Abstract
During evolution, genome reorganization includes large-scale events such as inversions, translocations, and segmental or even whole-genome duplications, as well as fine-scale events such as the relocation of individual genes. This latter category, which we will refer to as positionally relocated genes (PRGs), is the subject of this report. Assessment of the magnitude of such PRGs and of possible contributing mechanisms is aided by a comparative analysis of related genomes, where conserved chromosomal organization can aid in identifying genes that have acquired a new location in a lineage of these genomes. Here we utilize two methods to comprehensively identify relocated protein-coding genes in the recently sequenced genomes of 12 species of genus Drosophila. We use exceptions to the general rule of maintenance of chromosome arm (Muller element) association for most Drosophila genes to identify one major class of PRGs. We also identify a partially overlapping set of PRGs among “embedded genes,” located within the extents of other surrounding genes. We provide evidence that PRG movements have at least two different origins: Some events occur via retrotransposition of processed RNAs and others via a DNA-based transposition mechanism. Overall, we identify several hundred PRGs that arose within a lineage of the genus Drosophila phylogeny and provide suggestive evidence that a few thousand such events have occurred within the radiation of the insect order Diptera, thereby illustrating the magnitude of the contribution of PRG movement to chromosomal reorganization during evolution.
One of the primary goals in sequencing multiple Drosophila species was to enable the combination of the wealth of Drosophila evolutionary and morphological knowledge with genetic information in the process of whole-genome comparative analysis. Studying multiple genomes at varying levels of divergence provides the opportunity to analyze the patterns of evolutionary divergence that distinguish these species from each other. In this paper we focus on positionally relocated genes (PRGs), which we define as individual genes that have relocated to different chromosome arms. We distinguish this class of genes from those that are rearranged via large-scale events: paracentric and pericentric inversions, translocations, and fusion of chromosome arms. PRGs represent movement of genes at a much finer scale.
Previous studies to assess PRGs during evolution have either focused on a small set of genes or genomes (Neufeld et al. 1991; Llorente et al. 2000; Fischer et al. 2001; Coghlan and Wolfe 2002; Harrison et al. 2003; Zhang et al. 2003; Gonzalez et al. 2004) or on a particular mechanism for transposition (Brosius 1991; Betran and Long 2003; Langille and Clark 2007). Mechanisms such as retrotransposition or excision and insertion of genomic segments (Chia et al. 1985; Lovering et al. 1991) are thought to be responsible for transposition events (Gonzalez et al. 2004). Here, we present a comprehensive analysis of the 12 sequenced genus Drosophila genomes (Drosophila 12 Genomes Consortium 2007), leveraging the phylogeny (Powell 1997) and some well-established properties of chromosome organization in these flies to identify PRGs on a genome-wide scale.
The typical organization of the genome of any Drosophila species includes five major and one minor chromosome arms. The vast majority of the genes that are found on a single chromosome arm in one species of Drosophila are found on a single chromosome arm in any other species of Drosophila as well (Metz 1914; Muller 1940; Sturtevant and Novitski 1941; Ranz et al. 2003).This phenomenon, inferred from many small-scale studies, is now known to be true for the vast majority (95%) of Drosophila genes (A. Bhutkar, S. Schaeffer, S. Russo, M. Xu, T. Smith, and W. Gelbart, in prep.). These evolutionarily conserved arms have been termed Muller elements A through F (Muller 1940) (F being the minor arm). We exploited exceptions to arm conservation within genus Drosophila to identify PRGs and investigated possible mechanisms of movement. Additionally, we analyzed embedded gene relationships where a surrounding gene has another gene contained within its extent. This is synonymous with the term “nested genes” used elsewhere in the literature (Moriyama and Gojobori 1989; Rao and Sodja 1992; Kurzik-Dumke and Zengerle 1996; Kaymer et al. 1997; Pohar et al. 1999; Laundrie et al. 2003; Hudson et al. 2007). Studying the creation of embedded relationships, their conservation or loss, and their overlap with the set of PRGs shows this as an additional means to identify relocated genes.
The study of PRGs also sheds light on chromosomal reorganization between distant species during evolution. Extending our approach beyond genus Drosophila, we tested the hypothesis that PRGs might be significant contributors to differences in chromosomal organization between Drosophila and the mosquito Anopheles gambiae (Holt et al. 2002; Zdobnov et al. 2002). Additionally, using arm-level conservation between Drosophila and A. gambiae, we were able to show that the elimination of PRGs highlights arm-level conservation between these species and a member of the insect order Hymenoptera (the honeybee, Apis mellifera; Honeybee Genome Sequencing Consortium 2006).
Comparative analysis of multiple closely related genomes provides insights into the evolutionary process shaping different species and enhances our knowledge of the underlying biological mechanisms. We provide a window into one aspect of genomic change: the relocation of individual genes.
