Abstract
Insect genomes contain larger blocks of conserved gene order (microsynteny) than would be expected under a random breakage model of chromosome evolution. We present evidence that microsynteny has been retained to keep large arrays of highly conserved noncoding elements (HCNEs) intact. These arrays span key developmental regulatory genes, forming genomic regulatory blocks (GRBs). We recently described GRBs in vertebrates, where most HCNEs function as enhancers and HCNE arrays specify complex expression programs of their target genes. Here we present a comparison of five Drosophila genomes showing that HCNE density peaks centrally in large synteny blocks containing multiple genes. Besides developmental regulators that are likely targets of HCNE enhancers, HCNE arrays often span unrelated neighboring genes. We describe differences in core promoters between the target genes and the unrelated genes that offer an explanation for the differences in their responsiveness to enhancers. We show examples of a striking correspondence between boundaries of synteny blocks, HCNE arrays, and Polycomb binding regions, confirming that the synteny blocks correspond to regulatory domains. Although few noncoding elements are highly conserved between Drosophila and the malaria mosquito Anopheles gambiae, we find that A. gambiae regions orthologous to Drosophila GRBs contain an equivalent distribution of noncoding elements highly conserved in the yellow fever mosquito Aëdes aegypti and coincide with regions of ancient microsynteny between Drosophila and mosquitoes. The structural and functional equivalence between insect and vertebrate GRBs marks them as an ancient feature of metazoan genomes and as a key to future studies of development and gene regulation.
Long-range cis-regulation in vertebrates has recently been the focus of much attention, driven by the genome-wide discovery of highly conserved noncoding elements (HCNEs) found to span the loci of developmental regulatory genes. After a series of observations of high levels of conservation of individual developmental enhancers, whole-genome comparisons revealed an abundance of HCNEs that tend to cluster along chromosomes. The clusters most often coincide with genes encoding developmental and differentiation-related transcription factors. Many HCNEs have been characterized as long-range enhancers, first in studies of individual genes (Gottgens et al. 2000; Sumiyama and Ruddle 2003; Kimura-Yoshida et al. 2004; Milewski et al. 2004), followed by systematic studies in zebrafish, Xenopus, and mouse (de la Calle-Mustienes et al. 2005; Shin et al. 2005; Woolfe et al. 2005; Pennacchio et al. 2006). Genome-wide analyses of HCNE sequences have detected several overrepresented motifs that are believed to be associated with context-specific enhancer activity (Bailey et al. 2006; Pennacchio et al. 2007).
The emerging model is that an array of HCNEs defines a region of regulatory inputs of its target gene(s), and that the full complement of those inputs results in the actual expression pattern of the gene (Kimura-Yoshida et al. 2004; de la Calle-Mustienes et al. 2005; Woolfe et al. 2005; Pennacchio et al. 2006). It is plausible to speculate that the genes with the most complex spatiotemporal expression should have more complex regulatory inputs. This is in full agreement with the finding that the targets of the most elaborate arrays of HCNEs are genes encoding developmental regulators and genes for proteins that regulate axonal guidance and related processes in the central nervous system (Lindblad-Toh et al. 2005).
Many HCNE arrays span large gene-free regions—so-called “gene deserts”—around their target genes (Sandelin et al. 2004). However, very often the regions spanned by HCNEs contain genes whose biological functions and expression patterns are unrelated to those of the presumptive target genes. These unrelated genes, which we refer to as “bystander genes,” are independent of the regulatory input of HCNE arrays, but the pressure to maintain HCNE arrays have kept bystander and target genes together for hundreds of millions of years (Kikuta et al. 2007). We termed the HCNE-spanned regions containing such genes “genomic regulatory blocks” (GRBs) and found GRBs to correspond to the longest regions of conserved gene order across vertebrate genomes. In this paper, we use the term “microsynteny conservation” to denote the preservation of close proximity among genes through evolution, and we refer to chromosomal regions that have been largely maintained in evolution as “synteny blocks” (Zdobnov et al. 2002; Pevzner and Tesler 2003).
