Abstract
This article describes a set of alignments of 28 vertebrate genome sequences that is provided by the UCSC Genome Browser. The alignments can be viewed on the Human Genome Browser (March 2006 assembly) at http://genome.ucsc.edu, downloaded in bulk by anonymous FTP from http://hgdownload.cse.ucsc.edu/goldenPath/hg18/multiz28way, or analyzed with the Galaxy server at http://g2.bx.psu.edu. This article illustrates the power of this resource for exploring vertebrate and mammalian evolution, using three examples. First, we present several vignettes involving insertions and deletions within protein-coding regions, including a look at some human-specific indels. Then we study the extent to which start codons and stop codons in the human sequence are conserved in other species, showing that start codons are in general more poorly conserved than stop codons. Finally, an investigation of the phylogenetic depth of conservation for several classes of functional elements in the human genome reveals striking differences in the rates and modes of decay in alignability. Each functional class has a distinctive period of stringent constraint, followed by decays that allow (for the case of regulatory regions) or reject (for coding regions and ultraconserved elements) insertions and deletions.
The National Human Genome Research Institute is funding a number of vertebrate genome sequencing projects, primarily at the Broad Institute of MIT (Massachusetts Institute of Technology) and Harvard, the Human Genome Sequencing Center at the Baylor College of Medicine, and the Genome Sequencing Center at Washington University. Additional genome sequence data are being provided by other organizations, including the Sanger Center, the Department of Energy’s (DOE’s) Joint Genome Institute, and the National Institute of Genetics in Japan. (Supplemental Table S1 identifies the producers of the individual genome sequences.) The sequences are being immediately and freely released to the public to allow scientists to use the genomic information in their own research. (The scientific community is expected to postpone publications of large-scale analyses of the data until consortia organized by the data producers can publish their initial large-scale analyses, in the spirit of the Ft. Lauderdale agreement; http://www.wellcome.ac.uk/doc_wtd003208.html.)
While a few sophisticated users can mine the raw sequences, many prefer to wait until the data have been assembled into chromosomes, annotated, and aligned to other sequences. The sequencing centers work closely with several groups, including the UCSC Browser team, Ensembl, and the National Center for Biotechnology Information, who maximize data availability and in some cases align the genome sequences to each other so as to facilitate their direct comparison.
Alignments of genomic sequences have long been used as guides to help locate certain kinds of functional noncoding regions (e.g., Hardison 2000), and have more recently been used for finding protein-coding genes (Siepel and Haussler 2004; Gross and Brent 2006) and noncoding RNA genes (Pedersen et al. 2006). Indeed, the primary justification for much of the recent effort to sequence mammalian genomes was to more reliably identify functional elements via sequence alignments (Margulies et al. 2005). The alignments can also reveal similarities and differences between the sequences of humans and those of disease-model species, and thereby enhance the effectiveness of the model species for experiments aimed at improving human health (Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). Moreover, the alignments provide critical data for determining the course of evolution and for the computational reconstruction of ancestral genome sequences.
However, producing whole-genome alignments requires expertise and computational resources that are not easily available within a typical research group. Moreover, reproducibility of published results is facilitated if the alignments used by one group can be utilized directly by others, providing further impetus for creating a reliable, comprehensive, and up-to-date set of freely available alignments.
To meet these needs, the UCSC Genome Bioinformatics group produces browsers for a number of species, each containing genome-wide multiple alignments with conservation scoring (“conservation tracks”) in addition to other types of annotations. Alignments at the Human Genome Browser are constructed using all vertebrate species that have their own UCSC browser, plus a number of additional mammalian species for which low-coverage sequence is available (Margulies et al. 2005). For instance, although there is currently no browser devoted to the rabbit genome, the alignments at the Human Genome Browser include rabbit sequences.
We have recently performed a major update to the Conservation track on the latest UCSC Human Genome Browser. The purpose of this article is to describe the new alignments and to illustrate their use by exploring a few interesting biological issues. This article does not provide an overall analysis of the sequence data sets themselves, which will be the subject of various consortium papers in preparation.
Results and Discussion
The 28-way alignment
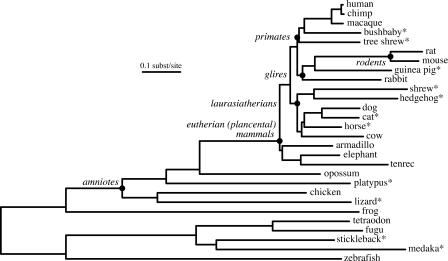
In April 2007, the UCSC Conservation track for the NCBI Build 36 (UCSC hg18) human genome sequence was updated from a 17-way to a 28-way vertebrate alignment (see Methods). The expanded conservation track now includes 11 new species and incorporates updated sequence assemblies for six of the old ones. Figure 1 shows the evolutionary relationships among the sequences that were assumed for the purpose of aligning them (see Methods). Table 1 provides data about the extent to which the individual sequences are represented in the alignment. Note that even for vertebrate species that are quite distant from humans and for mammals with only twofold shotgun sequence coverage, at least 79% of human protein-coding intervals are aligned, though the overall fraction of the human sequence that aligns is <2% for some species.
Figure 1.
A tree indicating assumed evolutionary relationships among the sequences in the 28-way alignment. Branch lengths are proportional to average number of substitutions per site. Species not previously available in our whole-genome alignments are indicated with an asterisk. Filled circles indicate named clades, such as amniotes and eutherians, that are mentioned in the text. A tree labeled with branch lengths is given in the Supplemental material.
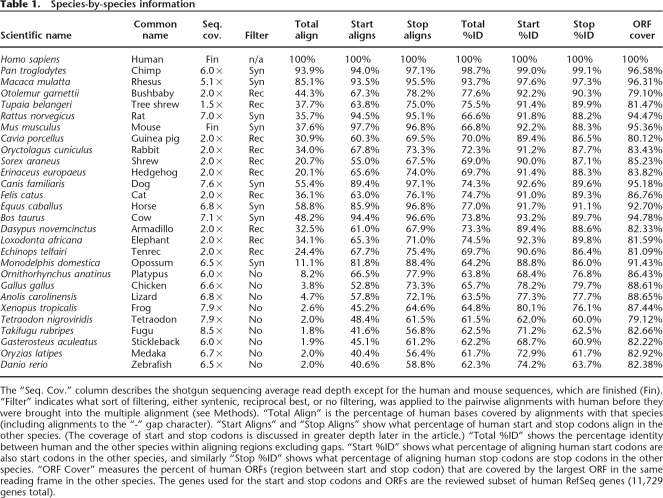
Table 1.
Species-by-species information
The “Seq. Cov.” column describes the shotgun sequencing average read depth except for the human and mouse sequences, which are finished (Fin). “Filter” indicates what sort of filtering, either syntenic, reciprocal best, or no filtering, was applied to the pairwise alignments with human before they were brought into the multiple alignment (see Methods). “Total Align” is the percentage of human bases covered by alignments with that species (including alignments to the “-” gap character). “Start Aligns” and “Stop Aligns” show what percentage of human start and stop codons align in the other species. (The coverage of start and stop codons is discussed in greater depth later in the article.) “Total %ID” shows the percentage identity between human and the other species within aligning regions excluding gaps. “Start %ID” shows what percentage of aligning human start codons are also start codons in the other species, and similarly “Stop %ID” shows what percentage of aligning human stop codons are stop codons in the other species. “ORF Cover” measures the percent of human ORFs (region between start and stop codon) that are covered by the largest ORF in the same reading frame in the other species. The genes used for the start and stop codons and ORFs are the reviewed subset of human RefSeq genes (11,729 genes total).