Results
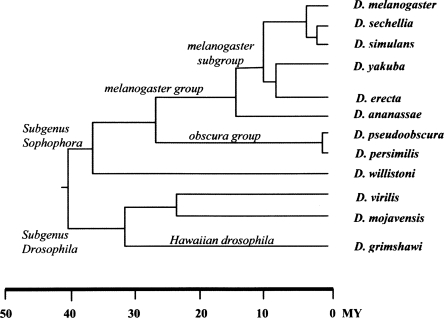
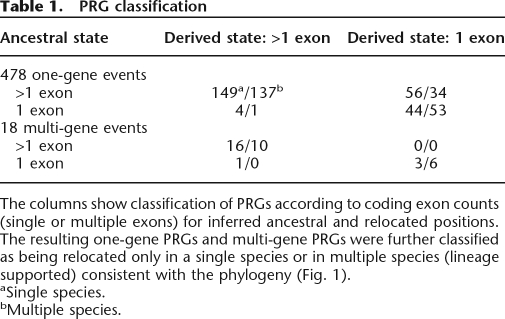
We identified candidate PRGs as putative orthologs that had no syntenic support and were, in fact, localized on a genome scaffold mapping to a nonsyntenic chromosome arm. A total of 1383 such candidate PRGs were initially identified. Cases that could be explained as matches to paralogous genes or that were not phylogenetically coherent were filtered out (see Supplemental material online). This left us with a high-confidence PRG set of 514 genes: 273 genes having relocated from the ancestral arm only in a single species and 241 genes in a lineage of multiple species consistent with the phylogeny (Fig. 1; Table 1). Within the high-confidence set, almost all of the PRGs involve the movement of only one gene (478), whereas the remainder involve multiple genes (18 events; 36 genes). In these latter cases, the genes are always adjacent and in preserved relative orientation in both the ancestral and relocated lineages. These are most simply explained as each resulting from a single multigene transposition event. The distribution of all inferred PRG events within the phylogeny is shown (Fig. 2A). Because of gaps that necessarily exist in the draft whole-genome shotgun assemblies of the 11 non-melanogaster species, such artifacts might contribute to single-species PRG estimates.
Figure 1.
Genus Drosophila phylogeny (Powell 1997).
Table 1.
PRG classification
The columns show classification of PRGs according to coding exon counts (single or multiple exons) for inferred ancestral and relocated positions. The resulting one-gene PRGs and multi-gene PRGs were further classified as being relocated only in a single species or in multiple species (lineage supported) consistent with the phylogeny (Fig. 1).
aSingle species.
bMultiple species.
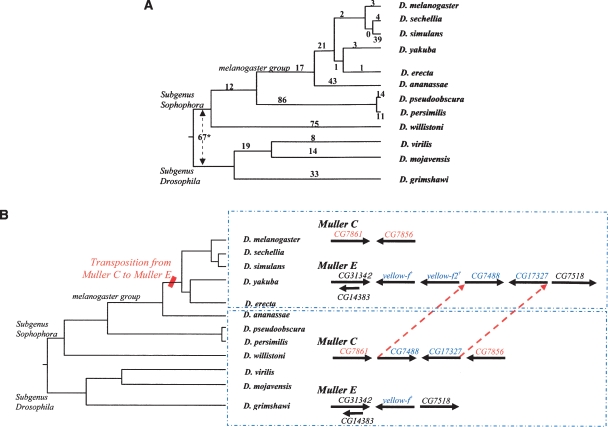
Figure 2.
PRGs identified by gene movement between chromosome arms. (A) Observed PRG counts across the Drosophila phylogeny. The set of 478 one-gene PRGs (Table 1) have been traced to events at various nodes of the phylogeny. (*) For the 67 PRGs at the genus root, ancestral states could not be determined. Five PRGs could not be placed unambiguously. (B) Multigene PRG relocation. Phylogenetic analysis of the original and new genomic location of this multigene PRG supports a DNA-based movement hypothesis. (†) There is a duplication of the gene yellow-f(f2) adjacent to the PRG relocation. Intron-exon structures are not shown. There are additional context changes in some species (D. persimilis, D. pseudoobscura).
We then compared gene structures in the ancestral and relocated lineages, making use of the consensus annotation sets available for each of the 11 non-melanogaster species (Drosophila 12 Genomes Consortium 2007; http://rana.lbl.gov/drosophila/) and the FlyBase D. melanogaster Release_4.3 annotation set (Crosby et al. 2007), subdividing them into multiexon and single-exon gene models. Since retrotransposition is one mechanism posited to produce PRGs, our goal was to determine whether there was enrichment for single-exon genes among our candidate PRG sets as a way of determining the validity of this approach. Of the 478 one-gene PRGs, 39% are single-exon genes in the relocated species or lineage. This is about a threefold enrichment over the 14% of single-exon genes among syntenically conserved (non-PRG) genes. Further, 24% of ancestral multiexon genes are relocated as single-exon genes (27% for the non–lineage-supported PRGs and 20% for the lineage-supported PRGs). In contrast, only 6 of the 478 one-gene PRGs have an ancestral single-exon gene and multiexon gene predictions in the relocated state; these few cases may be examples of genes acquiring new introns or may be inaccurate gene models produced by the automated gene prediction algorithms (Drosophila 12 Genomes Consortium 2007). These observations support our methodology to identify PRGs and provide a minimum estimate of the fraction of events (24%) that are likely due to retrotransposition of a processed mRNA. This may well be an underestimate since, of the 93 additional PRGs that are single exon in both the ancestral and relocated state, some might have relocated via retrotransposition. However, we lack the evidence to determine this as both states have a single exon.
Retrotranspositions of processed mRNAs cannot account for the majority of PRGs, however. Fully 60% of the one-gene PRGs are multiexon in both the ancestral and derived states. While the nature of the available gene models and the rates of divergence of the amino acid sequences of the proteins preclude a systematic analysis, it is our general impression that the intron locations in these multiexon genes are largely conserved between the ancestral and derived states. For example, in a set of 50 such genes with lineage support we found over 70% to have an equivalent number of exons in both states, despite the preliminary nature of gene models. These may represent retrotranspositions of pre-mRNAs and DNA-based relocation events. The contribution of DNA-based relocation events is supported by the 18 multigene PRG pairs. For these multigene PRGs, no unambiguous retrotransposition events were identified (Table 1). Rather, we find that these segment translocations show strong conservation of the gene structure; in all but one case, the ancestral and relocated genes were either both multiexon or were both single-exon genes (and that one exception may be an artifact of automated gene predictions as stated in the previous paragraph). Ancestral chromosome arm locations could be inferred in six of the eight multispecies cases and in all 10 of the single-species cases. We hypothesize that these multigene PRGs occur through transposition of DNA segments, without an RNA intermediate (Fig. 2B). While other mechanisms may well exist, we note that previous studies indicate that transposable elements similar to Foldback (FB) elements in D. melanogaster (Chia et al. 1985; Lovering et al. 1991; Casals et al. 2003, 2005) are capable of mediating such movement.
