Abstract
Concerted evolution maintains at near identity the hundreds of tandemly arrayed ribosomal RNA (rRNA) genes and their spacers present in any eukaryote. Few comprehensive attempts have been made to directly measure the identity between the rDNA units. We used the original sequencing reads (trace archives) available through the whole-genome shotgun sequencing projects of 12 Drosophila species to locate the sequence variants within the 7.8–8.2 kb transcribed portions of the rDNA units. Three to 18 variants were identified in >3% of the total rDNA units from 11 species. Species where the rDNA units are present on multiple chromosomes exhibited only minor increases in sequence variation. Variants were 10–20 times more abundant in the noncoding compared with the coding regions of the rDNA unit. Within the coding regions, variants were three to eight times more abundant in the expansion compared with the conserved core regions. The distribution of variants was largely consistent with models of concerted evolution in which there is uniform recombination across the transcribed portion of the unit with the frequency of standing variants dependent upon the selection pressure to preserve that sequence. However, the 28S gene was found to contain fewer variants than the 18S gene despite evolving 2.5-fold faster. We postulate that the fewer variants in the 28S gene is due to localized gene conversion or DNA repair triggered by the activity of retrotransposable elements that are specialized for insertion into the 28S genes of these species.
Eukaryotic ribosomal RNAs (rRNA) are encoded by hundreds of gene copies organized in tandem arrays (the rDNA loci) (Long and Dawid 1980; Hillis and Dixon 1991). Each unit within the array contains one copy of three major rRNA genes: 18S, 28S, and 5.8S (Fig. 1). The different copies of the rDNA units have high-sequence identity within species, but differ between species, a phenomenon called concerted evolution. While the sequence of the rRNA genes evolves slowly, the internal transcribed spacers (ITS1 and ITS2), the external transcribed spacer (ETS), and the intergenic spacer (IGS) evolve rapidly.
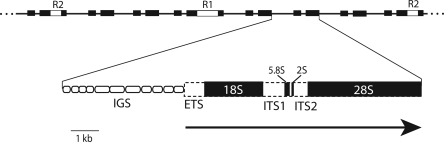
Figure 1.
Diagram of a typical Drosophila rRNA gene locus. Shown at the top is the tandemly repeated structure of the locus with only the 18S and 28S genes indicated. R1 and R2 elements (white boxes) are inserted into the 28S gene of 25%–50% of the units (Lathe et al. 1995; Lathe and Eickbush 1997). The expanded region shows a more detailed view of the organization of the complete rDNA unit found in all Drosophila species studied here. In addition to the 18S, 5.8S, and 28S genes, insect rDNA units also contain a 2S gene (Jordan et al. 1976). The transcribed region of the unit is indicated by the horizontal arrow at the bottom. The gene regions are shown as black boxes, the external transcribed spacer (ETS) and internal transcribed spacers (ITS1 and ITS2) as dotted boxes. The intergenic spacer region, IGS, is predominately composed of internal subrepeats (rounded boxes). The IGS of different units varies in length due to differences in the number of copies of each subrepeat. The sequence and length of these subrepeats vary dramatically between species (see Fig. 5).
Most models for the concerted evolution of the rDNA locus suggest that frequent recombination events involving unequal crossovers and gene conversions result in the high-sequence identity between units (Ohta 1980). In this process, a mutation originating in one unit is increased or decreased in number by recombination until it is either eliminated or present in all units. Substitutions in the coding regions are subject to selection for rRNA function and most are eliminated, while substitutions in the noncoding areas are under less selective pressure and are more often fixed by stochastic recombination events. Thus, concerted evolution is a dynamic balance between mutation, selection, and recombination.
Many studies have been conducted to follow the concerted evolution of the rDNA units in Drosophila melanogaster (Coen et al. 1982; Ohta and Dover 1983; Ohta 1984; Dover 1994; Schlötterer and Tautz 1994; Elder and Turner 1995; Polanco et al. 1998, 2000). Units in loci on different chromosomes within populations were shown to exhibit more sequence differences than units within a single locus, suggesting that intrachromosomal recombination occurred more often than interchromosomal recombination (Schlötterer and Tautz 1994; Polanco et al. 1998). The large variation in number of rDNA units in different individuals of a population (Lyckegaard and Clark 1991), and the rapid change in rDNA unit number in duplicate laboratory strains (Averbeck and Eickbush 2005) suggest that unequal sister chromatid exchange is likely to be the major mechanism involved in this intrachromosomal uniformity. The slower spread of sequences between chromosomes in a population is consistent with the low rates of crossovers observed between rDNA loci on different X chromosomes or between the X and Y chromosomes (Williams et al. 1989). Evidence for the role of gene conversion in the concerted evolution of the rDNA locus has been difficult to prove or disprove.
Evolution of the rDNA loci in Drosophila, as in many animals, is also influenced by retrotransposable elements, R1 and R2, which insert into specific sites of the 28S rRNA gene (Jakubczak et al. 1991). These elements have been stable components of the rDNA locus since the origin of arthropods (Malik et al. 1999). The fraction of 28S genes inserted by either of these elements can vary from 10% to >50% (Lathe et al. 1995; Lathe and Eickbush 1997). The extent to which these insertions affect the process of concerted evolution is not known.
A detailed view of the level and distribution of the sequence variation within the rDNA units of an organism would provide critical insights into the concerted evolution process. However, the dearth of distinguishing details between repeats has made this difficult. Even with the “complete genome sequences” now available for many species, the rDNA loci have not been assembled or the level of sequence variation quantified. While whole-genome shotgun sequencing does not provide a means to assemble rDNA loci, it does provide a wealth of information on the nucleotide variation that exists. In this report, the rDNA sequences from the original unassembled sequencing reads are used to characterize for the first time the nucleotide sequence variation that exists in the rDNA units of 11 Drosophila species. These data provide valuable insights into both the efficiency and the mechanism of the concerted evolution.
Results
Comparison of the consensus rDNA unit among species
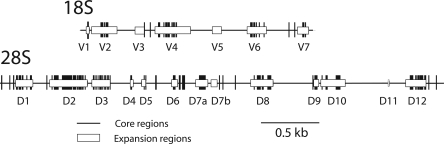
Consensus sequences for the transcribed portion of each rDNA unit were first assembled (Supplemental Fig. 1a–l). The small 5.8S and 2S genes were highly conserved with only the first position of the 5.8S gene having undergone a substitution in species of the melanogaster group. The 18S gene in all species was 1995 bp in length, while the 28S gene varied in length from 3948 to 3976 bp. The 18S and 28S rRNA sequences can be subdivided into the slower evolving “core regions,” which include the active sites, substrate binding sites, and contact points between subunits, and the “expansion regions,” which vary in sequence and account for most of the length differences among eukaryotic and prokaryotic rRNA genes (Clark et al. 1984; Hassouna et al. 1984). Figure 2 plots the sequence differences among the 12 species (vertical lines). The expansion regions (white boxes) contained most of the nucleotide differences and all but one of the indels found between Drosophila species, with most length variation in expansion regions D7a and D12. Most changes occurred in single-stranded or loop regions of the proposed secondary structure (Hancock and Dover 1988).
Figure 2.
Locations of fixed differences in the 18S and 28S genes among the 12 Drosophila species. The core (thin horizontal lines) and expansion (white boxes) regions of the genes are indicated and numbered as in Hancock et al. (1988). Sequence differences between the species are indicated by vertical bars.