The 28 genome sequences (Table 1) form a heterogeneous mix. They include two finished sequences, human and mouse, with an estimated coverage of >99% of the euchromatin and an error rate of one in 100,000 (International Human Genome Sequencing Consortium 2004). In addition, there are 16 high-quality draft sequences based on whole-genome shotgun assemblies with coverage from 5.1× to 8.5×. Finally, there are 10 whole-genome shotgun assemblies with coverage ∼2×. In theory, according to the Lander and Waterman (1988) formula, a 2× assembly should include 87.5% of the bases in the genome, and a 5× assembly 99.4%. In practice the base inclusion is somewhat less, due to cloning bias and related issues, and this bias is difficult to estimate precisely. Table 1 also shows the percentage of the human genome that aligns to the various other genomes. Because of coverage issues, the true total aligning portions once the genomes are finished will be likely be 10%–15% above what we currently see for the 2× assemblies, and 1%–4% above the current values for the higher coverage draft sequences.
Application 1: Three vignettes about insertions and deletions in protein-coding regions
To illustrate applications of the 28-way alignment for exploring biological issues and hypotheses, we look briefly at three uses of data about insertions and deletions (collectively called indels) in protein-coding regions. First, we test the hypothesis that coding indels have accumulated at a uniform rate during evolution of placental mammals. Second, we look for human-specific coding indels with phenotypic consequences. Finally, we consider the hypothesis that coding deletions are more likely to have adverse phenotypic consequences if the affected amino acids are well conserved over evolutionary time. These brief explorations illustrate the utility of the 28-way alignment for addressing a wide range of interesting questions, while highlighting several Browser features.
Do coding indels accumulate at a uniform rate?
Coding indels have been used for inferring phylogenetic relationships among species. For instance, Poux et al. (2002) identified a 6-bp deletion near the start of the prion protein gene, PRNP. The indel supports the claim that humans are more closely related to mice (and other glires) than to dogs (and other laurasiatherians). In brief, the reasoning is that the indel can be explained as resulting from a single evolutionary event if and only if primates and glires form a monophyletic clade; any other tree topology requires the unlikely situation that two or more evolutionary events combined to appear like a single event. Figure 2 shows this region of the 28-way alignment. Another use of coding indels to help understand the course of mammalian evolution is discussed by Murphy et al. (2007), supporting the assertion that elephants and armadillos separated from the human lineage in the same speciation event. The relationship between the alignments at the UCSC Browser and this use of coding indels has bidirectional synergy. Determination of the most likely history of mammalian evolution is needed to produce the most appropriate multiple alignments, and conversely, the pairwise and multiple alignments available at UCSC provide an excellent substrate for studying the mechanisms and history of vertebrate evolution.
Figure 2.
A 6-bp deletion near the start of PRNP. Species in the 28-way alignment lacking data for this region are not shown. This alignment can be seen in the current (hg18) UCSC Genome Browser at chr20:4,627,867–4,627,880. See Supplemental Figure S2 for an amino acid alignment of the deletion involving many more species.
Given that coding indels provide evidence concerning the topology (shape) of the phylogenetic tree, are they introduced at such a uniform rate that they can be used to estimate the time separating speciation events? To address this issue, we asked if accelerations or decelerations in the rate of insertions/deletions within protein-coding regions could be detected along certain branches of the phylogenetic tree. We started by searching the 28-way alignment for positions within annotated coding exons where humans have an insertion or deletion relative to both elephant and opossum, i.e., indels that appear to have been created during placental evolution along the human lineage. (The search used custom-built software; see Conclusions, below.) For each such indel and for each of the 16 other placental mammalian sequences in the alignment, we determined whether the species agrees with human, with elephant, or with neither. Finally, we retained the indels that could be unambiguously assigned to a currently sampled branch of the human lineage (relative to the phylogenetic tree that we used). For example, the deletion in Figure 2 would be assigned to the branch from the human–dog ancestor to the human–mouse ancestor (except that it fails a requirement for amount of flanking coding sequence imposed by our program). This process assigned 209 indels as shown in Figure 3.
Figure 3.
Number of inferred coding indels and number per million years on the placental branches leading to human. Estimated time elapsed on each branch is taken from Murphy et al. (2007).
We tested the hypothesis that coding indels were fixed in the human lineage at a uniform rate over time. A parametric bootstrap test showed that the observed indel frequencies differ significantly (P < 10−3) from the hypothesis of uniform distribution. Specifically, the indels detected by our procedure seem to have occurred at a rate of about four per million years in early placental evolution, but at less than two per million years in recent times.
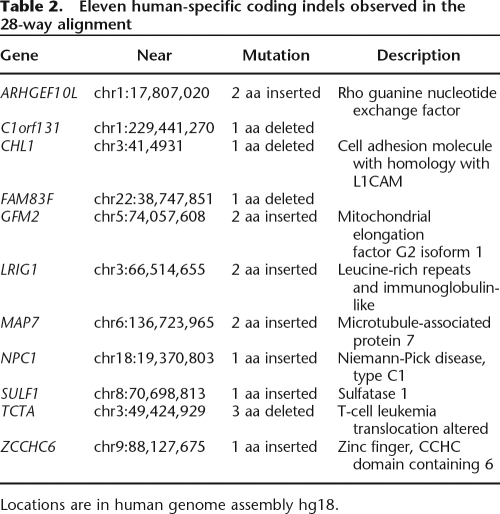
Human-specific protein indels
Among the assignments of coding indels to tree branches, described above, we identified 11 indels that arose after human–chimp divergence (Table 2). Much additional work is required before one can have any confidence that a particular mutation is related to an observable human trait; here we illustrate some appropriate kinds of bioinformatic analyses for two of these genes, SULF1 and GFM2. Laboratory or clinical evidence is required to make a convincing case for any phenotypic consequences of an indel from this list.
Table 2.
Eleven human-specific coding indels observed in the 28-way alignment
Locations are in human genome assembly hg18.
SULF1 has a human-specific 3-bp insertion in exon 11. The insertion adds a GAA codon (E, in the amino acid sequence) to a run of four identical codons. The repetitive nature of this region immediately suggests a mechanism for expansion and/or contraction by replication slippage, which makes the change in the human protein perhaps seem unremarkable. The insertion appears to be fixed in humans (i.e., not polymorphic). No reliable information is available about the three-dimensional structure of this part of the protein sequence, and we could find no published association of genetic disease with exon 11.
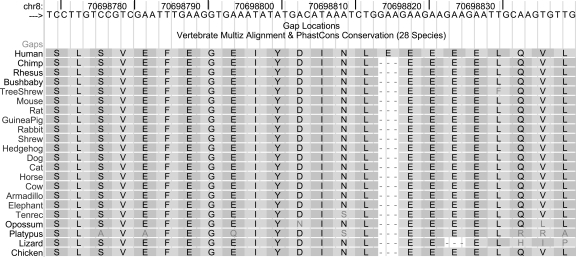
On the other hand, conservation of the sequence around the insertion site is extremely high (Fig. 4). Moreover, conservation of the four Es and the surrounding sequence was also observed in orangutan, marmoset, tarsier, squirrel, microbat, dolphin, pig, sloth, hyrax, and wallaby, according to sequences we located by searching public databases. For some reason, nature has retained the pattern of precisely four successive Es (except for a loss of one E in the lizard sequence) over a total of 2 billion years of evolution along the various lineages. This evidence that the change was heavily resisted over evolutionary time, despite existence of a putative mechanism that could easily make the change (replication slippage), raises the question of whether the extra amino acid in humans is beneficial or deleterious.
Figure 4.
