Abstract
Comprehensive approaches to detect protein–protein interactions (PPIs) have been most successful in the yeast model system. Here we present “Cross-and-Capture,” a novel assay for rapid, sensitive assessment of PPIs via pulldown of differently tagged yeast strain arrays. About 500 yeast genes that function in DNA replication, repair, and recombination and nuclear proteins of unknown function were chromosomally tagged with six histidine residues or triple VSV epitopes. We demonstrate that the assay can interrogate a wide range of previously known protein complexes with increased resolution and sensitivity. Furthermore, we use “Cross-and-Capture” to identify two novel protein complexes: Rtt101p–Mms1p and Sae2p–Mre11p. The Rtt101p–Mms1p interaction was subsequently characterized by genetic and functional analyses. Our studies establish the “Cross-and-Capture” assay as a novel, versatile tool that provides a valuable complement for the next generation of yeast proteomic studies.
Recent years have witnessed a tremendous increase in our understanding of how the proteomes of model organisms are organized. These advances have been driven, in large part, by the application of novel yeast-based technologies that permit experimental analysis in high-throughput and at increasing levels of resolution (Kumar and Snyder 2001). Studies that assess the entire yeast proteome for protein–protein interactions (PPIs), protein localization, and modification and that query the functional relationships between genes have provided a wealth of information (for review, see Suter et al. 2006). A well-established tool to identify PPIs is the yeast two-hybrid (YTH) method, where bait and prey proteins are expressed as fusions to DNA binding and activation domains of a transcription factor (Fields and Song 1989; Brent and Finley 1994). An interaction between bait and prey proteins is monitored by the activation of reporter genes, allowing rapid assessment of a PPI. Entire interactomes can be monitored in high-throughput format via YTH by creating and screening large sets of baits and preys and relying on automation for many of the screening steps. This approach has been fruitful for Saccharomyces cerevisiae (Uetz et al. 2000; Ito et al. 2001), Drosophila melanogaster (Giot et al. 2003), Caenorhabditis elegans (Li et al. 2004), and Homo sapiens (Rual et al. 2005; Stelzl et al. 2005). Numerous variations of the YTH methodology have been developed, such as the split-ubiquitin assay (Johnsson and Varshavsky 1994; Stagljar et al. 1998; Thaminy et al. 2003; Paumi et al. 2007), the reverse Ras (rRas) system (Hubsman et al. 2001), a system that exploits the well-characterized G-protein signaling pathway as a readout (Ehrhard et al. 2000), and the tethered catalysis two-hybrid system to map interactions that are dependent on post-translational protein modifications (Guo et al. 2004). A second important approach in high-throughput proteomics is the biochemical isolation of tagged multiprotein complexes, followed by identification of the components by mass spectrometry. For example, tandem affinity purification (TAP) methodology allows the enrichment of protein complexes under largely physiological conditions (Rigaut et al. 1999). Using TAP, protein complexes have been isolated for much of the yeast proteome (Gavin et al. 2006; Krogan et al. 2006). Despite these advances to the interactome of S. cerevisiae, important interactions may be missed for various reasons, including the stringent purification procedure employed. Furthermore, the requirement for large cultures and mass-spectrometric analysis is laborious and resource-intensive.
Considering the limitations of the current existing assays, we developed a method that combines elements of YTH (bait and prey protein fusions, high-throughput format) with a simple and reliable biochemical pulldown assay. This “Cross-and-Capture” assay is a novel method that permits rapid analysis of PPIs by using differentially tagged yeast arrays in the two haploid yeast mating types. In total, 506 ORFs that encode proteins involved in DNA repair, replication, and recombination, and a large cohort of nuclear proteins of unknown function were tagged with six histidine residues (6×HIS) and triple VSV epitopes (3×VSV). In the course of validating the Cross-and-Capture system, we detected a number of known and, equally important, several novel PPIs. Hence, our results demonstrate that the Cross-and-Capture system provides a useful tool to promote further extensive analysis of the yeast proteome.
Results
Principle of the Cross-and-Capture assay
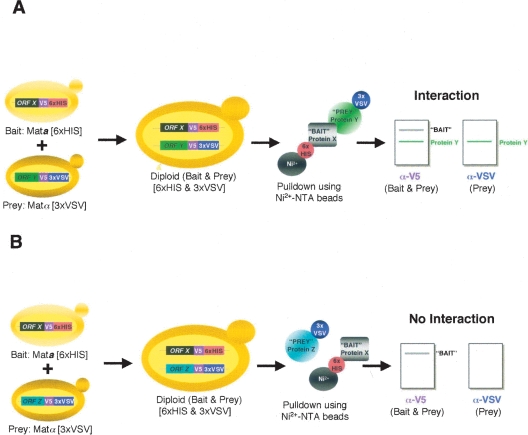
We generated two endogenously tagged haploid yeast arrays which are mated to one another to produce diploid yeast expressing two differentially tagged ORFs of interest (Fig. 1). In MATa cells, “bait” ORFs are tagged at the 3′ end with a sequence encoding six histidines (6×HIS), while “prey” ORFs in MATα cells are tagged with a sequence encoding a triple VSV tag (3×VSV). The combination of 6×HIS and 3×VSV tags was selected over other epitope tags (such as HA, myc, and FLAG) based on careful comparison of detection efficiency, reliability, and cost. Both tags also contain a V5 epitope to allow identification of both bait and prey proteins. To examine a particular PPI, a bait strain is crossed with a prey strain to generate a diploid expressing the desired bait- and prey-tagged proteins. Following diploid growth and cell lysis, extracts are incubated with nickel beads, allowing isolation of the 6×HIS-tagged bait and its associated proteins. Bound proteins are examined by immunoblot analysis for the presence of the bait and prey proteins using anti-V5 and anti-VSV antibodies. If the prey protein binds to the nickel beads in a bait-dependent manner, a PPI is inferred (Fig. 1A). Conversely, the absence of the prey protein in a pulldown reaction suggests that the two proteins fail to interact (Fig. 1B).