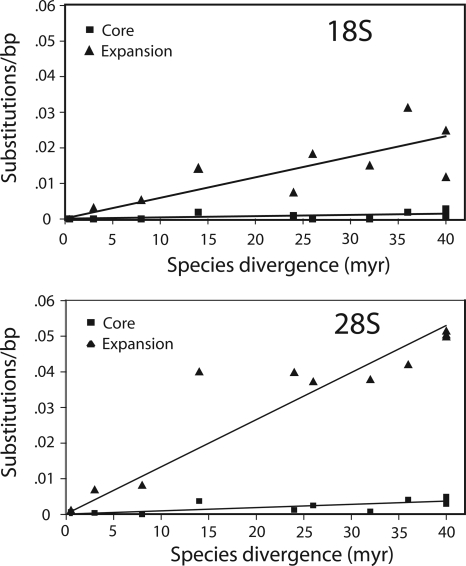
To calculate the rate at which the core and expansion regions accumulated nucleotide substitutions, the sequence divergences of these regions were determined for various species pairs. These divergences are plotted in Figure 3 as a function of the time estimates since separation of the species. The expansion regions of the 18S and 28S genes diverged about 15 times faster than their respective core regions. Both the core and expansion regions of the 28S gene diverged 2.5 times faster than the 18S gene.
Figure 3.
Nucleotide substitution rates for the core and expansion regions of the 18S and 28S genes. Divergence between sequences was calculated as the number of differences per aligned sequence (indels were not considered). Species pairs for which divergence was calculated were simulans vs. sechellia; melanogaster vs. each of sechellia, yakuba, ananassae, pseudoobscura, willistoni, and virilis; and finally, virilis vs. each of mojavensis, grimshawi, willistoni, and pseudoobscura. The species divergence times were those obtained from the Drosophila Species Genomes Web site (http://insects.eugenes.org/species/).
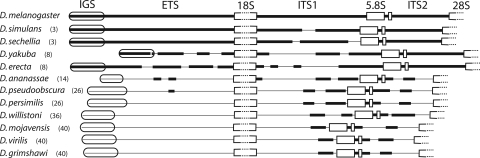
The transcriptional start site marking the beginning of the ETS has been determined for D. melanogaster and Drosophila virilis (Long et al. 1981; Murtif and Rae 1985). Because in these species the ETS starts near the last subrepeat in the IGS, the first nucleotide downstream of the last tandem subrepeat in the IGS was arbitrarily defined as the ETS boundary for the remaining species. The length of the ETS as well as the ITS1 and ITS2 regions in each species are shown in Figure 4. Regions within these spacers with >75% nucleotide identity to the corresponding D. melanogaster sequence are indicated by the thicker horizontal lines. Extensive sequence identities were only found between members of the melanogaster species subgroup (melanogaster, simulans, sechellia, yakuba, and erecta). However, as was noted previously by Schlötterer et al. (1994) the 3′ end of ITS1 and 5′ end of ITS2 are more conserved, showing sequence identity across all species. Sequence conservation in these two regions has been suggested to be a result of secondary structures needed for processing of the primary RNA transcript.
Figure 4.
Comparison of the transcribed spacer regions in the 12 Drosophila species. A schematic diagram of the rDNA unit in each species is shown with horizontal lines representing the ETS and ITS regions, rounded boxes the last subrepeat of the IGS, and boxes the gene regions (the 18S and 28S genes are not drawn to scale). Thicker lines indicate regions of the spacers from each species that have at least 75% sequence identity with the D. melanogaster sequence. Time of species divergence from D. melanogaster (in millions of years) is indicated next to the species names. All gene regions are >95% identical in all species. Sequence comparisons were done using BLAST 2 sequences (Tatusova and Madden 1999) and Pustell DNA matrix (MacVector v 7.2.3, Accelrys).
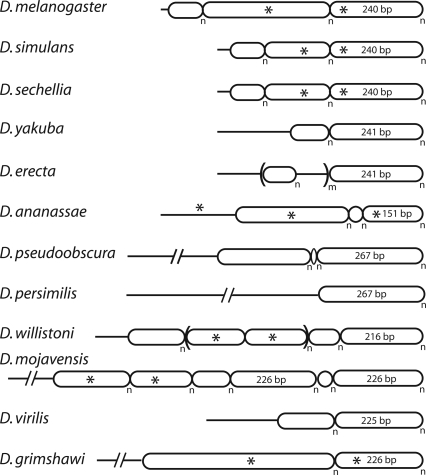
Simplified diagrams of the tandem subrepeat organization of the IGS regions in each of the 12 species are shown in Figure 5. These assembled IGS regions are not consensus sequences, because individual rDNA units within the same species differ in the numbers of each subrepeat (Coen et al. 1982; Polanco et al. 1998, 2000; Averbeck and Eickbush 2005). The only region of the IGS not organized into tandem repeats is the region immediately downstream of the 28S gene. An example of the primary sequence of each subrepeat and of the nonrepeated regions can be found in Supplemental Table 1.
Figure 5.
Organization of the intergenic spacer (IGS) region of the 12 Drosophila species. All subrepeats are represented as rounded boxes. The nonrepeated regions adjacent to the upstream 28S gene are shown as horizontal lines. Asterisks indicate sequence identity between adjacent regions of the IGS. The n subscript indicates that each subrepeat is duplicated a variable number of times in the different rDNA units of each species, while the m subscript indicates a higher ordered repeat. An example of the nonrepeated and subrepeat sequences of each species can be found in Supplemental Table 2.
A previous study of the IGS from four Drosophila species found the subrepeats and unique regions to change rapidly in length and sequence between species (Tautz et al. 1987). Our data confirm and extend these observations. The IGS regions are composed of from one to six subrepeats, with most species containing two or three subrepeats. Subrepeat lengths varied from only 6 nucleotides (Drosophila pseudoobscura) to almost 500 nucleotides (Drosophila grimshawi). All subrepeats were AT rich (median 71% AT), with neighboring subrepeats often sharing sequence motifs (asterisks in Fig. 5). While not diagrammed in Figure 5 for simplicity, the boundaries between different subrepeats often contained partial repeats, chimeras of adjacent repeats, or short nonrepeated sequences. These boundary sequences suggest that occasional recombination between subrepeats gives rise to the rapid changes in repeat lengths seen between species.
The only IGS feature commonly found between species was that the subrepeats closest to the ETS were in most cases 225–267 bp in length. The 240-bp repeat in D. melanogaster contains a partial copy of the promoter sequences associated with transcription of the rDNA unit (Kohorn and Rae 1983; Miller et al. 1983). However, sequence conservation of this repeat was not higher than most other noncoding sequences of the rDNA unit (Fig. 4).
Sequence variation within the rDNA units of each species
Because sequence variation within the rDNA locus is extremely low, sequencing errors in the trace archives were a significant problem. We estimated the sequencing errors in the D. melanogaster trace archives as 0.13% by scoring differences within the individual traces obtained for various single-copy genes in the genome (28 errors in 20,900 bp). The error rate in the final assembled sequences for the genome was much lower than this rate because the 12-fold coverage of the D. melanogaster genome allows a consensus sequence to be obtained from the individual reads (Adams et al. 2000). When dealing with large multigene families, separating authentic sequence variation from sequencing errors is complicated by recurring errors (i.e., the same error detected in multiple traces of the same sequence). Even a slight tendency for a particular sequence to be misidentified can result in multiple traces with the same sequencing error, because the hundreds of rRNA genes present in each species give rise to thousands of reads for each region of the rDNA unit. The strongest evidence for a recurring error is obtained when a nucleotide variant is greatly over-represented by traces derived from one sequencing direction (i.e., traces from the opposite strand sequence do not contain the variant). Therefore, the greater the total number of traces with a putative variant, the greater the reliability in differentiating a recurring sequencing error from an authentic variant.