Extreme conservation of the region around the 3-bp insertion in human SULF1. The symbol “-” indicates that there is no base in the aligning species that aligns to this location. Placement of the gap at the first human E results from tie-breaking rules in the alignment software. The second gap in the lizard sequence was positioned using nucleotide content of codons. hg18.chr8:70,698,769–70,698,840.
A 6-bp insertion in human GFM2 illustrates other aspects of the computational analysis of human-specific coding indels identified in the 28-way alignment. In this case, the region around the indel is not particularly well conserved. Indeed, the insertion appears to be found in only some humans (Fig. 5); i.e., some of the available human sequence data for this gene lack the extra 6 bp. We have some information about the three-dimensional conformation of this region of the GFM2 product, based on the known structure of a protein (Protein Data Bank [PDB] entry 2bm0) with 42% identity. This model of the protein structure does not suggest that the insertion site is constrained by secondary or tertiary structural interactions that would make the sequence change difficult to accommodate (see Supplemental materials). That is, the structural model does not suggest that there is likely to be any phenotypic consequence for humans.
Figure 5.
The 6-bp insertion in the human GFM2 gene, showing the location of a 6-bp interval that is absent in some people. The symbol “-” indicates that there is no base in the aligning species that aligns to this location, and “=” indicates that at this location in the aligning species there is a sequence of bases of such different length and/or sequence composition that it cannot be reliably aligned. Sequence for this interval is currently not available for shrew or tenrec. chr5:74,057,590–74,057,630.
Have positions in potentially disease-associated deletions resisted substitution over evolutionary time?
The number of known amino acid differences in the human population is rapidly increasing. There is much interest in developing computational tools to help predict which of these changes might have implications for human health, as summarized by Ng and Henikoff (2006). One type of clue that has been successfully employed comes from interspecies conservation. In particular, it is known that human replacement mutations resulting in disease are overabundant at amino acid positions that are most conserved throughout the long-term history of metazoans (Subramanian and Kumar 2006, and references cited therein).
One way to bring indels into the picture is to ask if disease-associated deletions in the human population tend to involve highly conserved amino acids. Our discussion differs somewhat from that of Subramanian and Kumar (2006) by focusing on potentially disease-associated deletions and by considering data from more species. We employed the data on human variants that are available in the PhenCode Locus Variants custom track (Giardine et al. 2007; www.bx.psu.edu/phencode) for the UCSC Human Genome Browser. In particular, we looked at the gene for PAH (phenylalanine hydroxylase); PAH deficiency causes PKU, the most common inborn error of amino acid metabolism in Caucasians. To keep everything simple, our measure of (lack of) conservation at each position was the number of distinct amino acids in that column of the 28-way alignment.
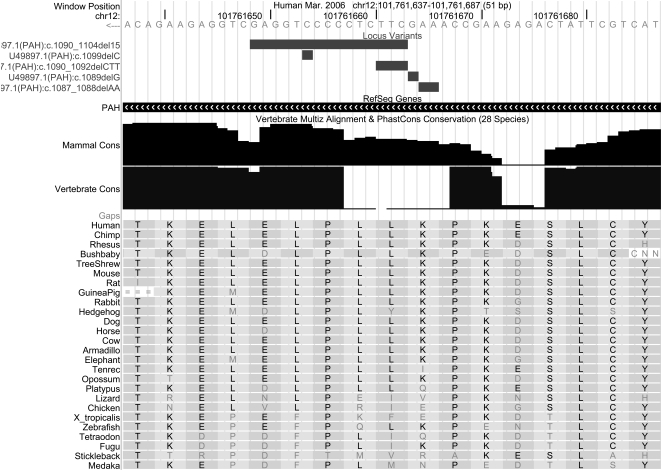
The distribution of these conservation scores at positions where a frame-preserving deletion (i.e., of length divisible by three) is annotated in the Locus Variants track was not strikingly different from the scores’ distribution at all positions of PAH, so this small experiment failed to support our hypothesis. (The P-value for the two-tailed permutation test based on 1000 iterations was ∼0.69.) However, the number of annotated PAH deletions is very small, so the result is heavily affected by one deletion where the amino acid has low conservation, shown in Figure 6.
Figure 6.
A segment of the gene for PAH, showing positions of some deletions that may be associated with the disease PKU. The CTT deletion (shown in reverse orientation) removes an amino acid whose column has six distinct letters and in that sense is not well conserved. The nucleotide symbol “N” represents an unsequenced base, and “=” indicates that at this location in the aligning species there is a sequence of bases of such different length and/or sequence composition that it cannot be reliably aligned. The two conservation tracks indicate that the deleted position is not well conserved among all vertebrates, but is fairly well conserved within mammals. chr12:101,761,637–101,761,687.
The PAH mutations in the Locus Variants track were obtained from the PAHdb knowledgebase (Scriver et al. 2003; www.pahdb.mcgill.ca), which documents known mutations in this gene. The majority of these deletions result in deficient enzyme activity but some do not, so one cannot assume that the mere presence of a particular mutation in the track implies that it causes disease. In this case, one can follow links from the Locus Variants track to the publication cited for the association of PKU with this deletion (namely, Svensson et al. 1991). That and earlier work referenced in the cited article present evidence from Swedish families carrying this mutation that the alteration likely results in a “profound loss of enzymatic activity” (Svensson et al. 1990). In any case, as this small example suggests, the Browser, together with the Locus Variants custom track, is a valuable resource for this kind of study, provided that appropriate caution is taken.
Application 2: Conservation of start and stop codons
In general, as one would expect, the sequence within human coding regions is much more likely to align, and when aligning much more likely to be identical to other species, than the human genome overall. This becomes more and more apparent the more distantly related the other organism is to human. While the overall amount of the coding region that aligns to human typically remains >80%, even in fish, the stop and particularly the start codons drift away at a much faster rate (Table 1). By the time we reach zebrafish, only 40.6% of start codons and 58.8% of stop codons align. The trend of more stop than start codons aligning is present in every sequence except for the one other finished sequence, mouse, where 97.7% of starts and 96.8% of stops align (P-value for one-tailed test for proportions < 10−3). This level of drift certainly makes the job of identifying precise gene boundaries based on comparative genomics data more difficult.
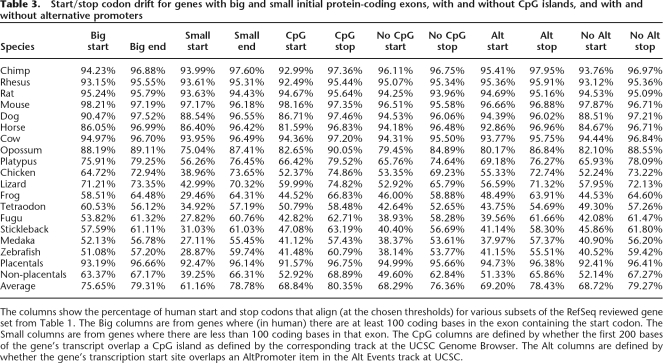
The greater rate of start codon drift compared with stop codon drift is puzzling. We advanced three hypotheses to explain this. Since the one finished sequence, mouse, was an exception to this rule, we hypothesized that it might be due to the CpG islands that are common near gene starts being more difficult to sequence. If we look at genes that do not start in a CpG island, indeed there is no statistically significant difference between the start and stop codon drift within placental mammals (P-value for the two-tailed test for proportions is 0.16). However at further evolutionary distances, the difference in drift is actually more pronounced in genes lacking CpG islands. For instance, for Tetraodon the difference between stop and start codon conservation is 58.48% − 50.79% = 7.71% for genes with a CpG island and 52.65% − 42.64% = 10.01% for those without (Table 3).