By tracking genes that have relocated from syntenic to nonsyntenic arms, we have identified substantial numbers of PRGs, but we do not believe that this represents all such PRG events that have arisen in the lineages of the 12 Drosophila species. One argument for this is that, while they are more difficult to identify unambiguously, we have observed examples of PRGs within a given chromosome arm (data not shown). Indeed, if there were no transpositional biases, we would expect about 80% of relocations to occur between arms and 20% within arms (given that the typical Drosophila genome principally consists of 5 major arms, all roughly equivalent in length). A second argument comes from examining so-called “embedded” genes, that is, genes that reside in introns of other genes. From prior studies involving the human and mouse genomes, retrotransposition is thought to be an important contributor to the formation of embedded genes (Yu et al. 2005) (Fig. 3A,B). In the best annotated Drosophila species, D. melanogaster, there are 763 embedded gene relationships involving a “surrounding gene” and an embedded gene contained within its extents (see Supplemental material). We find embedded genes in D. melanogaster to be transcribed in the opposite direction to that of the surrounding gene with a 2:1 preference, the same ratio as seen in the human genome (Yu et al. 2005). Fifty percent of the embedded genes in D. melanogaster have a single exon, as opposed to a background frequency of 14% of single-exon syntenically conserved euchromatin genes. This is consistent with a substantial contribution of retrotransposition to the formation of embedded genes and supports the idea that they can be considered PRGs. Further, there is about 53% nonoverlap between genes inferred to have been involved in the formation of embedded relationships in the D. melanogaster lineage (Fig. 3C) and PRGs identified by movement between chromosome arms (Table 1), suggesting that embedded genes offer an additional way to identify such relocation events. These events can be complex. For example, we depict the movement of the Rh4 gene (Neufeld et al. 1991) (Fig. 3A), which ancestrally was a multiexon gene with embedded genes within one of its introns. It has moved to become, in the subgenus Drosophila, an embedded gene elsewhere on the cognate chromosome arm.
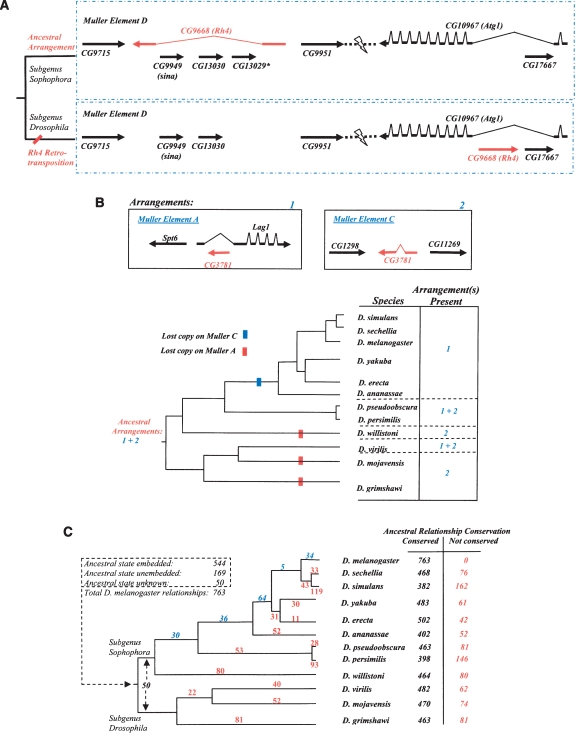
Figure 3.
PRGs identified among embedded genes. (A) Embedded gene relationships. Such relationships are observed to be labile. This example illustrates how embedded relationships can change. The gene Rh4 (CG9668) has three embedded genes in D. melanogaster. The embedded relationship involving Rh4 and one gene representing sina (CG9949) or CG13030 (which are duplicated genes) was ancestral, based on conservation in the outgroup mosquitoes (Aedes aegypti, A. gambiae). Previous analysis had shown the movement of Rh4 (Neufeld et al. 1991) to a different location on the same chromosome arm (Muller Element D). Our analysis reveals the ancestral arrangement of these genes, the position of the Rh4 transposition event within the Drosophila phylogeny, and the creation of a new embedded relationship as a result of this relocation. Movement of Rh4 into an embedded position within an intron of Atg1 (CG10967) on the same arm is most likely the result of a retrotransposition event as the relocated Rh4 is intronless in its new location. The loss of the ancestral copy of Rh4 caused sina and CG13030 to be unembedded. The third embedded gene in Rh4, CG13029, is not found outside of the D. melanogaster–D. ananassae lineage. Gene extents are not to scale and intron-exon structure has been shown for Rh4 and Atg1 only. (B) Changes to an ancestral embedded relationship via differential loss of retrotransposed or parent gene. Because of a previous retrotransposition, duplicated genes paralogous to CG3781 are thought to be present in the last common ancestor to the genus Drosophila, with the relocated copy hypothesized to have arisen as an embedded copy on Muller A, originating from gene CG3781 on Muller C. Three independent losses of the gene copy on Muller Element A and one on Muller Element C appear to have occurred, although there exists the possibility that some of the Muller Element C losses might be due to the gene being absent in one or more of the assemblies. Gene order and orientation is depicted in the diagram: Intron and exon structure is not shown for all genes and gene extents are not to scale. (C) Differential conservation of embedded gene relationships. The phylogeny shows the distribution of the inferred positions of occurrence of the remaining nonancestral embedded relationships in the lineage leading to D. melanogaster (blue numbers). Loss of ancestral embedded relationships in non-melanogaster lineages are shown (red numbers) and totals for each species are presented. Such losses can occur as a result of gene movement or gene loss in a given lineage. Because gene prediction methods are more error prone in regions containing embedded genes, this analysis was based on the alternative Synpipe methodology (Bhutkar et al. 2006) not requiring gene models in the test species. Briefly, translations of the genomic regions of the other 11 species were assessed for the embedded or unembedded state using TBLASTN and the D. melanogaster protein set as queries (see Methods).