Our ability to identify rDNA variants differed between the 12 Drosophila species for a number of reasons. First, the level and type of error may differ because different vectors were used and the sequencing was conducted at six centers. Second, the fold coverage of the shotgun sequencing varied from threefold (simulans, persimilis, sechellia) to 12-fold (melanogaster, pseudoobscura). Third, the number and location of the rDNA loci is not known for most species (Lohe and Roberts 2000; Roy et al. 2005). The presence of rDNA units on the sex chromosomes, particularly the Y, significantly reduces the number of trace reads from those units. Finally, the number of rRNA genes in the sequenced strain from each species is also not known. While estimates of 200–250 units per haploid content for most species have been made, this number could vary at least twofold (Long and Dawid 1980).
The following approach allowed us to test for the level at which variants in the rDNA locus could be reliably scored in the different species (see also Methods). The 7.8–8.2 kb consensus transcriptional rDNA unit was divided into 525-bp regions for screening the trace archives. Consecutive BLAST searches were spaced at 500-bp intervals to allow 25 bp of overlap between searches. Approximately 250 sequence traces were selected at random from the BLAST results for each screen. For a selected trace to be used, it had to contain the entire query sequence and no more than two undetermined positions (N) in the query region. This approach equally sampled all rDNA sequences, because no preference was given to those traces with greater sequence identity to the query. The initial ∼250 sequences were aligned and putative variants identified. Variants present in multiple traces were confirmed by BLAST searches using shorter (100 bp) query sequences incorporating each putative variant (see Methods).
Our approach was based on the assumption that the trace reads of each sequencing project representatively sampled the rDNA units. While biases are sometimes encountered in the cloning of DNA fragments, the following arguments suggest that such biases were not present. First, in all species but D. pseudoobscura, the number of trace reads from the rDNA locus was generally consistent with the fold-sequencing coverage and an estimated 100–400 rDNA units. In some species, a reduction in the number of traces corresponding to the 3′ ends of the transcription unit was detected and was assumed to be due to the greater instability of clones containing tandemly repeated IGS sequences. Second, in D. melanogaster we attempted to determine whether specific rDNA units were over- or under-represented in the trace archive by monitoring the number of reads corresponding to specific R2 5′ junctions on the X chromosome. R2 insertions with identifiable 5′ junctions have previously been shown to be predominately single copy (Perèz-Gonzalèz and Eickbush 2001; Averbeck and Eickbush 2005). The median number of traces, 8.5 (range 5–16), corresponded to the ninefold coverage expected of the X chromosome (Adams et al. 2000).
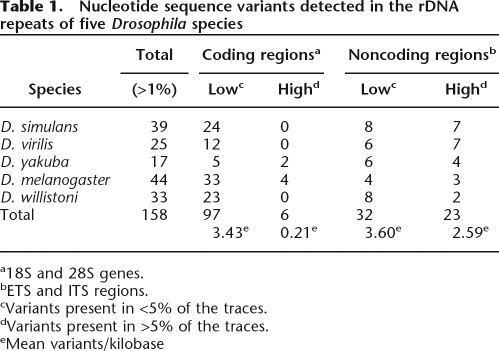
Initial analyses were conducted with the sequence reads obtained from melanogaster, simulans, virilis, willistoni, and yakuba. These analyses indicated that variants identified in only two traces from the original collection of 250 were often difficult to confirm, because insufficient numbers of total traces are present to allow resolution between sequencing errors and the stochastic recovery of traces from one orientation. Greater reliability was obtained by limiting our analyses to variant sites present in at least three trace reads in the original collection of 250 traces. Using this approach, from 17 (Drosophila yakuba) to 44 (D. melanogaster) variant nucleotide positions were detected in the rDNA unit of the five species (Table 1).
Table 1.
Nucleotide sequence variants detected in the rDNA repeats of five Drosophila species
a18S and 28S genes.
bETS and ITS regions.
cVariants present in <5% of the traces.
dVariants present in >5% of the traces.
eMean variants/kilobase
Models for the concerted evolution of the rDNA locus suggest that selective pressure to maintain a specific structure for the rRNA should eliminate many substitutions in the coding regions of the locus (Ohta 1980). Thus, one would expect to see lower levels of standing nucleotide variation in the coding regions compared with the noncoding regions of the rDNA unit. The distribution of the variants detected in the coding and noncoding regions of the transcription units of each species is shown in Table 1. To correct for the length of each region, the total number of variants within the coding and noncoding regions of all five species were divided by their lengths to give the number of variant positions per kilobase. The variants have been divided into two frequency classes: those variants that are present in <5% of the units (low frequency) and those variants present in >5% of the units (high frequency). This separation yielded a striking difference in the distribution of variants. The coding and noncoding regions had similar levels of variants present in the low-frequency class (3.4 and 3.6 variants/kilobase, respectively), but for the high-frequency variants, the noncoding regions had 12 times the number of variants present in the coding region (2.59 and 0.21 variants/kilobase, respectively). This distribution suggests that selection can prevent many variants in the coding region to expand beyond 5% of the rDNA units in the genome.
A larger number of low-frequency variants in each species could be scored by initially sampling more than 250 traces and using programs to monitor sequence reliability. However, this report concerns only those variants present in the rDNA units at frequencies in which the effects of selection could be measured. Thus, for the remaining Drosophila species we scored only those variants present eight or more times in the initial set of ∼250 trace sequences. By this approach, we sampled a large fraction of the moderate frequency variants in each species (those variants between 3% and 5%) and virtually all high-frequency variants. This approach was applied successfully to six additional species; however, as mentioned previously, too few rDNA traces were present in the D. pseudoobscura archive to reliably score variants.
Distribution of sequence variants within the rDNA units
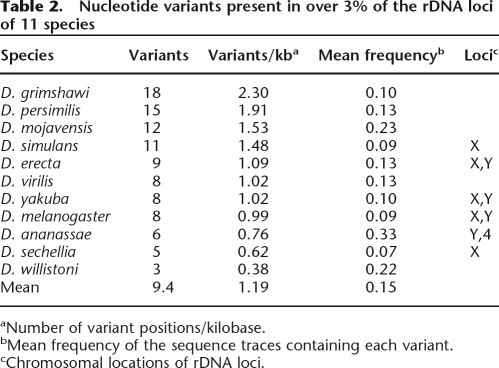
The number of variants identified by the above approach in the 11 Drosophila species is summarized in Table 2, while a description of each variant can be found in Supplemental Table 2. On average, 10 variants were found in each species (range from three in Drosophila willistoni to 18 in D. grimshawi). To correct for the different lengths of the noncoding regions of the rDNA units in each species, the number of variants per kilobase are also shown in Table 2. The average for all species was about one variant per kilobase (range from 0.38 variants/kilobase to 2.3 variants/kilobase). The species also differed in the abundance of each variant within the rDNA units. The mean frequency of variants for most species was from 0.07 to 0.13. However, in three species, Drosophila ananassae, Drosophila mojavensis, and D. willistoni, the mean variant frequency ranged from 0.22 to 0.33. There was no relationship between the number of variants in a species and their average frequency.
Table 2.
Nucleotide variants present in over 3% of the rDNA loci of 11 species
aNumber of variant positions/kilobase.
bMean frequency of the sequence traces containing each variant.
cChromosomal locations of rDNA loci.
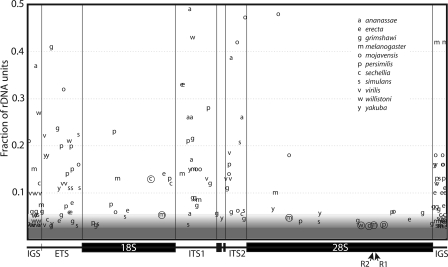
A summary diagram of the locations and frequencies of the 103 variants detected in the 11 species is presented in Figure 6. Variants in the 300 bp flanking the 5′ and 3′ ends of the transcription unit are also shown in Figure 6. However, because these flanking areas are part of tandem subrepeats, it is not possible to distinguish between mutations arising in the subrepeats and the scrambling of extant variation within the subrepeats. Thus, our subsequent discussions will not include these IGS region variants.