Table 3.
Start/stop codon drift for genes with big and small initial protein-coding exons, with and without CpG islands, and with and without alternative promoters
The columns show the percentage of human start and stop codons that align (at the chosen thresholds) for various subsets of the RefSeq reviewed gene set from Table 1. The Big columns are from genes where (in human) there are at least 100 coding bases in the exon containing the start codon. The Small columns are from genes where there are less than 100 coding bases in that exon. The CpG columns are defined by whether the first 200 bases of the gene’s transcript overlap a CpG island as defined by the corresponding track at the UCSC Genome Browser. The Alt columns are defined by whether the gene’s transcription start site overlaps an AltPromoter item in the Alt Events track at UCSC.
Another hypothesis was that selection at the start codon might be more relaxed in genes with multiple promoters, and this could account for some of the difference between the start and stop codon drift. However, this does not seem to be the case. For instance, the average difference for placental mammals was 96.38% − 94.73% = 1.65% for genes with alternate promoters, and 96.41% − 92.41% = 4.00% for other genes. The third hypothesis was that since the initial coding exon is often small, particularly at large evolutionary distances, our programs might not have enough surrounding conserved sequence to reliably align the region around the start codon. We did find that between humans and species more distant than placental mammals, there is a large increase in alignability of start codons that are part of exons with at least 100 coding bases (P-value < 10−3), though still some disparity remained with 67% of stop codons but only 63% of start codons aligning. For additional details on these experiments, see Table 3.
Overall, a bias against CpG islands in the draft sequence combined with difficulty in aligning small initial coding exons does explain a great deal of the observed unalignability of start codons compared with stop codons. Another hypothesis, which is harder to test, is that the start of a protein, since it is very often trimmed by proteases, is subject to less selective pressure than protein ends. Related to this, part of the reason that the smaller initial coding exons are particularly vulnerable to this drift may be that there is less selection against deletion of the entire exon when the protein-coding portion of it is small.
Regardless of the actual cause of the drift, in practical terms it is something that investigators building gene models based on an analysis of multiple genomic alignments need to be well aware of, as was noted in regards to start codons based on CAGE tag data (Frith et al. 2006). This suggests that the total branch length provided by several placental species may be superior for gene prediction to the branch length provided by a single nonplacental, and that finishing additional placental genomes may be worthwhile.
Application 3: Phylogenetic extent of alignment (alignability) of functional regions
A major observation of the recent article (Mikkelsen et al. 2007) on the genome sequence of Monodelphis domestica (short-tailed opossum) is that conservation in noncoding regions is much more subject to evolutionary turnover than in protein-coding regions. More specifically, conservation turnover refers to cases where an interval of human sequence shows signs of purifying selection compared with some species but not with others. The 28-way alignment provides an ideal resource for investigating conservation turnover in greater depth, as we now show.
Effective use of comparative genomics to find and better understand functional regions of genomes remains a challenge. Most coding exons show a strong signature of evolutionary constraint, but others are under positive selection for adaptive changes. A small fraction of cis-regulatory modules (CRMs) are deeply conserved from humans to fish (Woolfe et al. 2005), but others are present only in particular clades (Valverde-Garduno et al. 2004; King et al. 2007). The new multiple alignment provides the opportunity to examine the phylogenetic depth of conservation of different functional classes at a higher resolution, because of the larger number of species included. In this study, we examined 251,000 coding exons of RefSeq genes (Pruitt and Maglott 2001), 481 ultraconserved elements (UCEs) (Bejerano et al. 2004), and 94,000 predicted regulatory regions, which we call PRPs, characterized by both clusters of conserved transcription factor binding sites (PReMods) (Blanchette et al. 2006) and a strong signal for alignment patterns that discriminate between regulatory regions and neutral DNA (high RP; see Methods) (Taylor et al. 2006). We also included a collection of 3900 putative transcriptional regulatory regions (pTRRs) discovered by chromatin immunoprecipitation followed by hybridization to high-density microarray chips (ChIP-chip experiments) from the ENCODE pilot project (The ENCODE Project Consortium 2007; King et al. 2007). The latter data set is composed of functional regions identified by methods that do not depend on sequences or their alignments.
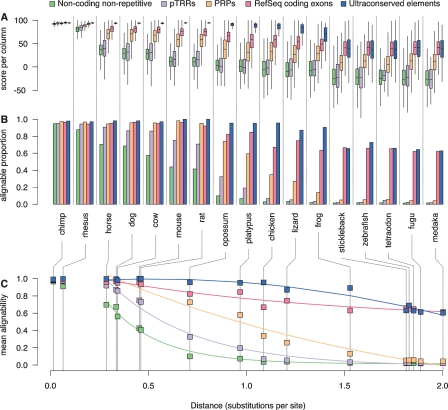
Determining the range of comparison species in which homologs of a DNA sequence are present is fundamental to studying its evolution. We define the alignability of a particular DNA segment (e.g., in human) as the fraction that aligns with a designated comparison species. In the current study, the 28-way alignment is the basis for computing alignability (see Methods). In Figure 7C, the alignability of the human functional classes with each comparison species is plotted as a function of phylogenetic distance estimated by substitutions per fourfold degenerate (4D) site in coding regions. The 4D sites were the ones established by detailed examination of protein-coding segments in ENCODE regions (Harrow et al. 2006; The ENCODE Project Consortium 2007). The substitutions per 4D site were determined using the REV model (Yang 1997). A set of tools have been added to the Galaxy workspace (Blankenberg et al. 2007) to facilitate similar analyses of other data sets and other phylogenetic ranges.
Figure 7.
Phylogenetic extent of the alignment of functional features. (A) The distributions of alignment scores per column for the subset of intervals in each feature set (coding exons, UCEs, putative transcriptional regulatory regions, and PRPs) and the background human genome (nonrepetitive, noncoding) that align with each comparison species. For these box plots, the center line of each box is the median, the box extends from the 25th to 75th percentiles, and the feathers extend to 1.5 times the interquartile distance. The boxes are colored by feature set according to the legend along the top. (B) Barplots showing the fraction of intervals with >50% alignability for each feature set and for the background. (C) Decay of mean alignability as a function of phylogenetic distance. The mean alignabilities of the background human genome and intervals in each feature set are plotted against the distance from human to each comparison species. The distance is measured as the total substitutions per 4D site on each of the branches connecting human to the comparison species. The common name for each comparison species is given below the barplots in B and is connected to the phylogenetic distance in C by dotted lines. The data are best fit by two decay curves, one for primates with a slow rate of change and the other for horse to medaka. The curves shown are the fits to the data points from horse to medaka. (Statistics and coefficients for these fits are in Supplemental Table S4.)
In order to establish a baseline for evaluation of the alignability of various feature sets, we examined the alignability of the nonrepetitive, noncoding portion of the human genome, which we will refer to as the background. Loss of alignability is likely to be driven by deletions in the lineage to the comparison species and insertions in the human lineage, with some contribution from substitutions. If these events occur independently, then the alignability should decay exponentially with increasing phylogenetic distance (Kimura 1969). This is indeed observed for the background alignability, especially for comparisons outside primates (Fig. 7C; Supplemental Table S4).
This predictable decline of the background alignability with phylogenetic distance provides a basis for comparison of the decay (if any) in alignability of functional regions. We examined several feature sets to determine the range of comparison species over which high alignability is maintained, the mode and rate of decay, the fraction of intervals that continue to align at a given distance, and the quality of alignments, and we found striking differences.