More than half of the D. melanogaster embedded relationships appear to be the products of events that occurred before the time of the last common ancestor to the genus Drosophila and the remaining have arisen subsequently along the D. melanogaster lineage (Fig. 3C). This was determined by analyzing the conservation of the embedded nature of the 763 embedded D. melanogaster relationships across the 12 species on the basis of Synpipe (Bhutkar et al. 2006) syntenic TBLASTN analysis (see Supplemental material). In addition to noting that embedding PRGs have arisen at various points along the insect lineage, these data also demonstrate that secondary events arise in sublineages, such as differential conservation of a retrotransposed embedded gene versus the original copy of the gene on a different arm (Fig. 3B). The results show that about 80%–85% of ancestral embedded gene relationships are conserved in a given species but only 34% are conserved across all 12 species (Fig. 3C). Embedded relationships at the root of the Drosophila phylogeny can be undone if the putative ortholog has relocated to an unembedded location (paralogous relationships mask some of these events) or if there is a gene loss event. For fewer than half of the 763 D. melanogaster embedded gene relationships, the ancestral state was unembedded, and the establishment of the embedded state occurs at any of several nodes along the lineage to D. melanogaster (Fig. 3C).
Overall, embedded genes appear to offer another partially complementary detection system for PRGs. We do not claim that every embedded gene is the result of a PRG, as there could be cases of exon recruitment or alternate splice misannotation (Manak et al. 2006), among other explanations. Additionally, the lineage-supported embedded gene PRGs (Fig. 3C) are more reliably determined than single-species events because of comparative evidence.
Our results raise some interesting questions about the nature of organizational relationships between more distant species. Here, we have had the opportunity to examine PRGs in a cluster of species that are separated by ∼50–60 million years. While we cannot be exact in our estimates, we can be confident that several hundred PRGs have occurred in the radiation of genus Drosophila. Based on this ballpark number, we would anticipate that a few thousand PRGs have arisen in deeper radiations, such as since the time of the last common ancestor between Drosophila and mosquitoes (both members of the order Diptera), generally estimated to have existed some 250 million years ago.
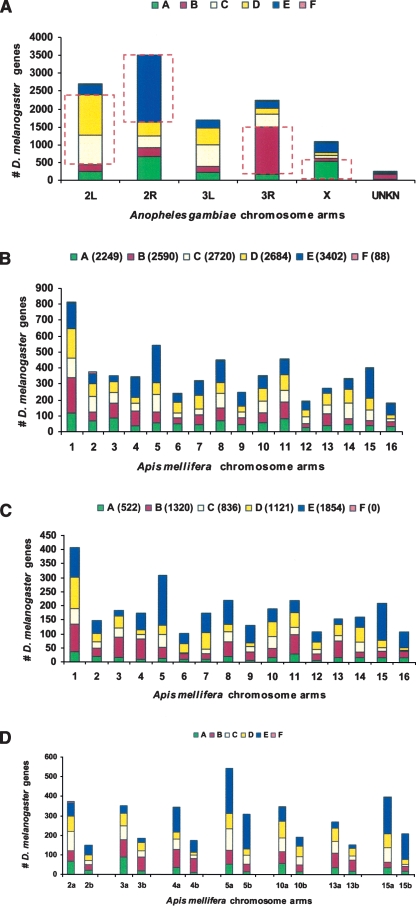
We indirectly tested the idea that there is a large population of PRGs that contribute to the differences in genome organization between Drosophila and mosquitoes. We carried out Synpipe TBLASTN analysis (Bhutkar et al. 2006) of the A. gambiae mosquito genome (Holt et al. 2002; Zdobnov et al. 2002) using the D. melanogaster protein query set and identified the major chromosome arm conservations between the two species (Fig. 4A), which account for ∼5700 of the ∼14,000 D. melanogaster genes (see Supplemental material). These arm-correspondence results agree with previous analyses (Zdobnov et al. 2002) comparing these genomes. We then compared the results of the following two analyses of an outgroup species, a member of the insect order Hymenoptera (the honeybee, A. mellifera; Honeybee Genome Sequencing Consortium 2006). One analysis used the entire D. melanogaster protein set (Fig. 4B). The other used the set of ∼5700 genes that are arm-conserved with A. gambiae (Fig. 4C)—we posited that this set eliminates most PRGs that arose during the Dipteran radiation (since mosquito and Drosophila are very distant members of the radiation). We asked whether by eliminating the PRGs we would observe increases in conservation of chromosome-level synteny between D. melanogaster and A. mellifera. Indeed, by comparing the results for individual chromosomes, we do see an increased gene association of D. melanogaster chromosome arms and individual honeybee chromosomes (Fig. 4D). We conclude that this supports the idea that the set of non–arm-conserved D. melanogaster genes eliminated in the latter analysis include many candidate PRGs and, hence, that PRGs are important contributors to the overall genome organizational differences that are seen between distantly related species.
Figure 4.