Figure 1.
Cross-and-Capture Assay. (A,B) A strain containing the bait ORFX tagged with a V5 epitope and six histidines (6×HIS) is crossed with strains that contain prey ORFY or ORFZ tagged with a V5 epitope followed by a triple VSV tag (3×VSV). Diploids, which express both tagged bait and prey, are grown on selective medium. Protein extracts from the diploids are then incubated with nickel beads (Ni2+-NTA), allowing isolation of bait (Protein X–6×HIS) and bait-associated prey protein (Protein Y–3×VSV) (A), whereas a non-interacting protein (Protein Z–3×VSV) will not bind (B). Proteins are then separated by SDS-PAGE, and blots are probed for bait and prey (anti-V5 antibody) and specifically for the prey (anti-VSV antibody) by immunoblot.
Generation and verification of tagged arrays
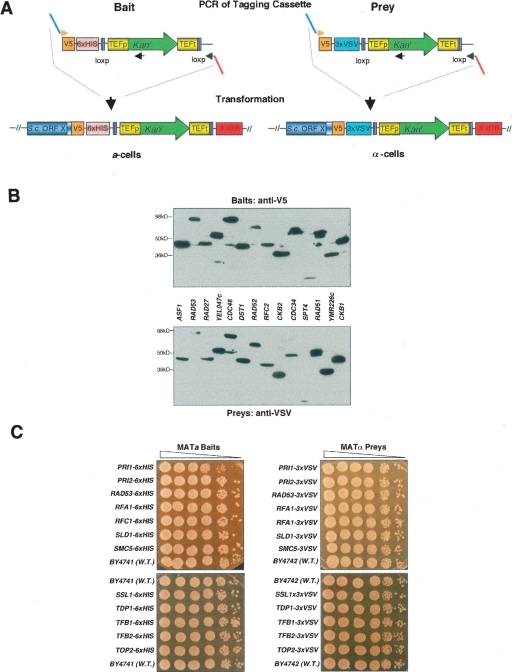
We applied the Cross-and-Capture system to a subset of 506 yeast ORFs (Supplemental Table 1): 258 of these ORFs encode proteins involved in DNA repair, replication, and recombination (Saccharomyces Genome Database, http://www.yeastgenome.org), and 248 ORFs encode proteins of unknown function that were assigned to the nucleus based on their localization patterns (Huh et al. 2003). PCR products containing the desired tags and the Kanr cassette were produced from bait- and prey-specific plasmids and transformed into MATa and MATα strains, respectively (Fig. 2A). Colony PCR products were sequenced over the ORF/tag junction.
Figure 2.
Generation and verification of tagged protein arrays. (A) To tag ORFX as bait (V5–6×HIS) and prey (V5–3×VSV), a set of primers is used that anneal to identical binding sites within the template plasmids and have flanking sequence homologous to ORFX. PCR products generated from the bait and prey templates are transformed into a- and α-cells, respectively. Homologous recombination occurs between the variable portion of the 5′ primer (light blue) and the 3′ terminus of the ORF, and between the variable portion of the 3′ primer (red) and the 3′ UTR) of ORFX. Transformants are selected on G418 plates, and colony PCR is performed to verify integration of the Kanr downstream of the desired ORF. Abbreviations: TEF, translational elongation factor; TEFp, TEF promoter; TEFt, TEF terminator: Kanr, kanamycin resistance; loxp, site for CRE specific homologous recombination. (B) Monitoring the quality of the arrays by Western analysis of 14 ORFs as baits and preys. We chose proteins with a high level of expression (>2000 molecules per cell) as judged by Ghaemmaghami et al. (2003). The asterisk (*) denotes possible misloading or protein degradation. Note in the RAD51 lane the multiple protein products. Expected protein sizes are listed in Supplemental Table 1. (C) Analysis of effects on cell growth by tagging essential genes. A total of 24 strains with essential genes tagged as baits (6×HIS) and preys (3×VSV) were grown to saturation and spotted in 10-fold dilutions on YPD. Pictures were taken after 2 d at 30°C.
We confirmed the successful tagging of all 506 ORFs by sequencing, and by immunoblotting ∼200 bait- and/or prey-tagged ORFs (see Fig. 2B; Supplemental Table 1). We found a very high success rate of detecting proteins in both backgrounds, and the observed levels of protein expression compared well to those reported previously (Ghaemmaghami et al. 2003). This correspondence of protein expression between two independently created endogenously tagged (bait and prey) strains also strongly suggests that the correct ORFs have been successfully tagged. Occasionally, the expression of low-abundance proteins could not be properly verified in whole-cell extracts with anti-V5. However, ∼90% of all bait proteins could be detected with anti-V5 in the pulldown. The few exceptions comprised very low copy number or meiotic proteins not expressed during vegetative growth. Of the 97 essential ORFs in our array, we successfully tagged 87 ORFs as both baits and preys, eight as either baits or preys, with only two refractory to tagging. No significant growth defects were found when 25 strains with essential proteins tagged as baits and preys were analyzed in spotting assays on YPD (Fig. 2C). Therefore, in most cases, tagging of essential genes revealed that our small tags (3–6 kDa) did not adversely affect cell growth or protein function (Fig. 2C).