The fruit fly Drosophila melanogaster (Dmel) has been used for a century as a model organism for studies of genetics, animal development, behavior, and many other aspects of biology. It is remarkable that most developmental regulatory genes in the fly have conserved orthologs in vertebrates, often with analogous functions (Carroll 2005), and that many of these genes are associated with HCNEs in both flies and vertebrates (Glazov et al. 2005; Vavouri et al. 2007). Although insect HCNEs have not been studied as extensively as vertebrate HCNEs, the trends described are similar, strongly suggesting that most HCNEs function as developmental regulatory elements in vertebrates and insects alike. In both vertebrate and insect genomes, most bases that are conserved above neutral evolution rates appear to be noncoding (Siepel et al. 2005). More than 20,000 intronic and intergenic elements are perfectly conserved over at least 50 bp between Dmel and the closely related D. pseudoobscura (Dpse), and most abundant in the vicinity of developmental transcription factor genes (Glazov et al. 2005). A recent search for HCNEs conserved between Dmel and the more distantly related D. virilis (Dvir) revealed several elements that coincide with characterized developmental enhancers (Papatsenko et al. 2006).
Regions of conserved microsynteny have been found between Dmel and the malaria mosquito Anopheles gambiae (Agam) although these organisms diverged ∼250 million years ago (Zdobnov et al. 2002). A recent comparison of 12 insect genomes demonstrated microsynteny conservation among more distantly related insects (Zdobnov and Bork 2007). This comparison also showed that the distribution of insect synteny block lengths is incompatible with a model where genes have been randomly shuffled in evolution, and would be better explained by the existence of rearrangement hotspots—regions that have been shuffled more than others in evolution. The same trend has been observed in comparisons of mammalian genomes (Kent et al. 2003; Pevzner and Tesler 2003; Murphy et al. 2005). In vertebrates, conserved microsynteny can at least in part be explained by the occurrence of GRBs (Kikuta et al. 2007).
In this study, we present evidence for the existence of GRBs in insects and their functional equivalence to those in vertebrates. We have identified 6779 HCNEs shared among five different Drosophila species, demonstrating that fly genomes contain an extensive core repertoire of HCNEs. We show that an equivalent organization can be observed in orthologous mosquito loci through comparisons of the genome sequences of Anopheles gambiae and Aëdes aegypti, and that the maintenance of HCNE clusters is likely to underlie preservation of microsynteny between flies and mosquitoes. The regions of HCNE arrays and microsynteny conservation also contain unrelated genes, probably in a similar way to bystander genes in vertebrate GRBs (Kikuta et al. 2007). We provide genome-wide evidence that these genes generally differ from target genes in their type of core promoter, which might for the first time explain on a genome-wide level why bystander genes do not specifically respond to long-range regulation in the region. Finally, we report a striking correspondence between Polycomb binding regions and several Drosophila GRBs, and discuss the occurrence of GRBs as an ancient and fundamental feature of metazoan genomes.
Results
We identified HCNEs in pairwise alignments between the euchromatic genome sequences of D. melanogaster (Dmel) and four other Drosophila species—D. ananassae (Dana), D. pseudoobscura (Dpse), D. virilis (Dvir), and D. mojavensis (Dmoj)—selected based on the state of their genome assemblies, availability of whole-genome sequence alignments to Dmel, and phylogenetic relationships (Supplemental Fig. S1). We required HCNEs to be conserved at 98% identity over at least 50 bp in all four pairwise comparisons. To focus on elements that are most likely to function in regulation of transcription, we discarded elements that partially or entirely overlapped exons (Bejerano et al. 2004; Glazov et al. 2005; Woolfe et al. 2005; Bailey et al. 2006). There were 6779 HCNEs, with a median size of 59 bp and a maximum of 157 bp. Consistent with earlier observations for flies (Glazov et al. 2005), nematodes (Vavouri et al. 2007), and vertebrates (Bejerano et al. 2004; Sandelin et al. 2004), we found regions of high HCNE density to be strongly enriched for genes encoding developmental transcriptional regulators (Supplemental Table S1).