Figure 6.
Sequence variants within the rDNA units of 11 Drosophila species. The X-axis shows the location of each variant within the unit. The Y-axis shows the fraction of the trace reads that contain the variant. The shading at the bottom of the figure indicates that variants present in <3% of the trace were not recovered by our approach. Above this 3% level, the probability of the recovery of specific variants increased with their frequency in the locus (see Methods). In all gene regions, the 3′ end of ITS1 and 5′ end of ITS2 could be aligned between species (see Fig. 4). Variant locations in the remaining regions of the ITS, the ETS, and the 300 bp of IGS flanking the ETS and 28S were based on their relative position within the region. Circled variants represent indels present in the coding regions. The location of the R1 and R2 insertion sites within the 28S gene are indicated with arrows.
Figure 6 suggests that a larger number of variants are present in the noncoding regions of the rDNA unit. The nature of the variants detected in the coding and noncoding regions also differed. Nearly 40% of the variants detected in the noncoding regions were indels typically 1–10 bp in length. Only 20% of the variants in the coding regions of the unit were indels, and their distribution was not random. Of the seven indels within the genes (circled letters in Fig. 6), one was at the R1 insertion site within the 28S gene, one was located 4 bp upstream of the R2 insertion site (at 33 bp, this deletion represented the largest indel detected), and two other indels were in the general area of the R1 and R2 sites. This clustering of indels within the 28S gene near the R1 and R2 insertion sites suggests that they were generated by the repair of DNA cleavages produced by the site-specific endonucleases encoded by these elements (see Discussion).
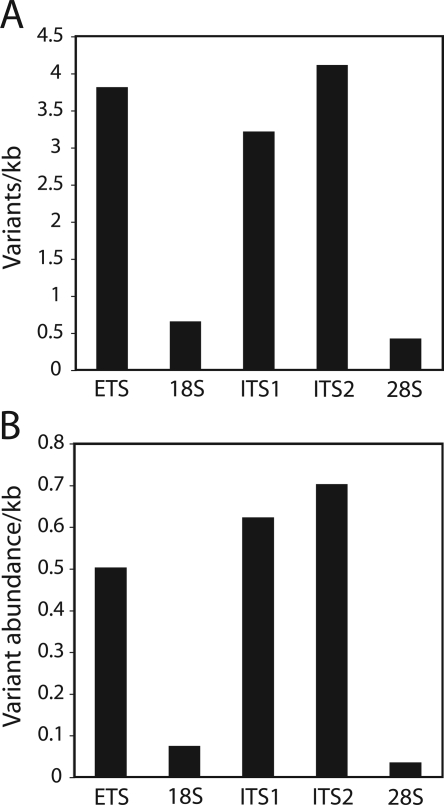
The mean number of variants/kilobase for each coding and noncoding region of the rDNA among the 11 species was plotted in Figure 7A. By evaluating the pooled data from all 11 species, we were unable to detect differences in the level of variation among the noncoding regions, ETS, ITS1, and ITS2 (P = 0.80, χ2 = 0.44, d.f. = 2). However, evaluation of the 18S and ETS (P < 0.0001, χ2 = 37.3, d.f. = 1), 18S and ITS1 (P < 0.0001, χ2 = 27.6, d.f. = 1), 28S and ETS (P = 0.0001, χ2 = 81.5, d.f. = 1), and 28S and ITS1 (P < 0.0001, χ2 = 61.2, d.f. = 1) strongly suggested that these regions do not harbor the same level of variants. In addition to being greater in number, the variants within the noncoding regions of the unit were also present at higher frequencies (Kolmogorov-Smirnov test, P = 0, D = 0.4602). Variants in the noncoding region averaged 16% of the total units, while those in the coding region averaged 8.5%. In Figure 7B, each variant was multiplied by its frequency to provide a measure of the total “abundance” of the variants in the different regions of the rDNA unit. The total abundance of variants in the noncoding regions was about 10 times that of the 18S genes and 20 times that of the 28S genes, consistent with models in which selective pressure prevents most variants in the coding regions of the rDNA from expanding to high frequency.
Figure 7.
Relative number and frequency of variants in each region of the rDNA unit. (A) Mean number of variants/kilobase found among the 11 species. (B) Mean “variant abundance” found among the 11 species. Variant abundance was calculated by multiplying each variant times its frequency in the genome (Fig. 6).
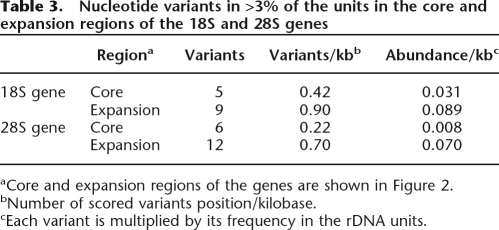
The association between the abundance of variants in a region of the rDNA and the level of selective pressure could also be detected within the coding regions of the genes. Comparison of the rDNA sequences from each species revealed that the expansion regions of the 18S and 28S genes diverged 15 times faster than the core regions (see Fig. 3). Consistent with their faster rate of evolution, significantly more variants were present in the expansion regions compared with the core regions (P = 0.003, χ2 = 8.57, d.f. = 1). As shown in Table 3, variants in the expansion regions of the 18S and 28S genes were two- and threefold higher than the core regions, respectively. If one factors in the frequency of each variant, then variants in the expansion regions are nearly three times more abundant than the core regions in the 18S genes and nearly nine times more abundant in the 28S genes. As will be discussed below, this association between the abundance of variants within a species and the rate of divergence between species was violated in only one comparison. The 28S rRNA gene, which diverges 2.5 times more rapidly than the 18S gene (Fig. 2) has lower than the expected level of variants (P < 0.0001, χ2 = 17.9, d.f. = 1).
Table 3.
Nucleotide variants in >3% of the units in the core and expansion regions of the 18S and 28S genes
aCore and expansion regions of the genes are shown in Figure 2.
bNumber of scored variants position/kilobase.
cEach variant is multiplied by its frequency in the rDNA units.
Discussion
Patterns of nucleotide change between species
The assembled consensus sequences of the rDNA units of 12 species revealed that the expansion regions of the 18S and 28S genes diverged 15-fold faster than the core regions, and the 28S gene diverged 2.5-fold faster than the 18S gene. The former is consistent with the higher rate of substitutions for the expansion regions in primates (Gonzalez et al. 1985), Xenopus (Ajuh et al. 1991), and diverse plants (Kuzoff et al. 1998). In the last study, the 26S gene was also found to diverge two times faster than the 18S gene. Thus, the relative rates of sequence change for the rRNA genes are similar across many eukaryotic taxa. The expansion regions of the Drosophila genes are composed of A-T-rich simple sequences that could presumably undergo rapid segmental changes (Hancock and Dover 1988); however, only expansion regions D7A and D12 of the 28S genes varied significantly.
In contrast to the slowly evolving rRNA genes, the noncoding regions of the Drosophila rDNA transcription unit changed rapidly in sequence. As previously noted (Schlötterer et al. 1994) only the 3′ end of the ITS1 and 5′ end of the ITS2 maintained significant levels of sequence identity across the 40–60 million years of divergence of the 12 species (Fig. 4). The IGS region of the rDNA unit also showed little conservation in sequence or length of the subrepeats. The only common feature was that the subrepeats immediately upstream of the ETS were typically around 240 bp. While these subrepeats have been shown to contain a partial copy of the promoter for the rDNA unit (Kohorn and Rae 1983; Tautz et al. 1987), sequence conservation of this subrepeat is difficult to detect outside of the melanogaster species subgroup. Schlötterer et al. (1994) estimated that the nonconserved regions of the ITS diverged at a level of 1.2% per million years, a rate similar to the neutral rate in Drosophila. From our results, it would appear that most noncoding regions of the rDNA unit are evolving near that rate. Meanwhile, the rates of change in the expansion regions of the 18S and 28S gene (calculated from Fig. 3) are 10–20 times slower, and the core regions 150–300 times slower than the noncoding regions.