Detection of protein–protein interactions using Cross-and-Capture
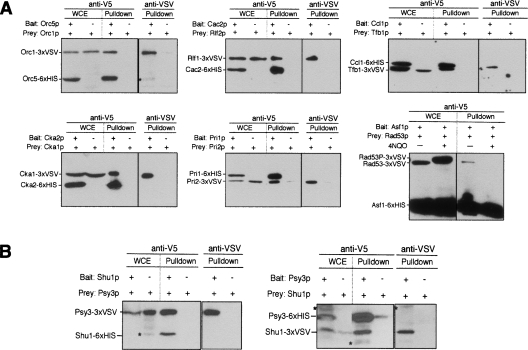
To test the ability of the Cross-and-Capture system to detect PPIs, we selected six high-confidence protein interactions (according to SGD) previously demonstrated using different methods, such as YTH and TAP purification (Fig. 3A). The first example considered was the origin recognition complex (ORC), a multiprotein complex regulating initiation of DNA replication (Bell and Dutta 2002). One diploid strain contained the Orc5 protein as bait and the Orc1 protein as prey, and the control diploid contained only the Orc1p prey. We detected both Orc5 and Orc1 proteins as prominent bands migrating at ∼55 and ∼100 kDa, respectively, with anti-V5 or anti-VSV. In total extracts from the control diploids, only the prey protein Orc1p was detected. When the pulldown reactions were analyzed, the Orc1p prey was detected along with the Orc5p bait. The control experiment shows that pulldown of Orc1p was dependent upon the presence of the Orc5p bait. In the same manner, interactions within the chromatin assembly complex (Cac2p–Rlf2p), the casein kinase complex (Cka2p–Cka1p), DNA primase (Pri1p–Pri2p), and the cyclin/TFIIH complex (Ccl1p–Tfb1p) were confirmed (Fig. 3A). Moreover, we reproduced the interaction between the Rad53p checkpoint kinase and the chromatin assembly factor Asf1p that occurs exclusively in the absence but not in the presence of DNA damage (Emili et al. 2001). All these interactions were also successfully confirmed by reciprocal pulldowns (data not shown). Hence, we successfully detected known constitutive and conditional PPIs using the Cross-and-Capture system.
Figure 3.
Proof-of-principle of Cross-and-Capture. (A) Detection of known protein complexes with Cross-and-Capture. Two diploids were examined, one expressing the bait and prey and the other expressing only the prey. Whole-cell extracts (WCEs) and pulldowns are probed with anti-V5. Pulldowns are additionally probed with anti-VSV. The origin-recognition complex (Orc1p–Orc5p), the chromatin assembly complex (Cac2p–Rlf2p), the casein kinase complex (Cka2p–Cka1p), DNA primase (Pri1p–Pri2p), the cyclin/TFIIH complex (Ccl1p–Tfb1p), and the Rad53p–Asf1p interaction are shown. For Rad53p–Asf1p interaction, growth for 1 h in 3 μg/mL 4NQO, is indicated by “+”. Note the band shift by phosphorylation (Rad53P–3×VSV) in presence of 4NQO. (B) Identification of the Shu1p–Psy3p complex. Pulldowns are done from diploids expressing Psy3 and Shu1 as both baits and preys. WCEs and pulldown are probed with anti-V5 and anti-VSV antibodies. Nonspecific background signals in the Western blots are indicated (*).
To show that Cross-and-Capture can characterize proteins that exist in low copy numbers or have small sizes, we selected Psy3p, which has an as yet undefined role in homologous recombination (Shor et al. 2002; Nislow and Giaever 2003). Psy3p is known to form a functional unit with Shu1p, Shu2p, and Csm2p, and although interactions between these proteins have been shown by YTH analysis (Ito et al. 2001; Shor et al. 2002), biochemical methods have, thus far, failed to identify these interactions. Probing extracts with anti-V5 and anti-VSV, we confirmed the expression of Shu1p and Psy3p (note that Shu1p could not be detected by V5 antibody in the whole cell extract due to its low copy number). Cross-and-Capture showed that the Psy3p prey interacts with the Shu1p bait (Fig. 3B, left panel). Furthermore, we confirmed the interaction by reciprocal pulldown with Shu1p as the prey and with Psy3p as the bait (Fig. 3B, right panel). We also tested and confirmed numerous other interactions by Cross-and-Capture (see Table 1 for all PPIs detected). In summary, we found a large number of known complexes but also a pair of small proteins (Psy3p–Shu1p) that previously escaped detection by TAP tag, possibly because of interference with protein function/interaction from the much larger TAP tag.
