani M., Wang Q., Ahmed A., Feneley M.R., Mackay A., Freeman A., Fenwick K., Iravani M., Ahmed A., Feneley M.R., Mackay A., Freeman A., Fenwick K., Iravani M., Feneley M.R., Mackay A., Freeman A., Fenwick K., Iravani M., Mackay A., Freeman A., Fenwick K., Iravani M., Freeman A., Fenwick K., Iravani M., Fenwick K., Iravani M., Iravani M., et al. Genome-wide screening for genetic changes in a matched pair of benign and prostate cancer cell lines using array CGH. Prostate Cancer Prostatic Dis. 2005;8:335–343. doi: 10.1038/sj.pcan.4500826. [DOI] [PubMed] [Google Scholar]
- Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K., Carter N.P., Hurles M.E., Pritchard J.K., Hurles M.E., Pritchard J.K., Pritchard J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- Emanuel B.S., Shaikh T.H., Shaikh T.H. Segmental duplications: An 'expanding' role in genomic instability and disease. Nat. Rev. Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- Festing M.F. Inbred strains of mice: C57BL. Jackson Laboratories; Leicester, UK: 1998. [Google Scholar]
- Feuk L., Carson A.R., Scherer S.W., Carson A.R., Scherer S.W., Scherer S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Graubert T.A., Cahan P., Edwin D., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Cahan P., Edwin D., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Edwin D., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Selzer R.R., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Richmond T.A., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Eis P.S., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Shannon W.D., Li X., McLeod H.L., Cheverud J.M., Li X., McLeod H.L., Cheverud J.M., McLeod H.L., Cheverud J.M., Cheverud J.M., et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3:e3. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupe A., Germer S., Usuka J., Aud D., Belknap J.K., Klein R.F., Ahluwalia M.K., Higuchi R., Peltz G., Germer S., Usuka J., Aud D., Belknap J.K., Klein R.F., Ahluwalia M.K., Higuchi R., Peltz G., Usuka J., Aud D., Belknap J.K., Klein R.F., Ahluwalia M.K., Higuchi R., Peltz G., Aud D., Belknap J.K., Klein R.F., Ahluwalia M.K., Higuchi R., Peltz G., Belknap J.K., Klein R.F., Ahluwalia M.K., Higuchi R., Peltz G., Klein R.F., Ahluwalia M.K., Higuchi R., Peltz G., Ahluwalia M.K., Higuchi R., Peltz G., Higuchi R., Peltz G., Peltz G. In silico mapping of complex disease-related traits in mice. Science. 2001;292:1915–1918. doi: 10.1126/science.1058889. [DOI] [PubMed] [Google Scholar]
- Hancock J.M. Gene factories, microfunctionalization and the evolution of gene families. Trends Genet. 2005;21:591–595. doi: 10.1016/j.tig.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Hinds D.A., Stuve L.L., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Stuve L.L., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R., Ballinger D.G., Frazer K.A., Cox D.R., Frazer K.A., Cox D.R., Cox D.R. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- Hsu F., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Kent W.J., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Clawson H., Kuhn R.M., Diekhans M., Haussler D., Kuhn R.M., Diekhans M., Haussler D., Diekhans M., Haussler D., Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Hupe P., Stransky N., Thiery J.P., Radvanyi F., Barillot E., Stransky N., Thiery J.P., Radvanyi F., Barillot E., Thiery J.P., Radvanyi F., Barillot E., Radvanyi F., Barillot E., Barillot E. Analysis of array CGH data: From signal ratio to gain and loss of DNA regions. Bioinformatics. 2004;20:3413–3422. doi: 10.1093/bioinformatics/bth418. [DOI] [PubMed] [Google Scholar]