Almost all coding exons align in all placental mammals, followed by a decay that is much slower than that of the background (Fig. 7B,C). Almost all UCEs align with species as distant as chicken, but a decline is seen with more distant species (Fig. 7B,C). Indeed, alignability of UCEs appears to decay as a function of the square of the phylogenetic distance. The decay equation (Supplemental material) predicts a substantial loss of recognizable UCEs at distances of 2.5 to 3 substitutions per site, which is consistent with the failure to detect UCEs outside vertebrates (Bejerano et al. 2004). The 60%–70% of UCEs conserved between humans and fish are distinct from the ones limited to mammals (see Supplemental materials). The pan-vertebrate UCEs are further from genes on average than are the pan-mammalian ones, reminiscent of the jungles of noncoding conserved sequences observed in human–chicken comparisons (Hillier et al. 2004). As expected, the deeply conserved UCEs are near genes enriched in GO categories for transcription factors and developmental regulatory genes (Woolfe et al. 2005). Both the coding exons and UCEs maintain a high conservation level in all comparisons, shown by the high values in the distributions of alignment scores per column (Fig. 7A).
The predicted CRMs (PReMods with high regulatory potential, or PRPs) show substantially elevated alignability above that of the background DNA (Fig. 7B,C). The PRPs were selected at least in part by their ability to align among mammalian species, and as expected, virtually all of them align to other eutherian species. In addition, a substantial fraction aligns to marsupials and monotremes. The alignability for comparisons outside primates decays exponentially but substantially more slowly than the background. The distributions of conservation-level scores are considerably higher than the background but substantially less than those for coding exons (Fig. 7A). Similar results are obtained for the decay in alignability of a small set of 93 curated known regulatory regions (Elnitski et al. 2003; data not shown).
Another set of genomic regions (pTRRs), which is implicated in transcriptional regulation by ChIP-chip biochemical analyses, shows a different pattern. The loss of alignability with phylogenetic distance for the ENCODE pTRRs is more rapid than the decay seen for PRPs (Fig. 7C). The shape of the decay curve is similar to that of the background, but shifted to the right. The rightward shift indicates a slower decay, implying that the pTRRs are constrained; i.e., as a class they are eroding more slowly than the background. This is also seen in the higher fraction of the pTRR intervals that pass an alignability threshold compared with that of the background (Fig. 7B). Interestingly, the similarity level of the alignments in the pTRR intervals overlaps substantially with that of the background, whereas the alignments of UCEs and coding exons have strikingly higher scores (Fig. 7A). Results similar to those for pTRRs are obtained for a genome-wide set of 13,000 segments occupied by the transcription factor CTCF (Kim et al. 2007; data not shown).
While the pTRRs as a class do have a higher alignability than the background for many comparisons, the number of pTRRs that are conserved drops dramatically between eutherian mammals (horse, dog, cow, and rodents) and the marsupial opossum (Fig. 7.B). This shows that 70% of pTRRs are conserved within eutherian mammals, while 70% are not conserved outside of eutherians. Smaller fractions are conserved in more distant comparisons. The pTRRs conserved out to chicken and fish are likely to have been under constraint over this phylogenetic span. Given the estimates for rates of neutral substitutions, sequences that still align between human and chicken, and more distant species, are all likely to be under constraint (Hillier et al. 2004).
This study illustrates the range and complexity of the relationships between the functionality of genomic sequences and their phylogenetic extent of alignability. The decays of alignability for some strongly constrained functional classes differ markedly from that of the genome background. Coding exons are largely conserved through eutherians, after which they decay much more slowly than the background. Almost all UCEs are conserved in amniotes, after which their alignability decays more rapidly than do coding exons. Alignability of functional classes associated with gene regulation presents exponential decay curves, but with lag times and slopes characteristic of the feature sets and distinctly different from the background. The alignability of PRPs begins to decline outside most eutherians whereas the alignability of pTRRs and CTCF binding sites begins to decay outside the available primates. This can be interpreted as stringent selection being exerted over different phylogenetic spans for the distinct types of regulatory regions, followed by decay in sequence similarity. The mode of decay in alignability is exponential both for the features associated with regulation and for the background. This could mean that the processes of sequence change are similar, with deletions and insertions allowed as in the background genome (albeit at a slower rate), whereas these events tend to be rejected in coding exons and UCEs.
Conclusion
To use alignments of placental-mammal genome sequences to identify small intervals (say, the size of a transcription-factor binding site) that are under strong negative selection, one needs sequence data from perhaps 40 well-chosen species (Eddy 2005; Margulies et al. 2005). Of course, one could obtain an equivalent total phylogenetic branch length by using a smaller number of more distant species, but as shown in Figure 7, many functional sites will then be lost to conservation turnover. The 28-way alignment comes the closest to date to attaining this 40-species goal. Here we illustrate the use of those alignments to explore hypotheses about vertebrate evolution. The individual results are interesting in their own right and worthy of further study, but they are merely a sampling of what we and others will discover with those alignments.
Some of the phenomena described here can be directly observed in the UCSC Browser. Others require the use of tools such as the UCSC Table Browser (Karolchik et al. 2004) or the Galaxy server (Giardine et al. 2005; Blankenberg et al. 2007) to help identify genome-wide trends. The most generally applicable approach is to write special purpose programs to analyze the alignments. For instance, C-language programs used for analyzing the rate of coding indels can be downloaded from http://www.bx.psu.edu/miller_lab/publications/. Other programs for manipulating the alignments are mentioned in the Methods section.
The analyses presented here were necessarily quite brief, and in each case, it would be possible to strengthen the analysis by using more complex approaches. For instance, our modeling of coding indels was quite simplistic, and much more sophisticated methods (e.g., Diallo et al. 2007) could be applied. Similarly, we frequently assumed that regions aligned to a functional human gene are themselves functional genes in the other species, though some of them may have become inactive pseudo-genes. To help ameliorate the (presumed minor) effects of such cases, one could incorporate gene-model data from the other species, such as that available from Ensembl (Hubbard et al. 2007).
While in practice we use the 28-way alignment to explore many issues, there are times when other alignments might be more appropriate. For instance, the 28-way alignment is based on a phylogenetic tree (Fig. 1) that is not universally accepted (e.g., see Wible et al. 2007). While this may not affect most conclusions, there are cases, such as the use of the alignment to support or reject certain evolutionary hypotheses, where the tree matters (Kumar and Filipski 2007). In such cases, it might be better to use a set of pairwise alignments that are agnostic about phylogenetic hypotheses (e.g., Rosenbloom et al. 2007). Also, our alignments are made with programs that implement only one approach and that use particular settings of a large number of thresholds and other parameters; others may prefer to use different software, such as the tools discussed by Margulies et al. (2007). In any case, users need to remain aware that there are genomic regions in which the 28-way alignment, or indeed an alignment computed by any means, is unreliable because evolutionary changes have saturated the sequences (Prakash and Tompa 2007).
A general theme running through our observations, and one that continues to enthrall many investigators, is to identify the best ways to use multispecies alignments to help predict the locations of functional genomic elements. In particular, are these elements more frequently revealed by reduced levels of substitutions, by particular patterns of substitutions, by reduced levels of small insertions and deletions, or by a reduced level of complete loss? Data summarized in Figure 7 can be interpreted as suggesting that resistance to complete loss is sometimes more informative than low substitution rates. Another, very preliminary, study reported here raises the question of whether substitution frequency can be used to predict whether deletion of an amino acid might cause human disease. Whole-genome alignments provide a valuable resource for investigating these and many other fascinating issues.
Methods
Phylogenetic tree
We used the tree topology (Fig. 1) that seemed in best agreement with our interpretation of the published literature. Branch lengths, which are used for quantifying sequence conservation (see below) but not for computing the alignments, were computed by phyloFit as described below.