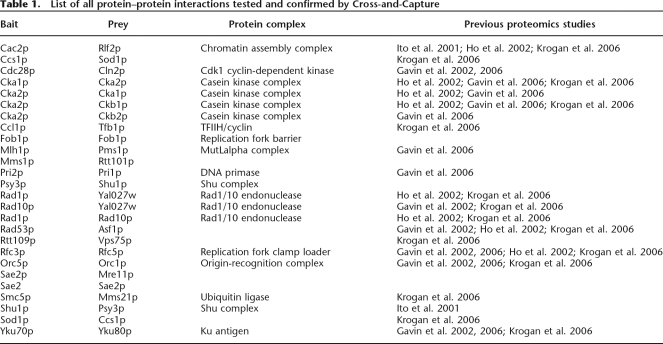
Table 1.
List of all protein–protein interactions tested and confirmed by Cross-and-Capture
Screening for novel protein–protein interactions using Cross-and-Capture
Besides detecting known or predicted PPIs, the Cross-and-Capture system can be adjusted to detect novel PPIs (Fig. 4). To assess many pulldown reactions in a screening procedure, we generated a series of diploids that contained only one selected prey but different baits, either from the whole bait array or from a subset thereof (Fig. 4A). Pulldown reactions are then run in parallel, thereby showing the amount of prey protein on the blot that is associated with individual baits. Detecting only one prey protein in a series of pulldowns minimizes the control pulldowns (no bait) that need to be performed for each prey to exclude nonspecific binding to the nickel beads. The candidate proteins for interaction screening were chosen either at random or when genetic evidence suggested a physical interaction. Since a stringent control for nonspecific association (prey only) is included in all our Western blots, the overall rate of false positives is very low. Furthermore, false positives that still show up in the sensitive anti-VSV Western blot are usually eliminated by comparison with the corresponding anti-V5 Western blot. Finally, we only scored an interaction as positive that could be reproduced at least three times.
Figure 4.
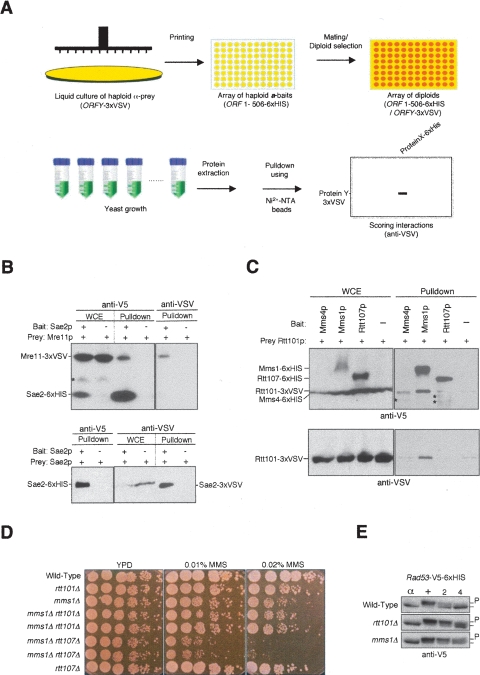
Using Cross-and-Capture to screen for novel PPIs. (A) Screening scheme. A strain containing a prey (ORFY–3×VSV) is combined with a set of different bait strains (ORF1-506–6×HIS) and diploids are selected. Extracts and Western blots are processed in parallel, and pulldowns of the prey protein are assessed. (B) Sae2p interacts with Mre11p and Sae2p. Pulldowns are from diploids expressing Mre11–3×VSV or Sae2–3×VSV as prey and Sae2–6×HIS as bait or no bait control (−). (C) Identification of the Rtt101p–Mms1p complex. Whole cell extracts (WCEs) and pulldowns probed with anti-V5 and anti-VSV antibodies are shown. Cells are treated with 0.02% MMS for 2 h before harvesting. Pulldowns are from diploids expressing Rtt101–3×VSV as prey and Mms1p, Mms4p, and Rtt107p as bait proteins (6×HIS) or no bait control (−). Nonspecific background signals in the Western blots are indicated (*). (D) Epistasis analysis for rtt101Δ and mms1Δ compared to rtt107Δ combined with mms1Δ. Cells are grown to mid-log phase and analyzed by spot assay in fivefold serial dilutions on YPD, YPD + 0.01% MMS, and YPD + 0.02% MMS. Pictures were taken after 2–3 d at 30°C. (E) Phosphorylation status of Rad53p (P) was determined by blotting Rad53–6×HIS from wild type, rtt101Δ, and mms1Δ strains with anti-V5 at indicated time points. Cells are arrested in G1 for 2 h with α-factor (α) and released into YPD containing 0.033% MMS for 2 h (+). Cells are then incubated with fresh YPD, and samples are collected after 2 and 4 h.
We first applied this scheme to screen for interactors of Sae2p, a protein involved in DNA double-strand break repair but for which no physical interactors are known (Rattray et al. 2001; Clerici et al. 2005). Screening for pulldown with ∼130 candidate proteins, we found that Sae2p associates with Mre11p (Fig. 4B, upper panel). Consistent with our results showing that Sae2p and Mre11p physically interact, Sae2p has been found to modulate the function of the Mre11p–Rad50p–Xrs2p complex in double-strand break (DSB) repair (Clerici et al. 2005, 2006). Interestingly, we found that Sae2p also interacts with itself, indicating dimer or multimer formation (Fig. 4B, lower panel).