A single population of rDNA units in each species
A high level of sequence identity among the many copies of the rDNA units present in all eukaryotes has long been noted (for review, see Eickbush and Eickbush 2007). However, few attempts have been made to quantitate this level of uniformity. In this report we used the large number of sequence reads generated by the whole-genome shotgun sequencing projects to identify the variants that exist in the rDNA units of 11 Drosophila species. Our approach sampled the variants present at frequencies from 3% to 5% of the rDNA units and recovered most variants above that level. Only 3–18 variants were detected in each species, and most of these variants were present in <15% of the units. Thus, while differences exist, the data suggest that in all species there is a single pool of rDNA units that are jointly undergoing the process of concerted evolution. No indications were found that the units are separating into diverging groups or subtypes as seen in the rDNA units of some other eukaryotes; e.g., planaria (Carranza et al. 1999), aphids (Fenton et al. 1998), and certain insects (Keller et al. 2006).
The uniformity of all rDNA units was somewhat surprising given that a significant fraction of the rDNA units in all Drosophila species are disrupted by R1 and R2 insertions (Lathe et al. 1995; Lathe and Eickbush 1997). The fraction of the rDNA units inserted with these elements is typically from 25% to 50%, but insertion levels over 75% have been observed (Hollocher and Templeton 1994; Malik and Eickbush 1999). We searched the trace archives using the junctions of the elements as queries and found the levels of insertions were consistent with previous estimates, suggesting that inserted rDNA units were not under-represented in the trace archives (data not shown). Thus, for most of the species in this study, from one-fourth to one-half of the traces surveyed were derived from units containing an R1 or R2 insertion.
The low levels of variants detected in all species studied here suggest that the abundant R1 and R2 elements present within the rDNA loci are not significantly disrupting the concerted evolution of the individual units. This finding is consistent with previous suggestions that R1 and R2 inserted units are rapidly lost from the rDNA locus, and the elements maintain their presence only by active retrotransposition (Jakubczak et al. 1992; Perèz-Gonzalèz and Eickbush 2001). To more directly test the impact of R1 and R2 insertion on the concerted evolution of the rDNA loci, it will be necessary to determine the frequency at which the variants detected in this report are associated with R1 and R2 inserted units.
Another factor that might affect the degree of concerted evolution of the rDNA units is the distribution of the rDNA units within the genome. Studies in several organisms have suggested that there is greater sequence homogeneity among units from the same locus compared with the units from loci on different chromosomes (Schlötterer and Tautz 1994; Polanco et al. 1998, 2000; Gonzalez and Sylvester 2001). Of the five species analyzed from the melanogaster species subgroup: D. melanogaster, D. yakuba, and Drosophila erecta have rDNA loci on both their X and Y chromosomes, while Drosophila simulans and Drosophila sechelia have a single rDNA locus on their X chromosome (Lohe and Roberts 2000; Roy et al. 2005). As shown in Table 2, a consistent difference in the number or frequency of variants was not detected in these species. Interestingly, the three variants detected in the ITS1 region of the D. melanogaster unit were originally identified by Schlötterer and Tautz (1994) and used to suggest a more rapid homogenization of units within one chromosome. Our data suggests that these and any additional variants differentially present in the X and Y loci of some species represent minor differences amid an overall high level of sequence uniformity.
However, two species did suggest that variants might be differentially fixed in different rDNA loci. As shown in Figure 6D, D. ananassae had a series of variants present in the ITS regions with frequencies near 30%, and D. mojavensis had variants present in the ETS, ITS, and 28S gene with frequencies near 40% of the units. These two species account for nine of the 14 total variants detected with frequencies >25%. Because the rDNA units in D. ananassae are on the Y and 4th chromosomes (Roy et al. 2005), we suggest that these variants represent sequence differences between the units on these chromosomes. Our data would predict that the rDNA units in D. mojanensis are also likely to be located on nonhomologous chromosomes, a situation proposed for several species of the repleta group (Hennig et al. 1982).
Distribution of variants across the rDNA unit and the mechanism of concerted evolution
The results in this report provide a first estimate of the abundance that most mutations in the gene regions of the rDNA unit can attain before being obviously influenced by selection. In the initial study of five species, the coding and noncoding regions of the unit were found to have similar numbers of variants with frequencies between 1% and 5%, but the noncoding regions had 12 times more variants at frequencies >5% (Table 1). Using the larger data set of variants collected from 11 species, variants with frequencies from only 3% to 6% were found in the noncoding regions at three times the level of variants for the coding regions. These findings reveal that many coding-region variants present in only a few percent of the total number of rDNA units are being selected against.
A number of different studies have suggested that only a small fraction of the 200–250 rRNA units present in most D. melanogaster strains are utilized. Deletion studies of the rDNA loci suggested that only 35–60 units are needed for normal viability in the laboratory (Ritossa 1968). Direct microscopic observations suggested that only 35 units were actively transcribed during early development, when the need for rRNA synthesis is high (McKnight and Miller 1976). Finally, various assays of transcriptionally active chromatin structure suggested that <10% of the rDNA units were being transcribed (Ye and Eickbush 2006). Two nonexclusive models could explain how transcription of only a small fraction of the rRNA genes could still lead to selection against most coding region variants if they expand by recombination to more than a few percent of the total number of units. The first model postulates that while only a small fraction of the units are transcribed at any one time, this activity is distributed over most regions of the locus in different cells or at different developmental periods. The second model postulates that new variants, even at low frequencies, are distributed throughout the expressed and nonexpressed regions of the rDNA locus. Resolution between these two models can be obtained by a better understanding of both the distribution of variants across the locus and the cellular processes that determine the regions of the loci to be transcribed.
Can the data in this report help to reveal the relative contributions of unequal crossovers and gene conversion to the concerted evolution of the rDNA locus? Unequal crossovers in the rDNA loci of Drosophila appear to account for the two- to fourfold differences in number of rDNA units associated with individuals of a population (Lyckegaard and Clark 1991; Zhang and Eickbush 2005). These crossovers occur frequently because replicate lines of a single rDNA locus can generate within a few hundred generations a distribution of unit numbers similar to that detected between individuals in a population (Averbeck and Eickbush 2005). The level of variants detected in this report across the different regions of the rDNA unit generally follows their rates of divergence between species (i.e., noncoding > coding [expansion regions] > coding [core regions]). Our data is therefore consistent with any model in which recombination is uniformly distributed over the rDNA unit, and the level of variants is determined by how many of the potential variants are eliminated by selection. Unequal crossovers, duplicating, or eliminating entire rDNA units from the chromosomes in a population can therefore readily explain this distribution of variants. If gene conversions are evenly distributed through the transcribed unit, they could also explain the pattern of variants observed. However, if gene conversions were localized to the conserved sequences of the transcription unit rather than the variable IGS regions, then the 5′ end of the ETS and the 3′ end of the 28S should accumulate more variants. While no such accumulation of variants at the edges of the transcription unit was found (Fig. 6), the role of gene conversion in the concerted evolution of Drosophila rDNA units remains frustratingly difficult to prove or disprove.