Alignments
Pairwise alignments with the human genome were generated for each species (BLASTZ; Schwartz et al. 2003) from repeat-masked genomic sequence (RepeatMasker, by A. Smit and R. Hubley [Institute for Systems Biology, Seattle, WA], or WindowMasker, by Morgulis et al. 2006), with lineage-specific repeats removed prior to alignment, then reinserted. The pairwise alignment coverage for all species is listed in Table 1. Pairwise alignments were then linked into chains using a dynamic programming algorithm that finds maximally scoring chains of gapless subsections of the alignments organized in a kd-tree (axtChain; Kent et al. 2003). The scoring matrix and parameters for pairwise alignment and chaining were tuned for each species based on its phylogenetic distance from humans. High-scoring chains were then placed along the genome, with gaps filled by lower-scoring chains (axtNet, by J. Kent), to produce an alignment net. Filtering of the component pairwise alignments was performed to reduce paralogs, pseudogenes, and suspect alignments from the 2× species. The alignments of high-quality mammalian sequences (placental and marsupial) were filtered based on synteny, while those for 2× mammalian genomes were filtered to retain only alignments that were best quality in both species (“reciprocal best”). The resulting best-in-genome pairwise alignments were progressively aligned following the tree topology of Figure 1, using the MULTIZ program (Blanchette et al. 2004). Alignments were post-processed to add annotations for alignment gaps and genomic breaks, indications of base quality in the component sequences (see Supplemental material), and information that permits prediction of amino acid sequences in all species.
Conservation scoring was performed using the phastCons package (Siepel et al. 2005), which computes conservation based on a two-state phylogenetic hidden Markov model (HMM). These measurements rely on a tree model containing the tree topology, branch lengths representing evolutionary distance at neutrally evolving sites, the background distribution of nucleotides, and a substitution rate matrix. The vertebrate tree model for this track was generated using the phyloFit program from the phastCons package (REV model, EM algorithm, medium precision) using multiple alignments of fourfold degenerate sites extracted from the 28-way alignment (msa_view). The 4D sites were derived from the October 2005 Gencode Reference Gene set (Harrow et al. 2006), which was filtered to select single-coverage long transcripts. A second tree model, including only placental mammals, was used to generate the placental mammal conservation scoring. The phastCons parameters were tuned to produce 5% conserved elements in the human genome for the vertebrate conservation measurement; this parameter set (expected-length, 45; target-coverage, 0.3; rho, 0.31) was then used to generate the placental mammal conservation scoring.
The phastCons program computes conservation scores based on a phylo-HMM, a type of probabilistic model that describes both the process of DNA substitution at each site in a genome and the way this process changes from one site to the next (Yang 1995; Felsenstein and Churchill 1996; Siepel and Haussler 2005). PhastCons uses a two-state phylo-HMM, with a state for conserved regions and a state for nonconserved regions. The value plotted at each site is the posterior probability that the corresponding alignment column was “generated” by the conserved state of the phylo-HMM. These scores reflect the phylogeny (including branch lengths) of the species in question, a continuous-time Markov model of the nucleotide substitution process, and a tendency for conservation levels to be autocorrelated along the genome (i.e., to be similar at adjacent sites). Unlike many conservation-scoring programs, phastCons does not rely on a sliding window of fixed size; therefore, short highly conserved regions and long moderately conserved regions can both obtain high scores. More information about phastCons can be found in work by Siepel et al. (2005).
PhastCons currently treats alignment gaps as missing data, which sometimes has the effect of producing undesirably high conservation scores in gappy regions of the alignment. We are looking at several possible ways of improving the handling of alignment gaps.
Extraction and analysis of MAF blocks
Local portions of the 28-way alignment are stored as so-called MAF (Multiple Alignment Format) blocks. In Galaxy, MAF-block extraction is implemented using an on-disk variation of the positional binning scheme described by Kent et al. (2002) to allow fast extraction of alignment blocks overlapping specific regions of the human sequence. Due to the size of these alignments, compression is essential; here we used LZO compression (http://www.lzop.org/). As a result of compression, it is no longer possible to seek directly to a particular alignment block in these files. However, because the LZO format compresses the data in independent chunks, accessing a particular alignment block only requires decompressing the containing chunk(s) of compressed data (i.e., semi-random access). This is a substantial benefit over the common GZIP format, for example, which requires decompressing all preceding data to access a particular location. We have implemented an indexing scheme for (1) identifying the locations of all MAF blocks that overlap a particular interval in some aligned species and (2) mapping those locations to their containing chunks of compressed data. Combined, these methods make working with alignments of this scale substantially easier. Command-line programs and Python modules for generating and using these indexes are available as part of the “bx-python” package (http://bx-python.trac.bx.psu.edu/).
PRPs
PRPs are a set of predicted CRMs that have both the properties of (1) clusters of conserved transcription factor binding sites (PReMods; Blanchette et al. 2006) and (2) high regulatory potential (high RP intervals; Taylor et al. 2006). PRP is a brief name denoting the combination of PReMod and RP.
The set of 118,402 PReMods (human genome assembly hg17) was collected from the Web server discussed by Blanchette et al. (2006). The RP scores were determined for a seven-way alignment of human (hg17), chimpanzee (panTro2), macaque (rheMac2), mouse (mm8), rat (rn4), dog (canFam2), and cow (bosTau2). Human DNA intervals with an RP score of at least 0.05 for at least 200 bp (i.e., the minimum score never goes below 0.05) were selected to obtain 314,020 high-RP intervals.
The next series of operations was conducted using online tools in Galaxy. KnownGenes exons obtained from the UCSC Table Browser (extended 15 bp on each side) were subtracted from the high RP intervals, and intervals shorter than 200 bp were removed to obtain a total of 282,639 nonexonic high-RP intervals. These were intersected with the PReMods, requiring an overlap of at least 10 bp, to produce 106,383 pieces present in both sets. Intervals within 100 bp of each other were then merged, the merged intervals were combined with the other intervals from the intersection, and those shorter than 50 bp were removed. This series of operations produced a set of 92,269 PRPs. Their average length is 350 bp, ranging from 50 bp to 3793 bp.
The ENCODE regions cover 1% of the human genome and have been examined experimentally for chromatin alterations, occupancy by a set of transcription factors, and transcription start sites (The ENCODE Project Consortium 2007; Margulies et al. 2007). Examination of the 1389 PRPs that lie within ENCODE regions supports the hypothesis that the PRPs are good predictors of regulatory function. About half are within 100 bp of an interval associated with transcriptional regulation (using the ENCODE pilot phase data available now at the UCSC Web site), which is about a twofold enrichment over the bulk DNA in the ENCODE regions. In some well-studied ENCODE regions, the support level for PRPs reaches 100%.
Alignability
The alignability of an interval in the human sequence with another species is the proportion of bases in that interval that are covered by any local pairwise alignment with the other species. Positions in the human interval are conservatively classified as aligned, not aligned, or potentially missing using the local alignments and alignment gap annotation, with the goal of excluding from analysis all positions in the human sequence that are not aligned due to missing data in the other species sequence. Specifically, (1) all positions covered by a local alignment are marked aligned; (2) if there is no local alignment or gap annotation covering the position, or the gap annotation indicates contiguity across the position but with missing data in the other species sequence, the position is marked potentially missing; and (3) if there is no local alignment but the gap annotation indicates contiguity in both species, the position is marked not aligned. Alignability is then the number of aligned positions divided by the number of aligned plus not aligned positions. For 10 of the 28 species, the alignability measures were markedly different from those seen for other species at a similar distance from human. This is likely a result of incomplete coverage, since all had ∼2× coverage or less. The program used to compute alignability, “maf_interval_alignability.py,” is available as part of the “bx-python” package (http://bx-python.trac.bx.psu.edu/).