Furthermore, we screened for interactors of Rtt101p, a cullin-based ubiquitin ligase that promotes DNA replication and functions in DNA damage repair (Luke et al. 2006). Based on analysis of synthetic lethal interactions, Rtt101p is predicted to form a functional module with Rtt107p, Mms1p, and Mms22p (Pan et al. 2006; Collins et al. 2007). Using Rtt101p as a prey, we found a clear association with Mms1p but not with Rtt107p, Mms4p, nor a large number (∼70) of other bait proteins (Fig. 4C and data not shown). The physical association between Mms1p and Rtt101p was also confirmed by a reciprocal pulldown experiment (data not shown). Since Mms1p and Rtt101p form a stable complex, one may suggest that the two proteins participate in the same functional pathway. Indeed, rtt101Δ did not further compromise growth of mms1Δ cells on methyl methanesulfonate (MMS), whereas the combination of mms1Δ and rtt107Δ and that of mms1Δ and rad52Δ led to increased MMS sensitivity (Fig. 4D; data not shown). Deletion of RTT101 compromises the recovery from checkpoint-mediated arrest following DNA damage, which manifests itself as persistent phosphorylation of Rad53p after cells have been removed from DNA damage (Luke et al. 2006). To test if mms1Δ shows the same phenotype, synchronized cells in G1-phase (α-factor) were subjected to DNA damage by release into S-phase in MMS-containing medium. This was followed by a 2-h release into fresh medium without MMS. Analysis of the electrophoretic mobility of the Rad53–6×HIS bait demonstrated that mms1Δ results in persistent phosphorylation of Rad53p during recovery from DNA damage, comparable to that seen in rtt101Δ (Fig. 4E). Hence, both the genetic epistasis and the functional assay underscore the biological significance of the Rtt101p–Mms1p interaction identified via Cross-and-Capture. Importantly, these two novel PPIs, identified by Cross-and-Capture (Sae2p–Mre11p and Rtt101p–Mms1p), could not be detected by the two comprehensive YTH screens (Uetz et al. 2000; Ito et al. 2001), and, in addition, Rtt101p causes a self-activation problem as a bait in a conventional YTH (data not shown).
Discussion
Herein we report on the development of a novel yeast-based PPI assay, the Cross-and-Capture system. Cross-and-Capture permits a rapid identification of PPIs that have previously escaped detection by other technologies. Detection of such “high-value” interactions is an important complement to existing yeastbased approaches, especially as we seek to refine and improve the resolution of the proteome. In particular, Cross-and-Capture should prove useful to validate results from YTH and mass-spectrometry–based approaches. Because the volume of cells required for Cross-and-Capture is relatively small, the technique is “user friendly” for examining constitutive, but also conditional PPIs, i.e., in presence of DNA damage. At its current stage of development, Cross-and-Capture, therefore, constitutes an ideal tool for “mid-sized” screens that may include 100–200 potential interactors. Refinement of our assay is focused on reducing the sample requirement even further and allowing more efficient screening on a larger scale. The small size of our tags ensures a minimum interference with normal protein function, especially in the case of small proteins. Cross-and-Capture can also be used to rapidly determine self-interaction of proteins (see Sae2p and Fob1p in Table 1). Therefore, our differentially tagged yeast arrays are efficient, versatile, and flexible and should prove ideal for future yeast-based proteomics studies.
Importantly, our differentially tagged yeast arrays can also interrogate other aspects of the yeast proteome besides determination of PPIs. For example, our system is well suited for measuring protein expression levels because the Cross-and-Capture tags are integrated at the chromosomal loci. Our tagged arrays may be used to characterize a variety of post-translational modifications (PTMs) under varying growth conditions. Previously, the O’Shea laboratory used the TAP collection in a systematic screen for sumoylated proteins by immunoprecipitating the epitope-tagged proteins with IgG, followed by detection of sumoylation by Western blot (Wykoff and O’Shea 2005). The authors considered their method superior to detect sumoylation of less-abundant proteins when compared with mass spectrometry. When a similar approach with our 6×HIS-tagged strain collection was taken, extracts could be prepared under denaturing conditions, thus allowing detection of labile PTMs such as sumoylation and ubiquitination.
As with any novel methodology, the Cross-and-Capture system has some practical constraints. Integrated tags avoid artifacts associated with protein overexpression, but such endogenous tagging presents a challenge for detecting poorly expressed proteins. Based on our experience, the sensitivity for protein detection can be maximized by increasing the amount of protein extract and by probing the Western blots with a maximum sensitivity reagent (e.g., Pierce’s “Femto” kit). Despite this limitation, the enrichment achieved during the pulldown allowed us to detect even rare protein species. Another consideration when using the cross and capture system is the occurrence of nonspecific protein binding to the nickel beads. Binding specificity can be altered by changing protein concentration, buffer conditions (imidazole), or the source of nickel beads. As an alternative to pulldown with nickel beads, the tagged arrays can be used for traditional immunoprecipitation, using an anti-VSV antibody or an anti-6×HIS antibody. Notably, our large collection of prey strains can be mated with the tandem affinity purification (TAP) lines produced by Ghaemmaghami et al. (2003), and hence the two methods can be combined to confirm or uncover protein interactions.
Potentially, the Cross-and-Capture system can be expanded by tagging all 6200 yeast ORFs. Further optimizing the extraction and pulldown procedures for sample processing and using direct detection of PPIs in 96-well plates will eventually allow us to advance our system to a true high-throughput methodology. Having established that differentially tagged arrays are an essential part of the toolkit for proteomics in S. cerevisiae, the concept and experimental design of Cross-and-Capture should be readily adaptable for screening the proteomes of other organisms.