Impact of PRGs on more distant genome comparisons. (A) Chromosome arm correspondence between D. melanogaster and A. gambiae. Arm correspondence was inferred based on majority of D. melanogaster orthologous genes (from Muller elements A–F) found on A. gambiae chromosome arms. For example, Drosophila Muller E corresponds to A. gambiae arm 2R. Other relationships were inferred as follows: Drosophila A and A. gambiae X, B and 3R, C and 2L, D and 2L. Genes in these sets (shown boxed) are inferred to colocalize on an ancestral chromosome arm that was the primary contributor to the extant arms in these species. (B) D. melanogaster orthology with A. mellifera (honeybee). D. melanogaster genes (number of genes in the query set are shown in parentheses for each Muller element) with orthologs are distributed across various arms of A. mellifera. (C) Orthology with inferred ancestral colocated set from A. The set of ancestrally colocated D. melanogaster genes shown boxed in A, are mapped across various arms of A. mellifera. Numbers in parentheses show the number of genes from each Drosophila Muller element that are part of this query set. This mapping filters out noise due to PRGs over this long evolutionary distance and highlights contributions of ancestral arms to the honeybee genome organization. For example, the ancestral arm corresponding to D. melanogaster Muller element E and A. gambiae arm 2R is the primary contributor to honeybee chromosome 5. (D) Removal of PRGs enhances correlated chromosomal locations between honeybee and Drosophila. A side-by-side comparison of orthology using all D. melanogaster genes versus the inferred colocated set from A shows how reducing the contributions of PRGs enriches the signal from primary arm contributors. For example, eliminating PRGs from A. mellifera chromosome 5 (5a vs. 5b) and chromosome 15 (15a vs. 15b) shows the ancestral arm corresponding to Drosophila Muller element E (A. gambiae 2R) as the primary contributor. Similarly, contributions of the ancestral arm corresponding to Drosophila Muller element B (A. gambiae 3R) stand out for A. mellifera chromosomes 3 and 4.
Discussion
We have presented a detailed study of PRGs based on a comparative genomics approach. While gross chromosomal rearrangements (paracentric and pericentric inversions, translocations, and arm fusions and fissions) are known to be major elements of genome reorganization, our studies point to the important role that PRGs play in this process. We see that such events can be classified according to the gene structures in the ancestral and derived locations. It is likely that some PRGs arise through retrotransposition mechanisms and others through direct transposition of genomic DNA, and it will be important to understand the underlying mechanisms. Although analysis of arm conservation across the phylogeny results in robust identification of PRGs, we believe the set of lineage-supported PRGs to be more reliable than those identified in only one species. The nature of draft shotgun genome assemblies could introduce assembly artifacts that would influence results in a single species. In addition to PRGs identified using the phylogeny and arm-conservation information, the overlap between PRGs and genes involved in the creation of embedded relationships in the D. melanogaster lineage point to another source of identifying PRGs. The labile nature of these relationships and their differential conservation (Fig. 3) across the phylogeny highlight the role of PRGs in the constant evolution of genome organization.
PRGs, especially those that move through mechanisms like retrotransposition, are most likely in a different regulatory environment compared to their original location. A cross-species comparison between PRGs and genes that have retained their ancestral genomic location shows that PRGs have a significantly higher rate of protein evolution (Drosophila 12 Genomes Consortium 2007). This suggests that PRGs undergo accelerated evolution due to a change in their genomic location. We also note traces of degraded genes via BLAST analysis (data not shown) where the ancestral location lacks a full-length gene model prediction.
We also sought any evidence to support the notion that male reproductive functions undergo accelerated molecular evolution (Swanson and Vacquier 2002; Swanson et al. 2003). We found that 42% (39 out of 94; see Supplemental material) of the PRGs that have a single exon in the derived state in the lineage-supported set are expressed in the D. melanogaster testes (50% in the case where both ancestral and derived states have a single exon; 25% of the genes in lineage supported DNA-based transposition events). This analysis shows a high percentage of PRGs to be expressed in the testes, based on the set of 2329 unique genes (18% or less of the more than 13,000 D. melanogaster euchromatic genes) currently known to be expressed in the testes (Chintapalli et al. 2007) (http://www.flymine.org; http://www.flyatlas.org).
The presence of PRGs also obscures the underlying arm-level conservation between distant species. This effect is more pronounced with species at a greater distance, as seen in the analysis with A. mellifera (honeybee). We find the results to be comparable starting with the D. melanogaster annotated protein set or with the A. gambiae annotated set (data not shown). The exclusion of PRGs improves the arm-level conservation signal in comparison with previous studies (Honeybee Genome Sequencing Consortium 2006), as seen in the case of honeybee chromosome 3 (Fig. 4D). In addition to segmentation, fission, and fusion of chromosome arms, we show that PRGs play a significant role in the differences in genome organization. We see similar results in comparisons with the red flour beetle, Tribolium castaneum (data not shown). As additional insect genomes become available, we expect to see similar results in arm conservation by identifying and removing putative PRGs from the analysis set.
PRGs should also provide profound opportunities for genes to acquire very different regulatory profiles through being subjugated to the regulatory controls of the chromosomal locations to which they relocate and other topological (Chen and Stein 2006) or transpositional (Nozawa et al. 2005) considerations. Finally, we should note that our studies only provide a glimpse into the number and frequency of successful transposition events. It will be important to develop ways to measure the genome-wide frequency and characteristics of all such relocation events, which undoubtedly will be far greater in magnitude than those that become fixed during evolution.