Highly conserved noncoding elements are enriched in large synteny blocks
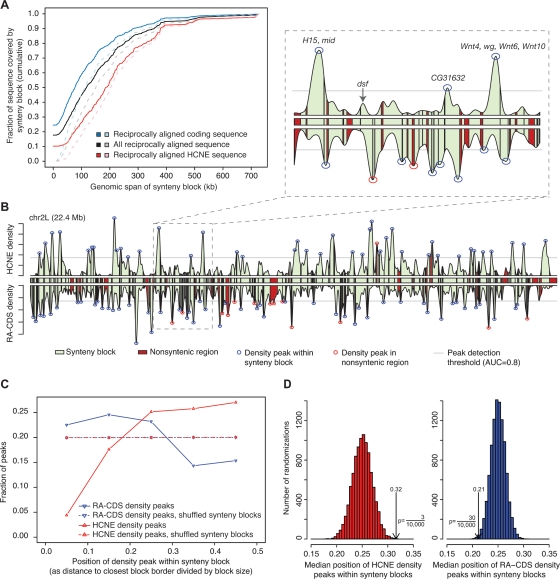
To study the distribution of HCNEs with respect to regions of microsynteny, we identified synteny blocks conserved among all five fly genomes as described in Methods. None of the four species that we compared to Dmel has a finished genome assembly. Nevertheless, our results indicate that reliable synteny blocks can be constructed because most of the sequence is in very large scaffolds. Although the synteny blocks included few scaffolds, they spanned 76% of the Dmel euchromatic sequence (Supplemental Table S2). We distinguish between the span of a synteny block, which we define as the entire genomic region between the extreme borders of the block, and its coverage, meaning the reciprocally aligned, syntenic bases in the block. Of the HCNEs, 94% were entirely spanned by synteny blocks, and 86% had at least 98% of their sequence covered by synteny blocks. We wished to compare the coverage of HCNE sequence by synteny blocks to the coverage of coding sequence (CDS) while controlling for the fact that the latter is less conserved overall. We therefore identified the bases in the Dmel sequence that were aligned in a reciprocal-best manner in all four pairwise genome comparisons (reciprocally best aligned [RA] sequence), and measured the fraction of them that was covered by synteny blocks. Remarkably, 90% of RA-HCNE sequence was covered by synteny blocks, compared to only 75% of RA-CDS. RA-HCNE sequence was enriched in large synteny blocks compared to RA-CDS (Fig. 1A).
Figure 1.
HCNE arrays are centrally positioned in large synteny blocks. (A) RA-HCNE sequence is enriched in large synteny blocks compared to RA-CDS. Dashed lines show the distributions when sequence not covered by any synteny block is excluded. (B) HCNE density, RA-CDS density, and synteny blocks on Dmel chromosome arm 2L. Synteny blocks (green boxes with black borders) are shown between the density curves and in the area under them. Density peaks were detected above a threshold (gray line) set to cover 80% of the area under the density curve for the chromosome arm. In the magnified section, HCNE density peaks are labeled with inferred regulatory target genes located in the same synteny block as the HCNE density peak. (C) Line histogram of position of density peaks within synteny blocks. For each density peak that was located within a synteny block, we computed the distance between the peak and the synteny break closest to it, and scaled the distances to [0, 0.5] by dividing with synteny block size. Dashed lines show distributions from 10,000 randomizations where synteny blocks were ordered independently of density peaks (Supplemental Fig. S3). (D) Histogram of median distance in each of the 10,000 randomizations. Arrows indicate medians for the nonrandomized data, and one-sided P-values indicate the fraction of randomizations having equal or more extreme medians.
HCNE arrays are centrally positioned in large synteny blocks that span multiple genes
We identified 164 peaks of HCNE density on Dmel chromosomes 2, 3, and X by first using a Gaussian kernel to compute local HCNE density at positions spaced 1 kb throughout the euchromatic sequence, and then locating peaks in the resulting density distribution. Many peaks of HCNE density are contained within single synteny blocks and are centrally positioned within those blocks (Fig. 1B,C; Supplemental Fig. S2). Only 5/164 HCNE density peaks were located outside synteny blocks. In contrast, RA-CDS density tends to peak near synteny breaks, confirming that the lower HCNE density in these regions is unlikely to be caused by variations in alignment quality (Fig. 1B,C; Supplemental Fig. S2). We confirmed the statistical significance of these trends by permutation tests (Fig. 1D) and found the trends to persist across a wide range of parameter settings (Supplemental Figs. S3, S4). RA-CDS density peaks were most frequent in small synteny blocks (Supplemental Fig. S3). This trend is consistent with the above findings but can also partially be explained by variability in intron size between genes. The frequency of HCNE density peaks per sequence length increased with synteny block size and nearly all synteny blocks that contained a large HCNE density peak also contained multiple genes (Supplemental Fig. S3). These findings strongly suggest that large regions containing multiple genes have maintained microsynteny in order to preserve arrays of HCNEs.