Methods
Construction of plasmids for tagging
To construct the bait plasmid (pV5–6×HIS), PCR was performed on the pYES2.1/V5–His/lacZ plasmid (Invitrogen) with a 5′ primer, which generated a BamHI site (5′-GCGGATCCGGTA AGCCTATCCCTAACCCTC-3′) and a 3′ primer, which generated a SalI site (5′-TGACGTCGACCTACACCGAACTGAGATAC-3′). This fragment was cloned into the BamHI/SalI sites of the pU6H2MYC plasmid (De Antoni and Gallwitz 2000). To eliminate a redundant primer binding site within the bait vector and minimize the PCR product generated for in vivo recombination, the newly generated bait vector was cut with SalI and BsrGI and the ends were filled in using Klenow and subsequently re-ligated. To construct the prey plasmid, PCR was performed on the pU6H3VSV plasmid (De Antoni and Gallwitz 2000) with a 5′ primer which generated a BamHI site and a V5 epitope tag (5′-GCGGATCCGGTAAGCCTATCCCTAACCCTCTCCTCGGT CTCGATTCTACGGGATACACCGATATCGAGATG-3′), and a 3′ primer located downstream of the PstI site within pU6H3VSV. The BamHI/PstI-cut PCR product was cloned back into the pU6H2MYC plasmid to generate pV5–3×VSV.
Yeast strains
BY4741 (MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0) was used as the background for the bait strains, and BY4742 (MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0) was used as the background for the prey strains. See Supplemental Table 2 for additional strains that were generated for this study.
PCR and yeast transformations
To generate a tagging cassette for homologous recombination, PCR was performed using bait and prey plasmids as templates. For both plasmids, identical forward and reverse primers (QIAGEN Operon) were used to generate the bait and prey ORF-specific tagging cassette. The forward primer contains 40 bases homologous to the 3′ end of the targeted gene up to, but not including, the gene’s stop codon and an additional 20 which anneals to the V5 epitope (5′homology-GGTAAGCCTATC CCTAACCC-3′). The reverse primer contains 40 bases homologous to the 3′ untranslated region (UTR) of the targeted gene which begins 110 bp downstream of the gene’s stop codon and an additional 20 which anneals 3′ to the Kanr cassette (5′homology-CCCCGCGCGTTGGCCGATTC-3′). Fifty-microliter PCR reactions were performed using a mixture of Taq and Pfu polymerase (6.6 U/1 U, respectively) with the following settings: 95°C, 1 min, (95°C, 15 sec; 60°C, 20 sec; 70°C, 2 min) for 35 cycles, and then 70°C, 5 min. The entire PCR product was used to transform yeast cells using the lithium acetate method (Gietz et al. 1992). Prior to transformation, competent yeast cells were frozen at −80°C using a NALGENE Cryo 1°C Freezing Container. After transformation, yeast cells with integrated PCR product were selected on G418 plates (200 μg/mL, Calbiochem) for resistance conferred by the integrated Kanr cassette. Transformants that grew on the G418 plates were restreaked and tested for proper integration of the tagging cassette via colony PCR. The forward primer for colony PCR is 20 bases and binds to a region located 300–400 bp from the targeted gene’s stop codon. The reverse primer for colony PCR anneals to the Kanr cassette (5′-GAGCGTTTCCCTGCTCGCAG-3′). Positive colony PCR products were sequenced (GATC Biotech), and glycerol stocks of each strain were prepared.
Immunoblot analysis
Samples were run on SDS–polyacrylamide gels and transferred to a PVDF membrane (Millipore, Immobilon-P) via tank transfer. Transfer buffer was prepared without methanol (25 mM Tris, 192 mM glycine). Membranes were probed either with an antibody directed against the V5 epitope (1:5000; mouse monoclonal, Invitrogen) or the 3×VSV epitope (1:800; mouse monoclonal, Roche). Anti-mouse secondary antibodies were horseradish peroxidase-conjugated and detected using either of the light-emitting substrates Supersignal West Dura or Supersignal West Femto (Pierce), depending upon the expression level of the proteins under consideration.
Mating, extract preparation, and pulldowns
Individual haploid bait and prey strains were mixed together overnight on YPD medium and diploids were selected on synthetic complete medium (SC) lacking methionine and lysine (−Met −Lys). Precultures of diploids were diluted to ∼2.0 × 106 cells/mL in 2–20 mL of SC −Met −Lys or YPD liquid medium and cultured until the cell density reached ∼2.0 × 107 cells/mL (mid-log phase). For some proteins expressed at very low copy numbers (<500 molecules/cell), the culture volume was increased to 50–100 mL to obtain optimal results. To induce DNA damage, cells were treated with 0.02% MMS for 1–2 generations before harvest. Cells were harvested, washed once with cold ddH2O, and resuspended in maximally 750 μL of lysis buffer (120 mM NaCl, 50 mM Tris-HCl at pH 7.5, 0.1% NP-40, 5 mM β-glycerol phosphate, 2 mM magnesium acetate, 10 mM imidazole, 2 mM PMSF, 1 μg/mL leupeptin, 1 μg/mL pepstatin, 1 μg/mL benzamidine). Cells were then vortexed for 10 min at 4°C in round-bottom tubes containing 400 μL (maximum) of glass beads (425–600 μM, Sigma). Lysed cells were spun at 4°C at maximum speed for 10 min. Supernatants were subsequently added to 50 μL of nickel beads prewashed with lysis buffer (Probond nickel-chelating resin beads, Invitrogen). The nickel beads plus supernatants were then incubated at 4°C with gentle rolling for 2 h. After the incubation, nickel beads were washed (4 × 3 mL) with lysis buffer utilizing a disposable polypropylene column with a 1-mL bead capacity (QIAGEN). During two wash steps, columns were agitated with gentle rolling for 5 min before the wash buffer was removed. After washes, the beads were collected and eluted with 50 μL of elution buffer (200 mM imidazole in PBS). The eluted proteins were mixed with 10 μL of SDS loading buffer and then heated at 95°C for 5 min before SDS-PAGE.