Methods
Gene orthology
Gene orthology information was derived from Synpipe (Bhutkar et al. 2006) synteny analysis and the GLEAN-R gene orthology set (Drosophila 12 Genomes Consortium 2007) (http://rana.lbl.gov/drosophila/). Synpipe uses TBLASTN results and synteny-aided placement criteria to determine gene homology assignment between a set of genes from a reference species and a candidate genome assembly for another species. Homology assignments for GLEAN-R gene predictions were determined using a multispecies extension of the reciprocal BLAST method (Tatusov et al. 1997; Drosophila 12 Genomes Consortium 2007). All gene names used in this study are based on FlyBase Release_4.3 (Crosby et al. 2007), and orthologs in other species are shown using these identifiers. Gene structure predictions from the GLEAN-R consensus annotation set (http://rana.lbl.gov/drosophila/) were used for species other than D. melanogaster. Assembly misjoins were identified using synteny analysis and experimental markers (Drosophila 12 Genomes Consortium 2007) and corrected for wherever applicable. Scaffolds were assigned to Muller elements based on majority hits from D. melanogaster orthologs (A. Bhutkar, S. Schaeffer, S. Russo, M. Xu, T. Smith, W. Gelbart, in prep.). Known rearrangement events within genus Drosophila resulting from arm fusion or large-scale pericentric inversions were filtered from these analyses, and genes involved in these events were not considered PRG candidates. Genes whose orthologs were inferred to be in assembly gaps based on Synpipe synteny analysis were not considered to be relocated PRGs in the presence of paralogous hits.
Positionally Relocated Genes (PRGs)
PRG identification was based on ortholog(s) found on different Muller elements in different species. Single-species PRGs include cases where the Muller assignment for a gene in all other species is the same (or the gene is not found in some species). Multispecies PRGs included cases where groups of species consistent with the phylogeny had the gene placed on a different Muller element. Both classes of PRG candidates included cases where either one or both of the original or transposed copies might exist in extant species. These correspond to conservative and duplicative transpositions, respectively (Gonzalez et al. 2004). Species that did not have an ortholog for a gene were excluded in the analysis of that gene. Multigene PRGs (units involving two or more adjacent genes) were identified based on the stringent criteria of both genes being identified in at least 9 of the 12 species, being maintained as an adjacent unit in 8 or more species, and being placed on scaffolds assigned to Muller elements with a high degree of confidence. PRG candidates with changes in Muller element location in groups of species inconsistent with the phylogeny (or in multiple lineages) were filtered from the high-confidence set. Further, both Synpipe and GLEAN-R orthology assignments were used to determine PRGs selected for the high-confidence set (single-species PRGs had to be supported by both and lineage moves were supported by one of the two). GLEAN-R orthology calls were refined using Synpipe placements (by the Eisen Laboratory, University of California, Berkeley) so these approaches agree in most cases. Further classification of PRGs based on exon counts in gene structure predictions was performed. Ancestral state (number of exons) was derived from phylogenetic distribution of the number of exons in orthologs across all species. Derived states were determined from the gene structure prediction for the relocated ortholog. The list of D. melanogaster genes expressed in the testes was obtained from FlyMine (http://www.flymine.org) with data sets from FlyAtlas (Chintapalli et al. 2007; http://www.flyatlas.org).
Embedded genes
D. melanogaster embedded gene relationships (763) were defined where a gene completely contained another gene within its extents (including UTR annotation). As gene prediction tools cannot always predict models for embedded genes cleanly (the surrounding gene might be broken up into multiple gene predictions, or the embedded gene might not have a prediction), a combination of TBLASTN and gene predictions were used for conservation inference. Additionally, gene predictions in other species do not include UTR annotations, so an adjacency criterion (±10 genes to allow for multiple embedded genes and break-up of the surrounding gene into multiple gene predictions) was used to supplement conservation inference. As a result, embedded gene conservation inference was based on a TBLASTN signal for the embedded gene within the gene model of the surrounding gene or a TBLASTN signal adjacent to it, to allow for preliminary gene predictions.
PRGs and genome organization
Using Synpipe orthology, matches from various D. melanogaster Muller element genes to each A. gambiae scaffold were counted. The set of genes that form the majority of hits to each A. gambiae arm (Fig. 4A) were then used and their Synpipe orthology assignments in A. mellifera (honeybee) were selected. This distribution was compared with the straightforward set of all D. melanogaster Muller element genes hitting various honeybee chromosomes to illustrate how removal of PRGs enhances correlated chromosomal locations between Drosophila and honeybee. Similar results were obtained with the A. gambiae protein coding genes as the starting point (data not shown).