The only unexpected feature of the distribution of variants in the rDNA loci of Drosophila was the different levels in the 18S and 28S genes. Because the rate of divergence of the 28S gene is 2.5-fold faster than that of the 18S gene (Fig. 3), the 28S gene appears to be under less sequence constraint, and thus selection against new substitutions should be less than for the 18S gene. However, fewer variants per kilobase were found in the 28S gene than in the 18S gene. This lower level of variants in the 28S gene was observed in both the core and expansion regions of the genes (Table 3). As just discussed, fewer variants within the 28S genes could be the result of localized gene conversions. However, the presence of R1 and R2 insertions in nearly one-half of the 28S genes of these species would be expected to inhibit recombination within the 28S gene, and we have found no evidence for gene-conversion-like events at the 5′ or 3′ ends of the R1 and R2 elements (Eickbush and Eickbush 1995; data not shown). An alternative explanation for the lower level of variants in the 28S gene is that it actually results from R1 and R2 activity. Both elements encode endonucleases that cleave their respective target sites (Eickbush and Eickbush 2007). In instances of aborted insertions (cleavages of the target site that do not give rise to insertions), cellular DNA machinery must repair this region using another uninserted unit as template. This DNA repair mechanism, in addition to the unequal crossovers driving the concerted evolution of the entire repeat, could lead to greater homogenization of the region surrounding the insertion sites. Further evidence for this model would be obtained if higher levels of variants are found in the 28S gene compared with the 18S gene in those organisms without transposable elements inserting into their 28S genes.
Finally, our data differ considerably from a similar analysis recently reported for the rDNA loci of five fungal species (Ganley and Kobayashi 2007). That study revealed almost no variants present in more than single units. This virtual absence of variants in both the coding and noncoding areas of the rDNA unit suggests a considerably more efficient process of concerted evolution than that seen here. The more rapid fixation or elimination of nucleotide variants in the rDNA of fungi could be a result of the smaller number of units and the higher fraction that is expressed, or the frequent gene conversions that are detected between any duplicated gene in fungi (Orr-Weaver and Szostak 1985).
Methods
Species and databases
The trace archives at GenBank, containing the original, unassembled sequencing reads generated by whole-genome shotgun (WGS) sequencing were used in this report. The Drosophila species analyzed were ananassae, erecta, grimshawi, melanogaster, mojavensis, persimilis, pseudoobscura, sechellia, simulans (white 501 strain), virilis, willistoni, and yakuba (http://www.ncbi.nlm.nih.gov/blast/mmtrace.shtml). All nucleotide sequences for this report were obtained from these trace archives by Mega BLAST (Zhang et al. 2000).
Assembling consensus rDNA sequences
To generate a consensus rDNA transcription unit for each species, the first 300 bp of the D. melanogaster 18S gene were used as the initial BLAST query. On average, 12 reads were collected and assembled in AssemblyLIGN (MacVector 7.2.3, Accelrys). Sequence extensions in either the 5′ or 3′ direction were obtained from these assembled sequences and used as the new BLAST queries until the repetitive IGS sequences were reached. BLAST parameters were default values except “percent identity” equaled 75, “hits computed” equaled 10,000, and the “low complexity” filter was unselected. Reads were chosen randomly from the first half of the BLAST results provided that they extended at least 200 bp beyond the end of the query in the direction being examined. Due to the high identity found among the rDNA units of each species, there was little ambiguity in the establishment of consensus sequences using these first small assemblies of sequence reads. The sequences are presented in Supplemental Figure 1a–l, and are available at the Web site http://www.rochester.edu/college/bio/thelab.
Because of the subrepeat structure and greater sequence variation, starting at the 3′ end of the 28S gene, at least 50 sequences were selected from each BLAST search and assembled using ClustalX (Thompson et al. 1997). BLAST parameters were as described above, except percent identity equaled 100 to facilitate the extension of the sequence through these repetitive regions. Sequence extensions from these initial assemblies usually revealed variable numbers of subrepeats. Analysis of the repeat structure of the IGS of each species was done using Tandem Repeats Finder (Benson 1999) and Pustell DNA Matrix (MacVector v 7.2.3, Accelrys). New searches with these subrepeat sequences typically revealed extensions that contained the next class of subrepeats. If no extensions were found into the next subrepeats, blocks of subrepeats not corresponding to the original query were used until extensions into the next subrepeat type or the ETS were encountered. The IGS assemblies are presented as Supplemental Table 2. This Table contains the single-copy sequence immediately downstream of the 28S gene of each species and an example of the most abundant subrepeat types present in that species. In each species, a few rDNA units may not have all subrepeat classes or may contain additional low-abundance subrepeat classes not shown here.
Identification of sequence variation within each species
Sequence variants within a species were scored across the rDNA transcriptional unit by sampling reads in the trace archives. Successive Mega BLAST searches (Zhang et al. 2000) using 525-bp queries were conducted, with each search overlapping the previous BLAST query by 25 bp. BLAST parameters were default values except “percent identity” equaled 75, “hits computed” equaled 10,000, and the “low complexity” filter was unselected. Approximately 250 reads, whose length spanned the query, were randomly selected from each search, except that trace reads with multiple undetermined positions (N’s) in the query region were eliminated. Putative variable sites were identified when the same substitution or indel was present in multiple traces of the ∼250 reads. Variant frequencies were calculated from the abundance found in the original ∼250 reads. To help identify the sequence variants, in-house script were designed and written in Java (M. Eickbush and D. Stage) to parse the BLAST results, link the output to ClustalX (v 1.83.1; Thompson et al. 1997) for sequence alignment, then to Jalview for minor alignment adjustments (v 1.8; Clamp et al. 2004), and finally to format the alignment to highlight differences found between sequences.
All putative sequence variants were confirmed by BLAST search using query sequences incorporating each change. Queries were 100-bp long (longer if necessary to encompass multiple linked variants) and matches had to be 100% identical in sequence. Cases where the initially identified sequence change occurred predominantly on sequencing reads of one orientation were assumed to be recurrent sequencing error and were excluded from the analysis. To be kept in the data set, there had to be at least two identical reads of 100 bp in each orientation with each orientation constituting at least 20% of the reads. All variants detected in at least three traces of the original set of 250 were tested in five species (melanogaster, simulans, virilis, willistoni, and yakuba). Based on the findings from these species, only variants present in a least eight traces in the initial set of 250 were tested in ananassae, erecta, grimshawi, mojavensis, persimilis, and sechellia. Variants in the rDNA units of D. pseudoobscura were not determined because rDNA traces were greatly under-represented in the database.
Statistical tests for the analysis of the distribution and frequency of variants were conducted using χ2 (http://www.graphpad.com/quickcalcs/chisquared1.cfm) and Kolmogorov-Smirnov (K-S) (http://www.physics.csbsju.edu/stats/KS-test.html).