Acknowledgments
We thank Charlie Boone, Grant Brown, Dawn Edmonds, Stanley Fields, Takashi Ito, Steve Jackson, Susan Michaelis, Tania Roberts, and Raffi Tonikian for advice and support during the experimental stages of this study and for assistance in the writing of the manuscript. The Stagljar laboratory is supported by grants from the Canadian Foundation for Innovation (CFI), Canadian Institute for Health Research (CIHR), National Cancer Institute of Canada (NCIC), Genome Canada and the Ontario Genomics Institute, Gebert Rüf Foundation, Genentech, and Novartis.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6667007
References
- Bell S.P., Dutta A., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Brent R., Finley R.L., Finley R.L. Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc. Natl. Acad. Sci. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M., Mantiero D., Lucchini G., Longhese M.P., Mantiero D., Lucchini G., Longhese M.P., Lucchini G., Longhese M.P., Longhese M.P. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- Clerici M., Mantiero D., Lucchini G., Longhese M.P., Mantiero D., Lucchini G., Longhese M.P., Lucchini G., Longhese M.P., Longhese M.P. The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep. 2006;7:212–218. doi: 10.1038/sj.embor.7400593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S.R., Miller K.M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Miller K.M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., Schuldiner M., Gebbia M., Recht J., Shales M., Gebbia M., Recht J., Shales M., Recht J., Shales M., Shales M., et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- De Antoni A., Gallwitz D., Gallwitz D. A novel multi-purpose cassette for repeated integrative epitope tagging of genes in Saccharomyces cerevisiae. Gene. 2000;246:179–185. doi: 10.1016/s0378-1119(00)00083-4. [DOI] [PubMed] [Google Scholar]
- Ehrhard K.N., Jacoby J.J., Fu X.Y., Jahn R., Dohlman H.G., Jacoby J.J., Fu X.Y., Jahn R., Dohlman H.G., Fu X.Y., Jahn R., Dohlman H.G., Jahn R., Dohlman H.G., Dohlman H.G. Use of G-protein fusions to monitor integral membrane protein–protein interactions in yeast. Nat. Biotechnol. 2000;18:1075–1079. doi: 10.1038/80274. [DOI] [PubMed] [Google Scholar]
- Emili A., Schieltz D.M., Yates J.R., Hartwell L.H., Schieltz D.M., Yates J.R., Hartwell L.H., Yates J.R., Hartwell L.H., Hartwell L.H. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- Fields S., Song O., Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Rick J.M., Michon A.M., Cruciat C.M., Michon A.M., Cruciat C.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dumpelfeld B., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dumpelfeld B., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dumpelfeld B., Krause R., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dumpelfeld B., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dumpelfeld B., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dumpelfeld B., Rau C., Jensen L.J., Bastuck S., Dumpelfeld B., Jensen L.J., Bastuck S., Dumpelfeld B., Bastuck S., Dumpelfeld B., Dumpelfeld B., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S., Belle A., Dephoure N., O’Shea E.K., Weissman J.S., Dephoure N., O’Shea E.K., Weissman J.S., O’Shea E.K., Weissman J.S., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R.A., Schiestl R.H., St Jean A., Woods R.A., Schiestl R.H., Woods R.A., Schiestl R.H., Schiestl R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Hao Y.L., Ooi C.E., Godwin B., Vitols E., Ooi C.E., Godwin B., Vitols E., Godwin B., Vitols E., Vitols E., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Guo D., Hazbun T.R., Xu X.J., Ng S.L., Fields S., Kuo M.H., Hazbun T.R., Xu X.J., Ng S.L., Fields S., Kuo M.H., Xu X.J., Ng S.L., Fields S., Kuo M.H., Ng S.L., Fields S., Kuo M.H., Fields S., Kuo M.H., Kuo M.H. A tethered catalysis, two-hybrid system to identify protein–protein interactions requiring post-translational modifications. Nat. Biotechnol. 2004;22:888–892. doi: 10.1038/nbt985. [DOI] [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Millar A., Taylor P., Bennett K., Boutilier K., Taylor P., Bennett K., Boutilier K., Bennett K., Boutilier K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hubsman M., Yudkovsky G., Aronheim A., Yudkovsky G., Aronheim A., Aronheim A. A novel approach for the identification of protein–protein interaction with integral membrane proteins. Nucleic Acids Res. 2001;29:e18. doi: 10.1093/nar/29.4.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K., Howson R.W., Weissman J.S., O’Shea E.K., Weissman J.S., O’Shea E.K., O’Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Yoshida M., Hattori M., Sakaki Y., Hattori M., Sakaki Y., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N., Varshavsky A., Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Li J., Pu S., Datta N., Tikuisis A.P., Pu S., Datta N., Tikuisis A.P., Datta N., Tikuisis A.P., Tikuisis A.P., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kumar A., Snyder M., Snyder M. Emerging technologies in yeast genomics. Nat. Rev. Genet. 2001;2:302–312. doi: 10.1038/35066084. [DOI] [PubMed] [Google Scholar]