Acknowledgments
We thank the AAA (Assembly/Alignment/Annotation) coordinating committee for the 12 Drosophila genomes project (all Drosophila data are part of the AAA CAF1 freeze [http://rana.lbl.gov/drosophila/] and has been used in accordance with AAA guidelines for a companion paper); FlyBase for D. melanogaster data sets and computing support; Agencourt Biosciences for sequence data for D. erecta, D. ananassae, D. virilis, D. mojavensis, D. grimshawi genome assemblies; Baylor College of Medicine HGSC for D. pseudoobscura and Tribolium castaneum (data not shown); Washington University GSC for assemblies of D. yakuba and D. simulans; Broad Institute and TIGR for D. sechellia, D. persimilis; JCVI for D. willistoni; Venky Iyer (Eisen Laboratory, University of California, Berkeley), and AAA for releasing GLEAN-R gene predictions; the research computing group in the Division of Life Sciences at Harvard University for systems support; and BMERC (Boston U.) computing and support staff. This work was supported by a subcontract to Boston University from Harvard University under NIH grant HG000739.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7062307
References
- Betran E., Long M., Long M. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics. 2003;164:977–988. doi: 10.1093/genetics/164.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutkar A., Russo S., Smith T.F., Gelbart W.M., Russo S., Smith T.F., Gelbart W.M., Smith T.F., Gelbart W.M., Gelbart W.M. Techniques for multi-genome synteny analysis to overcome assembly limitations. Genome Inform. 2006;17:152–161. [PubMed] [Google Scholar]
- Brosius J. Retroposons—seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Casals F., Caceres M., Ruiz A., Caceres M., Ruiz A., Ruiz A. The foldback-like transposon Galileo is involved in the generation of two different natural chromosomal inversions of Drosophila buzzatii. Mol. Biol. Evol. 2003;20:674–685. doi: 10.1093/molbev/msg070. [DOI] [PubMed] [Google Scholar]
- Casals F., Caceres M., Manfrin M.H., Gonzalez J., Ruiz A., Caceres M., Manfrin M.H., Gonzalez J., Ruiz A., Manfrin M.H., Gonzalez J., Ruiz A., Gonzalez J., Ruiz A., Ruiz A. Molecular characterization and chromosomal distribution of Galileo, Kepler and Newton, three foldback transposable elements of the Drosophila buzzatii species complex. Genetics. 2005;169:2047–2059. doi: 10.1534/genetics.104.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Stein L.D., Stein L.D. Conservation and functional significance of gene topology in the genome of Caenorhabditis elegans. Genome Res. 2006;16:606–617. doi: 10.1101/gr.4515306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia W., McGill S., Karp R., Gubb D., Ashburner M., McGill S., Karp R., Gubb D., Ashburner M., Karp R., Gubb D., Ashburner M., Gubb D., Ashburner M., Ashburner M. Spontaneous excision of a large composite transposable element of Drosophila melanogaster. Nature. 1985;316:81–83. doi: 10.1038/316081a0. [DOI] [PubMed] [Google Scholar]
- Chintapalli V.R., Wang J., Dow J.A., Wang J., Dow J.A., Dow J.A. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Coghlan A., Wolfe K.H., Wolfe K.H. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 2002;12:857–867. doi: 10.1101/gr.172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby M.A., Goodman J.L., Strelets V.B., Zhang P., Gelbart W.M., Goodman J.L., Strelets V.B., Zhang P., Gelbart W.M., Strelets V.B., Zhang P., Gelbart W.M., Zhang P., Gelbart W.M., Gelbart W.M. FlyBase: Genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007 doi: 10.1038/nature06341. (in press) [DOI] [PubMed] [Google Scholar]
- Fischer G., Neuveglise C., Durrens P., Gaillardin C., Dujon B., Neuveglise C., Durrens P., Gaillardin C., Dujon B., Durrens P., Gaillardin C., Dujon B., Gaillardin C., Dujon B., Dujon B. Evolution of gene order in the genomes of two related yeast species. Genome Res. 2001;11:2009–2019. doi: 10.1101/gr.212701. [DOI] [PubMed] [Google Scholar]
- Gonzalez J., Casals F., Ruiz A., Casals F., Ruiz A., Ruiz A. Duplicative and conservative transpositions of larval serum protein 1 genes in the genus Drosophila. Genetics. 2004;168:253–264. doi: 10.1534/genetics.103.025916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P.M., Milburn D., Zhang Z., Bertone P., Gerstein M., Milburn D., Zhang Z., Bertone P., Gerstein M., Zhang Z., Bertone P., Gerstein M., Bertone P., Gerstein M., Gerstein M. Identification of pseudogenes in the Drosophila melanogaster genome. Nucleic Acids Res. 2003;31:1033–1037. doi: 10.1093/nar/gkg169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt R.A., Subramanian G.M., Halpern A., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Subramanian G.M., Halpern A., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Halpern A., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Sutton G.G., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Charlab R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Nusskern D.R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Wincker P., Clark A.G., Ribeiro J.M., Wides R., Clark A.G., Ribeiro J.M., Wides R., Ribeiro J.M., Wides R., Wides R., et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson S., Garrett M., Micklem G., Goldstein E., Newfeld S., Garrett M., Micklem G., Goldstein E., Newfeld S., Micklem G., Goldstein E., Newfeld S., Goldstein E., Newfeld S., Newfeld S. Phylogenetic and genome-wide analyses suggest a functional relationship between kayak the Drosophila Fos homolog and fig a predicted PP2C phosphatase nested within a kayak intron. Genetics. 2007 doi: 10.1534/genetics.107.071670. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaymer M., Debes A., Kress H., Kurzik-Dumke U., Debes A., Kress H., Kurzik-Dumke U., Kress H., Kurzik-Dumke U., Kurzik-Dumke U. Sequence, molecular organization and products of the Drosophila virilis homologs of the D. melanogaster nested genes lethal(2) tumorous imaginal discs [1(2)tid] and lethal(2) neighbour of tid [1(2)not] Gene. 1997;204:91–103. doi: 10.1016/s0378-1119(97)00528-3. [DOI] [PubMed] [Google Scholar]
- Kurzik-Dumke U., Zengerle A., Zengerle A. Identification of a novel Drosophila melanogaster gene, angel, a member of a nested gene cluster at locus 59F4,5. Biochim. Biophys. Acta. 1996;1308:177–181. doi: 10.1016/0167-4781(96)00108-x. [DOI] [PubMed] [Google Scholar]