Acknowledgments
Support for this work was provided by the National Science Foundation (MCB-0544071) (T.H.E.) and a Caspari Fellowship from the University of Rochester (D.E.S.). We thank Michael Eickbush for his programming prowess, and the following sequencing centers for making the trace sequences publicly available prior to publication: Genome Sequencing Center, Washington University (D. simulans, D. yakuba); Broad Institute (D. sechellia, D. persimilis); Agencourt Bioscience Corporation (D. erecta, D. ananassae, D. mojavensis, D. virilis, D. grimshawi); J. Craig Venter Institute (D. willistoni). Finally, we thank Xian Zhang, Danna Eickbush, and Bill Burke for their suggestions and comments on the manuscript.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6376807
References
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., Li P.W., Hoskins R.A., Galle R.F., Hoskins R.A., Galle R.F., Galle R.F., et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Ajuh P.M., Heeney P.A., Madden B.E.H., Heeney P.A., Madden B.E.H., Madden B.E.H. Xenopus borealis and Xenopus laevis 28S ribosomal DNA and the complete 40S ribosomal precursor RNA coding units of both species. Proc. R. Soc. Lond. B. Biol. Sci. 1991;245:65–71. doi: 10.1098/rspb.1991.0089. [DOI] [PubMed] [Google Scholar]
- Averbeck K.T., Eickbush T.H., Eickbush T.H. Monitoring the tempo and mode of concerted evolution in the Drosophila melanogaster rDNA locus. Genetics. 2005;171:1837–1846. doi: 10.1534/genetics.105.047670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranza S., Baguna J., Riutort M., Baguna J., Riutort M., Riutort M. Origin and evolution of paralogous rRNA gene clusters within the flatworm family Dugesiidae (Platyhelminthes, Tricladdid) J. Mol. Evol. 1999;49:250–259. doi: 10.1007/pl00006547. [DOI] [PubMed] [Google Scholar]
- Clamp M., Cuff J., Searle S.M., Barton G.J., Cuff J., Searle S.M., Barton G.J., Searle S.M., Barton G.J., Barton G.J. The Jalview Java alignment editor. Bioinformatics. 2004;12:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Clark C.G., Tague B.W., Ware V.C., Gerbi S.A., Tague B.W., Ware V.C., Gerbi S.A., Ware V.C., Gerbi S.A., Gerbi S.A. Xenopus laevis 28S ribosomal RNA: A secondary structure model and its evolutionary and functional implications. Nucleic Acids Res. 1984;12:6197–6220. doi: 10.1093/nar/12.15.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E.S., Thoday J.M., Dover G., Thoday J.M., Dover G., Dover G. Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature. 1982;295:564–568. doi: 10.1038/295564a0. [DOI] [PubMed] [Google Scholar]
- Dover G. Concerted evolution, molecular drive and natural selection. Curr. Biol. 1994;4:1165–1166. doi: 10.1016/s0960-9822(00)00265-7. [DOI] [PubMed] [Google Scholar]
- Eickbush D.G., Eickbush T.H., Eickbush T.H. Vertical transmission of the retrotransposable elements R1 and R2 during the evolution of the Drosophila melanogaster species subgroup. Genetics. 1995;139:671–684. doi: 10.1093/genetics/139.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush T.H., Eickbush D.G., Eickbush D.G. Finely orchestrated movements: Evolution of the ribosomal RNA genes. Genetics. 2007;175:477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elder J.F., Turner B.J., Turner B.J. Concerted evolution of repetitive DNA sequences in eukaryotes. Q. Rev. Biol. 1995;70:297–320. doi: 10.1086/419073. [DOI] [PubMed] [Google Scholar]
- Fenton B., Malloch G., Germa F., Malloch G., Germa F., Germa F. A study of variation in rDNA ITS regions shows that two haplotypes coexist within a single aphid genome. Genome. 1998;41:337–345. [PubMed] [Google Scholar]
- Ganley A.R.D., Kobayashi T., Kobayashi T. Highly efficient concerted evolution in the ribosomal DNA repeats: Total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 2007;17:184–191. doi: 10.1101/gr.5457707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I.L., Sylvester J.E., Sylvester J.E. Human rDNA: Evolutionary patterns within the genes and tandem arrays derived from multiple chromosomes. Genomics. 2001;73:255–263. doi: 10.1006/geno.2001.6540. [DOI] [PubMed] [Google Scholar]
- Gonzalez I.L., Gorski L.L., Campden T.J., Dorney D.J., Erickson J.M., Sylvester J.E., Schmickel R.D., Gorski L.L., Campden T.J., Dorney D.J., Erickson J.M., Sylvester J.E., Schmickel R.D., Campden T.J., Dorney D.J., Erickson J.M., Sylvester J.E., Schmickel R.D., Dorney D.J., Erickson J.M., Sylvester J.E., Schmickel R.D., Erickson J.M., Sylvester J.E., Schmickel R.D., Sylvester J.E., Schmickel R.D., Schmickel R.D. Variation among human 28S ribosomal RNA genes. Proc. Natl. Acad. Sci. 1985;82:7666–7670. doi: 10.1073/pnas.82.22.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J.M., Dover G.A., Dover G.A. Molecular coevolution among cryptically simple expansion segments of eukaryotic 26S/28S rRNAs. Mol. Biol. Evol. 1988;5:377–391. doi: 10.1093/oxfordjournals.molbev.a040505. [DOI] [PubMed] [Google Scholar]
- Hancock J.M., Tautz D., Dover G.A., Tautz D., Dover G.A., Dover G.A. Evolution of the secondary structures and compensatory mutations of the ribosomal RNAs of Drosophila melanogaster. Mol. Biol. Evol. 1988;5:393–414. doi: 10.1093/oxfordjournals.molbev.a040501. [DOI] [PubMed] [Google Scholar]
- Hassouna N., Michot B., Bachellerie J.P., Michot B., Bachellerie J.P., Bachellerie J.P. The complete nucleotide sequence of mouse 28S rRNA gene: Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984;12:3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig W., Vogt P., Jacob G., Siegmund I., Vogt P., Jacob G., Siegmund I., Jacob G., Siegmund I., Siegmund I. Nucleolus organizer regions in Drosophila species of the replete group. Chromosoma. 1982;87:279–292. [Google Scholar]
- Hillis D.M., Dixon M.T., Dixon M.T. Ribosomal DNA: Molecular evolution and phylogenetic inference. Q. Rev. Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- Hollocher H., Templeton A.R., Templeton A.R. The molecular through ecological genetics of abnormal abdomen in Drosophila mercatorim. VI. The non-neutrality of the Y chromosome rDNA polymorphism. Genetics. 1994;136:1373–1384. doi: 10.1093/genetics/136.4.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczak J.L., Burke W.D., Eickbush T.H., Burke W.D., Eickbush T.H., Eickbush T.H. Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc. Natl. Acad. Sci. 1991;88:3295–3299. doi: 10.1073/pnas.88.8.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczak J.L., Zenni M.K., Woodruff R.C., Eickbush T.H., Zenni M.K., Woodruff R.C., Eickbush T.H., Woodruff R.C., Eickbush T.H., Eickbush T.H. Turnover R1 (type I) and R2 (type II) retrotransposable elements in the ribosomal DNA of Drosophila melanogaster. Genetics. 1992;131:129–142. doi: 10.1093/genetics/131.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B.R., Jourdan R., Jacq B., Jourdan R., Jacq B., Jacq B. Late steps in the maturation of Drosophila 26 S ribosomal RNA: Generation of 5.8 S and 2 S RNAs by cleavages occurring in the cytoplasm. J. Mol. Biol. 1976;101:85–105. doi: 10.1016/0022-2836(76)90067-x. [DOI] [PubMed] [Google Scholar]
- Keller I., Chintauan-Marquier I.C., Veltsos P., Nichols R.A., Chintauan-Marquier I.C., Veltsos P., Nichols R.A., Veltsos P., Nichols R.A., Nichols R.A. Ribosomal DNA in the grasshopper Podisma pedestris: Escape from concerted evolution. Genetics. 2006;174:863–874. doi: 10.1534/genetics.106.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn B.D., Rae P.M., Rae P.M. A component of Drosophila RNA polymerase I promoter lies within the rRNA transcription unit. Nature. 1983;304:179–181. doi: 10.1038/304179a0. [DOI] [PubMed] [Google Scholar]