- Li S., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Vidalain P.O., Han J.D., Chesneau A., Hao T., Han J.D., Chesneau A., Hao T., Chesneau A., Hao T., Hao T., et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B., Versini G., Jaquenoud M., Zaidi I.W., Kurz T., Pintard L., Pasero P., Peter M., Versini G., Jaquenoud M., Zaidi I.W., Kurz T., Pintard L., Pasero P., Peter M., Jaquenoud M., Zaidi I.W., Kurz T., Pintard L., Pasero P., Peter M., Zaidi I.W., Kurz T., Pintard L., Pasero P., Peter M., Kurz T., Pintard L., Pasero P., Peter M., Pintard L., Pasero P., Peter M., Pasero P., Peter M., Peter M. The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr. Biol. 2006;16:786–792. doi: 10.1016/j.cub.2006.02.071. [DOI] [PubMed] [Google Scholar]
- Nislow C., Giaever G., Giaever G. “Chemogenomics: Tools for protein families” and “Chemical genomics: Chemical and biological integration”. Pharmacogenomics. 2003;4:15–18. doi: 10.1517/phgs.4.1.15.22579. [DOI] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D., Yuan D.S., Wang X., Bader J.S., Boeke J.D., Wang X., Bader J.S., Boeke J.D., Bader J.S., Boeke J.D., Boeke J.D. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Paumi C.M., Menendez X., Arnoldo A., Engels K., Iyer R.K., Thaminy S., Georgiev O., Barral Y., Michaelis S., Stagljar I., Menendez X., Arnoldo A., Engels K., Iyer R.K., Thaminy S., Georgiev O., Barral Y., Michaelis S., Stagljar I., Arnoldo A., Engels K., Iyer R.K., Thaminy S., Georgiev O., Barral Y., Michaelis S., Stagljar I., Engels K., Iyer R.K., Thaminy S., Georgiev O., Barral Y., Michaelis S., Stagljar I., Iyer R.K., Thaminy S., Georgiev O., Barral Y., Michaelis S., Stagljar I., Thaminy S., Georgiev O., Barral Y., Michaelis S., Stagljar I., Georgiev O., Barral Y., Michaelis S., Stagljar I., Barral Y., Michaelis S., Stagljar I., Michaelis S., Stagljar I., Stagljar I. Mapping protein–protein interactions for the yeast ABC transporter Ycf1p by integrated split-ubiquitin membrane yeast two-hybrid (iMYTH) analysis. Mol. Cell. 2007;26:15–25. doi: 10.1016/j.molcel.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Rattray A.J., McGill C.B., Shafer B.K., Strathern J.N., McGill C.B., Shafer B.K., Strathern J.N., Shafer B.K., Strathern J.N., Strathern J.N. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: A role for SAE2/COM1. Genetics. 2001;158:109–122. doi: 10.1093/genetics/158.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B., Rutz B., Wilm M., Mann M., Seraphin B., Wilm M., Mann M., Seraphin B., Mann M., Seraphin B., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., Dreze M., Ayivi-Guedehoussou N., Ayivi-Guedehoussou N., et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Shor E., Gangloff S., Wagner M., Weinstein J., Price G., Rothstein R., Gangloff S., Wagner M., Weinstein J., Price G., Rothstein R., Wagner M., Weinstein J., Price G., Rothstein R., Weinstein J., Price G., Rothstein R., Price G., Rothstein R., Rothstein R. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagljar I., Korostensky C., Johnsson N., te Heesen S., Korostensky C., Johnsson N., te Heesen S., Johnsson N., te Heesen S., te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc. Natl. Acad. Sci. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Zenkner M., Schoenherr A., Koeppen S., Schoenherr A., Koeppen S., Koeppen S., et al. A human protein–protein interaction network: A resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Suter B., Auerbach D., Stagljar I., Auerbach D., Stagljar I., Stagljar I. Yeast-based functional genomics and proteomics technologies: The first 15 years and beyond. Biotechniques. 2006;40:625–644. doi: 10.2144/000112151. [DOI] [PubMed] [Google Scholar]
- Thaminy S., Auerbach D., Arnoldo A., Stagljar I., Auerbach D., Arnoldo A., Stagljar I., Arnoldo A., Stagljar I., Stagljar I. Identification of novel ErbB3-interacting factors using the split-ubiquitin membrane yeast two-hybrid system. Genome Res. 2003;13:1744–1753. doi: 10.1101/gr.1276503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Lockshon D., Narayan V., Srinivasan M., Pochart P., Narayan V., Srinivasan M., Pochart P., Srinivasan M., Pochart P., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Wykoff D.D., O’Shea E.K., O’Shea E.K. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol. Cell. Proteomics. 2005;4:73–83. doi: 10.1074/mcp.M400166-MCP200. [DOI] [PubMed] [Google Scholar]