- Langille M.G., Clark D.V., Clark D.V. Parent genes of retrotransposition-generated gene duplicates in Drosophila melanogaster have distinct expression profiles. Genomics. 2007;90:334–343. doi: 10.1016/j.ygeno.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Laundrie B., Peterson J.S., Baum J.S., Chang J.C., Fileppo D., Thompson S.R., McCall K., Peterson J.S., Baum J.S., Chang J.C., Fileppo D., Thompson S.R., McCall K., Baum J.S., Chang J.C., Fileppo D., Thompson S.R., McCall K., Chang J.C., Fileppo D., Thompson S.R., McCall K., Fileppo D., Thompson S.R., McCall K., Thompson S.R., McCall K., McCall K. Germline cell death is inhibited by P-element insertions disrupting the dcp-1/pita nested gene pair in Drosophila. Genetics. 2003;165:1881–1888. doi: 10.1093/genetics/165.4.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B., Malpertuy A., Neuveglise C., de Montigny J., Aigle M., Artiguenave F., Blandin G., Bolotin-Fukuhara M., Bon E., Brottier P., Malpertuy A., Neuveglise C., de Montigny J., Aigle M., Artiguenave F., Blandin G., Bolotin-Fukuhara M., Bon E., Brottier P., Neuveglise C., de Montigny J., Aigle M., Artiguenave F., Blandin G., Bolotin-Fukuhara M., Bon E., Brottier P., de Montigny J., Aigle M., Artiguenave F., Blandin G., Bolotin-Fukuhara M., Bon E., Brottier P., Aigle M., Artiguenave F., Blandin G., Bolotin-Fukuhara M., Bon E., Brottier P., Artiguenave F., Blandin G., Bolotin-Fukuhara M., Bon E., Brottier P., Blandin G., Bolotin-Fukuhara M., Bon E., Brottier P., Bolotin-Fukuhara M., Bon E., Brottier P., Bon E., Brottier P., Brottier P., et al. Genomic exploration of the hemiascomycetous yeasts: 18. Comparative analysis of chromosome maps and synteny with Saccharomyces cerevisiae. FEBS Lett. 2000;487:101–112. doi: 10.1016/s0014-5793(00)02289-4. [DOI] [PubMed] [Google Scholar]
- Lovering R., Harden N., Ashburner M., Harden N., Ashburner M., Ashburner M. The molecular structure of TE146 and its derivatives in Drosophila melanogaster. Genetics. 1991;128:357–372. doi: 10.1093/genetics/128.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak J.R., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Cheng J., Bell I., Ghosh S., Piccolboni A., Bell I., Ghosh S., Piccolboni A., Ghosh S., Piccolboni A., Piccolboni A., et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Metz C.W. Chromosome studies in the Diptera I. A preliminary survey of five different types of chromosome groups in the genus Drosophila. J. Exp. Zool. 1914;17:45–59. [Google Scholar]
- Moriyama E.N., Gojobori T., Gojobori T. Evolution of nested genes with special reference to cuticle proteins in Drosophila melanogaster. J. Mol. Evol. 1989;28:391–397. doi: 10.1007/BF02603074. [DOI] [PubMed] [Google Scholar]
- Muller H.J. The New Systematics. Clarendon Press; Oxford, UK: 1940. [Google Scholar]
- Neufeld T.P., Carthew R.W., Rubin G.M., Carthew R.W., Rubin G.M., Rubin G.M. Evolution of gene position: Chromosomal arrangement and sequence comparison of the Drosophila melanogaster and Drosophila virilis sina and Rh4 genes. Proc. Natl. Acad. Sci. 1991;88:10203–10207. doi: 10.1073/pnas.88.22.10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M., Aotsuka T., Tamura K., Aotsuka T., Tamura K., Tamura K. A novel chimeric gene, siren, with retroposed promoter sequence in the Drosophila bipectinata complex. Genetics. 2005;171:1719–1727. doi: 10.1534/genetics.105.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohar N., Godenschwege T.A., Buchner E., Godenschwege T.A., Buchner E., Buchner E. Invertebrate tissue inhibitor of metalloproteinase: Structure and nested gene organization within the synapsin locus is conserved from Drosophila to human. Genomics. 1999;57:293–296. doi: 10.1006/geno.1999.5776. [DOI] [PubMed] [Google Scholar]
- Powell J.R. Progress and prospects in evolutionary biology: The Drosophila model. Oxford University Press; Oxford, UK: 1997. [Google Scholar]
- Ranz J.M., Gonzalez J., Casals F., Ruiz A., Gonzalez J., Casals F., Ruiz A., Casals F., Ruiz A., Ruiz A. Low occurrence of gene transposition events during the evolution of the genus Drosophila. Evolution Int. J. Org. Evolution. 2003;57:1325–1335. doi: 10.1111/j.0014-3820.2003.tb00340.x. [DOI] [PubMed] [Google Scholar]
- Rao J.P., Sodja A., Sodja A. Further analysis of a transcript nested within the actin 5C gene of Drosophila melanogaster. Biochem. Biophys. Res. Commun. 1992;184:400–407. doi: 10.1016/0006-291x(92)91207-7. [DOI] [PubMed] [Google Scholar]
- Sturtevant A.H., Novitski E., Novitski E. The homologies of the chromosome elements in the genus Drosophila. Genetics. 1941;26:517–541. doi: 10.1093/genetics/26.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson W.J., Vacquier V.D., Vacquier V.D. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swanson W.J., Nielsen R., Yang Q., Nielsen R., Yang Q., Yang Q. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 2003;20:18–20. doi: 10.1093/oxfordjournals.molbev.a004233. [DOI] [PubMed] [Google Scholar]
- Tatusov R.L., Koonin E.V., Lipman D.J., Koonin E.V., Lipman D.J., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Yu P., Ma D., Xu M., Ma D., Xu M., Xu M. Nested genes in the human genome. Genomics. 2005;86:414–422. doi: 10.1016/j.ygeno.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Zdobnov E.M., von Mering C., Letunic I., Torrents D., Suyama M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., von Mering C., Letunic I., Torrents D., Suyama M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Letunic I., Torrents D., Suyama M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Torrents D., Suyama M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Suyama M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Thomasova D., Holt R.A., Subramanian G.M., Holt R.A., Subramanian G.M., Subramanian G.M., et al. Comparative genome and proteome analysis of Anopheles gambiaeand Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Harrison P.M., Liu Y., Gerstein M., Harrison P.M., Liu Y., Gerstein M., Liu Y., Gerstein M., Gerstein M. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]