- Kuzoff R.K., Sweere J.A., Soltis D.E., Soltis P.S., Zimmer E.A., Sweere J.A., Soltis D.E., Soltis P.S., Zimmer E.A., Soltis D.E., Soltis P.S., Zimmer E.A., Soltis P.S., Zimmer E.A., Zimmer E.A. The phylogenetic potential of entire 26S rDNA sequences in plants. Mol. Biol. Evol. 1998;15:251–263. doi: 10.1093/oxfordjournals.molbev.a025922. [DOI] [PubMed] [Google Scholar]
- Lathe W.C., Eickbush T.H., Eickbush T.H. A single lineage of R2 retrotransposable elements is an active, evolutionarily stable component of the Drosophila rDNA locus. Mol. Biol. Evol. 1997;14:1232–1241. doi: 10.1093/oxfordjournals.molbev.a025732. [DOI] [PubMed] [Google Scholar]
- Lathe W.C., Burke W.D., Eickbush D.G., Eickbush T.H., Burke W.D., Eickbush D.G., Eickbush T.H., Eickbush D.G., Eickbush T.H., Eickbush T.H. Evolutionary stability of the R1 retrotransposable element in the genus Drosophila. Mol. Biol. Evol. 1995;12:1094–1105. doi: 10.1093/oxfordjournals.molbev.a040283. [DOI] [PubMed] [Google Scholar]
- Lohe A.R., Roberts P.A., Roberts P.A. Evolution of DNA in heterochromatin: The Drosophila melanogaster sibling species subgroup as a resource. Genetica. 2000;109:125–130. doi: 10.1023/a:1026588217432. [DOI] [PubMed] [Google Scholar]
- Long E.O., Dawid I.B., Dawid I.B. Repeated genes in eukaryotes. Annu. Rev. Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Long E.O., Rebbert M.L., Dawid I.B., Rebbert M.L., Dawid I.B., Dawid I.B. Nucleotide sequence of the initiation site for ribosomal RNA transcription in Drosophila melanogaster: Comparison of genes with and without insertions. Proc. Natl. Acad. Sci. 1981;78:1513–1517. doi: 10.1073/pnas.78.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyckegaard E.M., Clark A.G., Clark A.G. Evolution of ribosomal RNA copy number on the sex chromosomes of Drosophila melanogaster. Mol. Biol. Evol. 1991;8:458–474. doi: 10.1093/oxfordjournals.molbev.a040664. [DOI] [PubMed] [Google Scholar]
- Malik H.S., Eickbush T.H., Eickbush T.H. Retrotransposable elements R1 and R2 in the rDNA units of Drosophila mercatorum: Abnormal abdomen revisited. Genetics. 1999;151:653–665. doi: 10.1093/genetics/151.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik H.S., Burke W.D., Eickbush T.H., Burke W.D., Eickbush T.H., Eickbush T.H. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- McKnight S.L., Miller O.L., Miller O.L. Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976;8:305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- Miller J.R., Hayward D.C., Glover D.M., Hayward D.C., Glover D.M., Glover D.M. Transcription of the ‘non-transcribed’ spacer of Drosophila melanogaster rDNA. Nucleic Acids Res. 1983;11:11–19. doi: 10.1093/nar/11.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtif V.L., Rae P.M., Rae P.M. In vivo transcription of rDNA spacers in Drosophila. Nucleic Acids Res. 1985;13:3221–3239. doi: 10.1093/nar/13.9.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Evolution and variation of multigene families. Springer-Verlag; Berlin, Germany: 1980. [Google Scholar]
- Ohta T. Some models of gene conversion for treating the evolution of multigene families. Genetics. 1984;106:517–528. doi: 10.1093/genetics/106.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Dover G.A., Dover G.A. Population genetics of multigene families that are dispersed into two or more chromosomes. Proc. Natl. Acad. Sci. 1983;80:4079–4083. doi: 10.1073/pnas.80.13.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T.L., Szostak J.W., Szostak J.W. Fungal recombination. Microbiol. Rev. 1985;49:33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-González C.E., Eickbush T.H., Eickbush T.H. Dynamics of R1 and R2 elements in the rDNA locus of Drosophila simulans. Genetics. 2001;158:1557–1567. doi: 10.1093/genetics/158.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanco C., González A.I., de la Fuente Á., Dover G.A., González A.I., de la Fuente Á., Dover G.A., de la Fuente Á., Dover G.A., Dover G.A. Multigene family of ribosomal DNA in D. melanogaster reveals contrasting patterns of homogenization for IGS and ITS spacer regions: A possible mechanism to resolve this paradox. Genetics. 1998;149:243–256. doi: 10.1093/genetics/149.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanco C., González A.I., Dover G.A., González A.I., Dover G.A., Dover G.A. Patterns of variation in the intergenic spacers of ribosomal DNA in Drosophila melanogaster support a model for genetic exchanges during X-Y pairing. Genetics. 2000;155:1221–1229. doi: 10.1093/genetics/155.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa F. Unstable redundancy of genes for ribosomal RNA. Proc. Natl. Acad. Sci. 1968;60:509–516. doi: 10.1073/pnas.60.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy V., Monti-Dedieu L., Chaminade N., Siljak-Yakovlev S., Aulard S., Lemeunier F., Montchamp-Moreau C., Monti-Dedieu L., Chaminade N., Siljak-Yakovlev S., Aulard S., Lemeunier F., Montchamp-Moreau C., Chaminade N., Siljak-Yakovlev S., Aulard S., Lemeunier F., Montchamp-Moreau C., Siljak-Yakovlev S., Aulard S., Lemeunier F., Montchamp-Moreau C., Aulard S., Lemeunier F., Montchamp-Moreau C., Lemeunier F., Montchamp-Moreau C., Montchamp-Moreau C. Evolution of the chromosomal location of rDNA genes in two Drosophila species subgroups: ananassae and melanogaster. Heredity. 2005;94:388–395. doi: 10.1038/sj.hdy.6800612. [DOI] [PubMed] [Google Scholar]
- Schlötterer C., Tautz D., Tautz D. Chromosomal homogeneity of Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Curr. Biol. 1994;4:777–783. doi: 10.1016/s0960-9822(00)00175-5. [DOI] [PubMed] [Google Scholar]
- Schlötterer C., Hauser M.T., von Hauseler A., Tautz D., Hauser M.T., von Hauseler A., Tautz D., von Hauseler A., Tautz D., Tautz D. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol. Biol. Evol. 1994;11:513–522. doi: 10.1093/oxfordjournals.molbev.a040131. [DOI] [PubMed] [Google Scholar]
- Tatusova T.A., Madden T.L., Madden T.L. BLAST 2 sequences—A new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Tautz D., Tautz C., Webb D., Dover G.A., Tautz C., Webb D., Dover G.A., Webb D., Dover G.A., Dover G.A. Evolutionary divergence of promoters and spacers in the rDNA family of four Drosophila species. J. Mol. Biol. 1987;195:525–542. doi: 10.1016/0022-2836(87)90181-1. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G., Plewniak F., Jeanmougin F., Higgins D.G., Jeanmougin F., Higgins D.G., Higgins D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.M., Kennison J.A., Robbins L.G., Strobeck C., Kennison J.A., Robbins L.G., Strobeck C., Robbins L.G., Strobeck C., Strobeck C. Reciprocal recombination and the evolution of the ribosomal gene family of Drosophila melanogaster. Genetics. 1989;122:617–624. doi: 10.1093/genetics/122.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Eickbush T.H., Eickbush T.H. Chromatin structure and transcription of the R1- and R2-inserted rRNA genes of Drosophila melanogaster. Mol. Cell. Biol. 2006;23:8781–8790. doi: 10.1128/MCB.01409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Eickbush T.H., Eickbush T.H. Characterization of active R2 retrotransposition in the rDNA locus of Drosophila simulans. Genetics. 2005;170:195–205. doi: 10.1534/genetics.104.038703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Schwartz S., Wagner L., Miller W., Schwartz S., Wagner L., Miller W., Wagner L., Miller W., Miller W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]