- Hurst C.D., Fiegler H., Carr P., Williams S., Carter N.P., Knowles M.A., Fiegler H., Carr P., Williams S., Carter N.P., Knowles M.A., Carr P., Williams S., Carter N.P., Knowles M.A., Williams S., Carter N.P., Knowles M.A., Carter N.P., Knowles M.A., Knowles M.A. High-resolution analysis of genomic copy number alterations in bladder cancer by microarray-based comparative genomic hybridization. Oncogene. 2004;23:2250–2263. doi: 10.1038/sj.onc.1207260. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L., Nickerson D.A., Nickerson D.A. Variation is the spice of life. Nat. Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- Kumar K.G., Poole A.C., York B., Volaufova J., Zuberi A., Richards B.K., Poole A.C., York B., Volaufova J., Zuberi A., Richards B.K., York B., Volaufova J., Zuberi A., Richards B.K., Volaufova J., Zuberi A., Richards B.K., Zuberi A., Richards B.K., Richards B.K. Quantitative trait loci for carbohydrate and total energy intake on mouse chromosome 17: Congenic strain confirmation and candidate gene analyses (Glo1, Glp1r) Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;292:R207–R216. doi: 10.1152/ajpregu.00491.2006. [DOI] [PubMed] [Google Scholar]
- Li J., Jiang T., Mao J.H., Balmain A., Peterson L., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Jiang T., Mao J.H., Balmain A., Peterson L., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Mao J.H., Balmain A., Peterson L., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Balmain A., Peterson L., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Peterson L., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Harris C., Rao P.H., Havlak P., Gibbs R., Cai W.W., Rao P.H., Havlak P., Gibbs R., Cai W.W., Havlak P., Gibbs R., Cai W.W., Gibbs R., Cai W.W., Cai W.W. Genomic segmental polymorphisms in inbred mouse strains. Nat. Genet. 2004;36:952–954. doi: 10.1038/ng1417. [DOI] [PubMed] [Google Scholar]
- Liu P., Wang Y., Vikis H., Maciag A., Wang D., Lu Y., Liu Y., You M., Wang Y., Vikis H., Maciag A., Wang D., Lu Y., Liu Y., You M., Vikis H., Maciag A., Wang D., Lu Y., Liu Y., You M., Maciag A., Wang D., Lu Y., Liu Y., You M., Wang D., Lu Y., Liu Y., You M., Lu Y., Liu Y., You M., Liu Y., You M., You M. Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat. Genet. 2006;38:888–895. doi: 10.1038/ng1849. [DOI] [PubMed] [Google Scholar]
- Mouritzen P., Noerholm M., Nielsen P.S., Jacobsen N., Lomholt C., Pfundheller H.M., Tolstrup N., Noerholm M., Nielsen P.S., Jacobsen N., Lomholt C., Pfundheller H.M., Tolstrup N., Nielsen P.S., Jacobsen N., Lomholt C., Pfundheller H.M., Tolstrup N., Jacobsen N., Lomholt C., Pfundheller H.M., Tolstrup N., Lomholt C., Pfundheller H.M., Tolstrup N., Pfundheller H.M., Tolstrup N., Tolstrup N. ProbeLibrary: A new method for faster design and execution of quantitative real-time PCR. Nat. Methods. 2005;2:313–316. [Google Scholar]
- Navarro M., de Rodriquez Fonseca F., Alvarez E., Chowen J.A., Zueco J.A., Gomez R., Eng J., Blazquez E., de Rodriquez Fonseca F., Alvarez E., Chowen J.A., Zueco J.A., Gomez R., Eng J., Blazquez E., Alvarez E., Chowen J.A., Zueco J.A., Gomez R., Eng J., Blazquez E., Chowen J.A., Zueco J.A., Gomez R., Eng J., Blazquez E., Zueco J.A., Gomez R., Eng J., Blazquez E., Gomez R., Eng J., Blazquez E., Eng J., Blazquez E., Blazquez E. Colocalization of glucagon-like peptide-1 (GLP-1) receptors, glucose transporter GLUT-2, and glucokinase mRNAs in rat hypothalamic cells: Evidence for a role of GLP-1 receptor agonists as an inhibitory signal for food and water intake. J. Neurochem. 1996;67:1982–1991. doi: 10.1046/j.1471-4159.1996.67051982.x. [DOI] [PubMed] [Google Scholar]
- Nei M., Rooney A.P., Rooney A.P. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Gu X., Sitnikova T., Gu X., Sitnikova T., Sitnikova T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. 1997;94:7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Collins C., Kuo W.L., Chen C., Zhai Y., Kuo W.L., Chen C., Zhai Y., Chen C., Zhai Y., Zhai Y., et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Fiegler H., Shapero M.H., Carson A.R., Chen W., Shapero M.H., Carson A.R., Chen W., Carson A.R., Chen W., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lundin P., Maner S., Massa H., Walker M., Chi M., Maner S., Massa H., Walker M., Chi M., Massa H., Walker M., Chi M., Walker M., Chi M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Seburn K. Mouse Phenome Database Web Site. The Jackson Laboratory; Bar Harbor, ME: 2001. Metabolic characterization (Seburn1). Accession Number MPD:92. [Google Scholar]
- Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Pertz L.M., Clark R.A., Schwartz S., Segraves R., Clark R.A., Schwartz S., Segraves R., Schwartz S., Segraves R., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw C.J., Lupski J.R., Lupski J.R. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum. Mol. Genet. 2004;13:57–64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P.B. Chemosensation and genetic individuality. Reproduction. 2001;121:529–539. doi: 10.1530/rep.0.1210529. [DOI] [PubMed] [Google Scholar]
- Snijders A.M., Nowak N.J., Huey B., Fridlyand J., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Nowak N.J., Huey B., Fridlyand J., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Huey B., Fridlyand J., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Fridlyand J., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Law S., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Conroy J., Tokuyasu T., Demir K., Chiu R., Mao J.H., Tokuyasu T., Demir K., Chiu R., Mao J.H., Demir K., Chiu R., Mao J.H., Chiu R., Mao J.H., Mao J.H., et al. Mapping segmental and sequence variations among laboratory mice using BAC array CGH. Genome Res. 2005;15:302–311. doi: 10.1101/gr.2902505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenson K.L., Von Smith R., Magnani P.A., Suetin H.R., Paigen B., Naggert J.K., Li R., Churchill G.A., Peters L.L., Von Smith R., Magnani P.A., Suetin H.R., Paigen B., Naggert J.K., Li R., Churchill G.A., Peters L.L., Magnani P.A., Suetin H.R., Paigen B., Naggert J.K., Li R., Churchill G.A., Peters L.L., Suetin H.R., Paigen B., Naggert J.K., Li R., Churchill G.A., Peters L.L., Paigen B., Naggert J.K., Li R., Churchill G.A., Peters L.L., Naggert J.K., Li R., Churchill G.A., Peters L.L., Li R., Churchill G.A., Peters L.L., Churchill G.A., Peters L.L., Peters L.L. Multiple trait measurements in 43 inbred mouse strains captures the phenotypic diversity characteristic of human populations. J. Appl. Physiol. 2007;102:2369–2378. doi: 10.1152/japplphysiol.01077.2006. [DOI] [PubMed] [Google Scholar]
- Turton M.D., O’Shea D., Gunn I., Beak S.A., Edwards C.M., Meeran K., Choi S.J., Taylor G.M., Heath M.M., Lambert P.D., O’Shea D., Gunn I., Beak S.A., Edwards C.M., Meeran K., Choi S.J., Taylor G.M., Heath M.M., Lambert P.D., Gunn I., Beak S.A., Edwards C.M., Meeran K., Choi S.J., Taylor G.M., Heath M.M., Lambert P.D., Beak S.A., Edwards C.M., Meeran K., Choi S.J., Taylor G.M., Heath M.M., Lambert P.D., Edwards C.M., Meeran K., Choi S.J., Taylor G.M., Heath M.M., Lambert P.D., Meeran K., Choi S.J., Taylor G.M., Heath M.M., Lambert P.D., Choi S.J., Taylor G.M., Heath M.M., Lambert P.D., Taylor G.M., Heath M.M., Lambert P.D., Heath M.M., Lambert P.D., Lambert P.D., et al. A role for glucagon-like peptide-1 in the central regulation of feeding. Nature. 1996;379:69–72. doi: 10.1038/379069a0. [DOI] [PubMed] [Google Scholar]
- Wang J., Liao G., Usuka J., Peltz G., Liao G., Usuka J., Peltz G., Usuka J., Peltz G., Peltz G. Computational genetics: From mouse to human? Trends Genet. 2005;21:526–532. doi: 10.1016/j.tig.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weng J., Mata N.L., Azarian S.M., Tzekov R.T., Birch D.G., Travis G.H., Mata N.L., Azarian S.M., Tzekov R.T., Birch D.G., Travis G.H., Azarian S.M., Tzekov R.T., Birch D.G., Travis G.H., Tzekov R.T., Birch D.G., Travis G.H., Birch D.G., Travis G.H., Travis G.H. Insights into the function of Rim protein in photoreceptors and etiology of Stargardt's disease from the phenotype in abcr knockout mice. Cell. 1999;98:13–23. doi: 10.1016/S0092-8674(00)80602-9. [DOI] [PubMed] [Google Scholar]
- Wong K.K., deLeeuw R.J., Dosanjh N.S., Kimm L.R., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., deLeeuw R.J., Dosanjh N.S., Kimm L.R., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., Dosanjh N.S., Kimm L.R., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., Kimm L.R., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., Cheng Z., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., Horsman D.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., MacAulay C., Ng R.T., Brown C.J., Eichler E.E., Ng R.T., Brown C.J., Eichler E.E., Brown C.J., Eichler E.E., Eichler E.E., et al. A comprehensive analysis of common copy-number variations in the human genome. Am. J. Hum. Genet. 2007;80:91–104. doi: 10.1086/510560. [DOI] [PMC free article] [PubMed] [Google Scholar]