Acknowledgments
This project has been funded in part by grants from NHGRI (HG002238 to W.M., and 1P41HG02371 to W.J.K.), NIDDK (DK65806 to R.C.H.), and NCI (Contract no. NO1-CO-12400). Sudhir Kumar and the reviewers provided many useful suggestions. Additional help was provided by Cathy Riemer and Heather Lawson.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6761107
References
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Stephen S., Kent W.J., Mattick J.S., Haussler D., Kent W.J., Mattick J.S., Haussler D., Mattick J.S., Haussler D., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Rosenbloom K., Clawson H., Green E.D., Clawson H., Green E.D., Green E.D., et al. Aligning multiple genomic sequences with the Threaded Blockset Aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., Deblois G., Giguere V., Ferretti V., Bergeron D., Giguere V., Ferretti V., Bergeron D., Ferretti V., Bergeron D., Bergeron D., et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D., Taylor J., Schenck I., He J., Zhang Y., Ghent M., Veeraaghavan N., Albert I., Miller W., Makova K., Taylor J., Schenck I., He J., Zhang Y., Ghent M., Veeraaghavan N., Albert I., Miller W., Makova K., Schenck I., He J., Zhang Y., Ghent M., Veeraaghavan N., Albert I., Miller W., Makova K., He J., Zhang Y., Ghent M., Veeraaghavan N., Albert I., Miller W., Makova K., Zhang Y., Ghent M., Veeraaghavan N., Albert I., Miller W., Makova K., Ghent M., Veeraaghavan N., Albert I., Miller W., Makova K., Veeraaghavan N., Albert I., Miller W., Makova K., Albert I., Miller W., Makova K., Miller W., Makova K., Makova K., et al. A framework for collaborative analysis of ENCODE data: Making large-scale analyses biologist-friendly. Genome Res. 2007;17:960–964. doi: 10.1101/gr.5578007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo A.B., Makarenkov V., Blanchette M., Makarenkov V., Blanchette M., Blanchette M. Exact and heuristic algorithms for the Indel Maximum Likelihood problem. J. Comput. Biol. 2007;14:446–461. doi: 10.1089/cmb.2007.A006. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. A model of the statistical power of comparative genome sequence analysis. PLoS Biol. 2005;3:e10. doi: 10.1371/journal.pbio.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L., Hardison R.C., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Hardison R.C., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Li J., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Yang S., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Kolbe D., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Eswara P., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., O’Connor M.J., Schwartz S., Miller W., Chiaromonte F., Schwartz S., Miller W., Chiaromonte F., Miller W., Chiaromonte F., Chiaromonte F. Distinguishing regulatory DNA from neutral sites. Genome Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium The ENCODE pilot project: Identification and analysis of functional elements in 1% of the human genome. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J., Churchill G.A., Churchill G.A. A hidden Markov model approach to variation among sites in rate of evolution. Mol. Biol. Evol. 1996;13:93–104. doi: 10.1093/oxfordjournals.molbev.a025575. [DOI] [PubMed] [Google Scholar]
- Frith M.C., Ponjavic J., Fredman D., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Ponjavic J., Fredman D., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Fredman D., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Kai C., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Kawai J., Carninci P., Hayashizaki Y., Sandelin A., Carninci P., Hayashizaki Y., Sandelin A., Hayashizaki Y., Sandelin A., Sandelin A. Evolutionary turnover of mammalian transcription start sites. Genome Res. 2006;16:713–722. doi: 10.1101/gr.5031006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Riemer C., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Hardison R.C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Zhang Y., Blankenberg D., Albert I., Taylor J., Blankenberg D., Albert I., Taylor J., Albert I., Taylor J., Taylor J., et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B., Riemer C., Hefferon T., Thomas D., Hsu F., Zielenski J., Sang Y., Elnitski L., Cutting G., Trumbower H., Riemer C., Hefferon T., Thomas D., Hsu F., Zielenski J., Sang Y., Elnitski L., Cutting G., Trumbower H., Hefferon T., Thomas D., Hsu F., Zielenski J., Sang Y., Elnitski L., Cutting G., Trumbower H., Thomas D., Hsu F., Zielenski J., Sang Y., Elnitski L., Cutting G., Trumbower H., Hsu F., Zielenski J., Sang Y., Elnitski L., Cutting G., Trumbower H., Zielenski J., Sang Y., Elnitski L., Cutting G., Trumbower H., Sang Y., Elnitski L., Cutting G., Trumbower H., Elnitski L., Cutting G., Trumbower H., Cutting G., Trumbower H., Trumbower H., et al. PhenCode: Connecting ENCODE data with mutations and phenotype. Hum. Mutat. 2007;28:554–562. doi: 10.1002/humu.20484. [DOI] [PubMed] [Google Scholar]
- Gross S.S., Brent M.R., Brent M.R. Using multiple alignments to improve gene prediction. J. Comput. Biol. 2006;13:379–393. doi: 10.1089/cmb.2006.13.379. [DOI] [PubMed] [Google Scholar]
- Hardison R.C. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Harrow J., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Denoeud F., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Frankish A., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Reymond A., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chen C.K., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Chrast J., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Lagarde J., Gilbert J.G., Storey R., Swarbreck D., Gilbert J.G., Storey R., Swarbreck D., Storey R., Swarbreck D., Swarbreck D., et al. GENCODE: Producing a reference annotation for ENCODE. Genome Biol. 2006;7:S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L.W., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Miller W., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Birney E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Warren W., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Hardison R.C., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Ponting C.P., Bork P., Burt D.W., Groenen M.A., Delany M.E., Bork P., Burt D.W., Groenen M.A., Delany M.E., Burt D.W., Groenen M.A., Delany M.E., Groenen M.A., Delany M.E., Delany M.E., et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Hubbard T.J., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Clarke L., Coates G., Cunningham F., Cutts T., Coates G., Cunningham F., Cutts T., Cunningham F., Cutts T., Cutts T., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Sugnet C.W., Haussler D., Kent W.J., Haussler D., Kent W.J., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D., Baertsch R., Hinrichs A., Miller W., Haussler D., Hinrichs A., Miller W., Haussler D., Miller W., Haussler D., Haussler D. Evolution’s cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Zhang M.Q., Lobanenkov V.V., Ren B., Lobanenkov V.V., Ren B., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The rate of molecular evolution considered from the standpoint of population genetics. Proc. Natl. Acad. Sci. 1969;63:1181–1188. doi: 10.1073/pnas.63.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D.C., Taylor J., Zhang Y., Cheng Y., Martin J., Chiaromonte F., Miller W., Hardison R.C., Taylor J., Zhang Y., Cheng Y., Martin J., Chiaromonte F., Miller W., Hardison R.C., Zhang Y., Cheng Y., Martin J., Chiaromonte F., Miller W., Hardison R.C., Cheng Y., Martin J., Chiaromonte F., Miller W., Hardison R.C., Martin J., Chiaromonte F., Miller W., Hardison R.C., ENCODE groups for Transcriptional Regulation and Multispecies Sequence Analysis. Chiaromonte F., Miller W., Hardison R.C., Miller W., Hardison R.C., Hardison R.C. Finding cis-regulatory elements using comparative genomics: Some lessons from ENCODE data. Genome Res. 2007;17:775–786. doi: 10.1101/gr.5592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Filipski A., Filipski A. Multiple sequence alignment: In pursuit of homologous DNA positions. Genome Res. 2007;17:127–135. doi: 10.1101/gr.5232407. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Waterman M.S., Waterman M.S. Genomic mapping by fingerprinting random clones: A mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- Margulies E.H., Vinson J.P., Miller W., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Vinson J.P., Miller W., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., NISC Comparative Sequencing Program. Miller W., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Jaffe D.B., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Lindblad-Toh K., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Chang J.L., Green E.D., Lander E.S., Mullikin J.C., Green E.D., Lander E.S., Mullikin J.C., Lander E.S., Mullikin J.C., Mullikin J.C., et al. An initial strategy for the systematic identification of functional elements in the human genome by low-redundancy comparative sequencing. Proc. Natl. Acad. Sci. 2005;102:4795–4800. doi: 10.1073/pnas.0409882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies E.H., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M., Birney E., Keefe D., Schwartz A.S., Hou M., Keefe D., Schwartz A.S., Hou M., Schwartz A.S., Hou M., Hou M., et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Wakefield M.J., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Aken B., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Amemiya C.T., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Chang J.L., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Duke S., Garber M., Gentles A.J., Goodstadt L., Heger A., Garber M., Gentles A.J., Goodstadt L., Heger A., Gentles A.J., Goodstadt L., Heger A., Goodstadt L., Heger A., Heger A., et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequence. Nature. 2007;447:167–178. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Pringle T.H., Crider T.A., Springer M.S., Miller W., Pringle T.H., Crider T.A., Springer M.S., Miller W., Crider T.A., Springer M.S., Miller W., Springer M.S., Miller W., Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S., Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Pedersen J.S., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent W.J., Miller W., Haussler D., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent W.J., Miller W., Haussler D., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent W.J., Miller W., Haussler D., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent W.J., Miller W., Haussler D., Lindblad-Toh K., Lander E.S., Kent W.J., Miller W., Haussler D., Lander E.S., Kent W.J., Miller W., Haussler D., Kent W.J., Miller W., Haussler D., Miller W., Haussler D., Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux C., van Rheede T., Madsen O., de Jong W.W., van Rheede T., Madsen O., de Jong W.W., Madsen O., de Jong W.W., de Jong W.W. Sequence gaps join mice and men: phylogenetic evidence from deletions in two proteins. Mol. Biol. Evol. 2002;19:2035–2037. doi: 10.1093/oxfordjournals.molbev.a004028. [DOI] [PubMed] [Google Scholar]
- Prakash A., Tompa M., Tompa M. Measuring the accuracy of genome-size multiple alignments. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-6-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Maglott D.R., Maglott D.R. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Rosenbloom K., Taylor J., Schaeffer S., Kent W.J., Haussler D., Miller W., Taylor J., Schaeffer S., Kent W.J., Haussler D., Miller W., Schaeffer S., Kent W.J., Haussler D., Miller W., Kent W.J., Haussler D., Miller W., Haussler D., Miller W., Miller W. Phylogenomic resources at the UCSC Genome Browser. In: Murphy W.J., editor. Methods and molecular biology: Phylogenomics. Humana Press; Totowa, NJ: 2007. (in press) [DOI] [PubMed] [Google Scholar]
- Schwartz S., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Kent W.J., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Smit A., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Zhang Z., Baertsch R., Hardison R.C., Haussler D., Miller W., Baertsch R., Hardison R.C., Haussler D., Miller W., Hardison R.C., Haussler D., Miller W., Haussler D., Miller W., Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriver C.R., Hurtubise M., Konecki D., Phommarinh M., Prevost L., Erlandsen H., Stevens R., Waters P.J., Ryan S., McDonald D., Hurtubise M., Konecki D., Phommarinh M., Prevost L., Erlandsen H., Stevens R., Waters P.J., Ryan S., McDonald D., Konecki D., Phommarinh M., Prevost L., Erlandsen H., Stevens R., Waters P.J., Ryan S., McDonald D., Phommarinh M., Prevost L., Erlandsen H., Stevens R., Waters P.J., Ryan S., McDonald D., Prevost L., Erlandsen H., Stevens R., Waters P.J., Ryan S., McDonald D., Erlandsen H., Stevens R., Waters P.J., Ryan S., McDonald D., Stevens R., Waters P.J., Ryan S., McDonald D., Waters P.J., Ryan S., McDonald D., Ryan S., McDonald D., McDonald D., et al. PAHdb 2003: What a locus-specific knowledgebase can do. Hum. Mutat. 2003;21:333–344. doi: 10.1002/humu.10200. [DOI] [PubMed] [Google Scholar]
- Siepel A., Haussler D., Haussler D. Proceedings of the 8th annual international conference on research in computational molecular biology. ACM Press; New York: 2004. Computational identification of evolutionarily conserved exons; pp. 177–186. [Google Scholar]
- Siepel A., Haussler D., Haussler D. Phylogenetic hidden Markov models. In: Nielsen R., editor. Statistical methods in molecular evolution. Springer; New York: 2005. pp. 325–351. [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Kumar S., Kumar S. Evolutionary anatomies of positions and types of disease-associated and neutral amino acid mutations in the human genome. BMC Genomics. 2006;7:306. doi: 10.1186/1471-2164-7-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson E., Andersson B., Hagenfeldt L., Andersson B., Hagenfeldt L., Hagenfeldt L. Two mutations within the coding sequence of the phenylalanine hydroxylase gene. Hum. Genet. 1990;85:300–304. doi: 10.1007/BF00206750. [DOI] [PubMed] [Google Scholar]
- Svensson E., von Döbeln U., Hagenfeldt L., von Döbeln U., Hagenfeldt L., Hagenfeldt L. Polymorphic DNA haplotypes at the phenylalanine hydroxylase locus and their relation to phenotype in Swedish phenylketonuria families. Hum. Genet. 1991;87:11–17. doi: 10.1007/BF01213084. [DOI] [PubMed] [Google Scholar]
- Taylor J., Tyekucheva S., King D.C., Hardison R.C., Miller W., Chiaromonte F., Tyekucheva S., King D.C., Hardison R.C., Miller W., Chiaromonte F., King D.C., Hardison R.C., Miller W., Chiaromonte F., Hardison R.C., Miller W., Chiaromonte F., Miller W., Chiaromonte F., Chiaromonte F. ESPERR: Learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde-Garduno V., Guyot B., Anguita E., Hamlett I., Porcher C., Vyas P., Guyot B., Anguita E., Hamlett I., Porcher C., Vyas P., Anguita E., Hamlett I., Porcher C., Vyas P., Hamlett I., Porcher C., Vyas P., Porcher C., Vyas P., Vyas P. Differences in the chromatin structure and cis-element organization of the human and mouse GATA1 loci: Implications for cis-element identification. Blood. 2004;104:3106–3116. doi: 10.1182/blood-2004-04-1333. [DOI] [PubMed] [Google Scholar]
- Wible J.R., Rougier G.W., Novacek M.J., Asher R.J., Rougier G.W., Novacek M.J., Asher R.J., Novacek M.J., Asher R.J., Asher R.J. Cretaceous eutherians and Laurasian origin for placental mammals near the K/T boundary. Nature. 2007;447:1003–1006. doi: 10.1038/nature05854. [DOI] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Smith S.F., North P., Callaway H., Kelly K., North P., Callaway H., Kelly K., Callaway H., Kelly K., Kelly K., et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. A space-time process model for the evolution of DNA sequences. Genetics. 1995;139:993–1005. doi: 10.1093/genetics/139.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]