HCNE-associated genes are in large blocks of conserved microsynteny between fly and mosquito
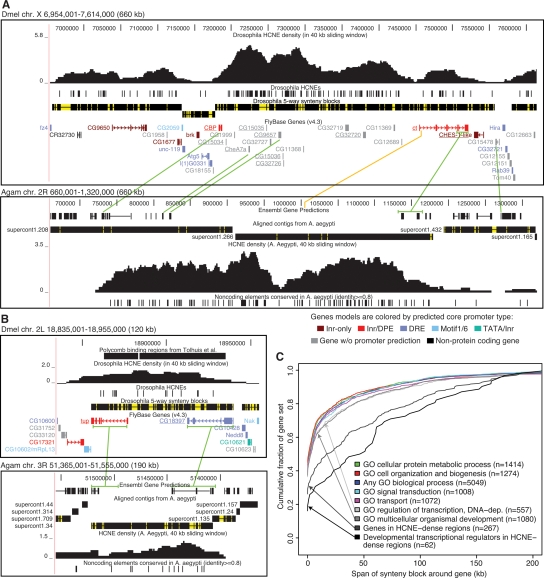
The ct locus (Fig. 2A) is one of the more extreme examples of the genome-wide trends described above. This synteny block contains the highest HCNE density peak on the Dmel X chromosome (Supplemental Fig. S2) and HCNE densities are high throughout most of the block. The block is flanked by regions of higher gene density than within the block. The ct gene encodes the homeodomain protein Cut, which regulates cell-fate decisions in multiple lineages (Nepveu 2001). ct has been maintained in microsynteny with at least nine other genes (underlined in Fig. 2A) throughout the five Drosophila genomes investigated here. There is little evidence of a functional relationship between ct and any of these nine genes: Five are unannotated, and the remaining four encode a sarcoplasmic calcium-binding protein (CBP), a putative protein phosphatase (CG15035), a putative Na+/solute symporter (CG9657), and a putative forkhead transcription factor (CHES-1-like). CHES-1-like may have a regulatory role, but its function is unknown. ct has been maintained in microsynteny with four genes (CBP, CheA7a, CG9657, and CG15478) between Dmel and Agam. Strikingly, these genes delimit roughly the same Dmel region as the five-way fly synteny block: the region spanned by the HCNE-cluster (Fig. 2A).
Figure 2.
Genes associated with HCNE arrays tend to be in large fly-mosquito synteny blocks. (A,B) Examples of synteny blocks. Gaps within synteny blocks are colored yellow. Green lines connect genes in conserved microsynteny between Dmel and Agam. Microsynteny conservation between Dmel and Agam was determined by examining chained BLASTZ and TBLASTN alignments in the UCSC Genome Browser (Kent et al. 2002). Sometimes only parts of genes could be matched (e.g., in the case of ct). Aaeg contigs aligned to the Agam assembly are shown with regions having ≥50% identity over 50 bp in black and other regions in yellow. HCNE densities were computed as the fraction of bases in HCNEs in sliding windows of 40 kb. The UCSC Genome Browser (Kent et al. 2002) was used in making the images. (A) The ct locus in Dmel (upper panel) and Agam (lower panel). ct and nine other genes (underlined) show strong evidence of being in conserved microsynteny among the five flies. The orange line indicates a noncoding BLASTZ match between Dmel and Agam and hints at the location of the first ct exon in Agam. Comparison of HCNE density curves also supports that the first ct exon in Agam is in the area indicated by the orange line. Supporting a common origin of the HCNE clusters at the ct loci in flies and mosquitoes, the HCNE density curves have similar shapes. Two density peaks are visible in both organisms: one between CG9657 and ct, and the other within the borders of ct. The developmental transcriptional regulator brk (Moser and Campbell 2005) is centrally positioned in an adjacent synteny block. CG9650, which dominates a neighboring HCNE-rich synteny block, is expressed in developing CNS and PNS and encodes a putative C2H2 zinc finger protein (McGovern et al. 2003). (B) The tailup (tup) locus in Dmel (upper panel) and Agam (lower panel). tup is in conserved microsynteny with CG18397 among the five flies, Agam and Aaeg. tup encodes a homeodomain transcription factor involved in development (Thor and Thomas 1997). CG18397 is predicted to encode a protein with an AMP-dependent synthetase and ligase domain. In both flies and mosquitoes, HCNEs are found throughout the synteny block. Some HCNEs are within introns of tup and CG18397. This, combined with the lack of evidence for a functional relationship between the two genes, indicates that they have been kept in proximity in order to maintain the HCNE array. (C) For each gene that we could assign to a synteny block, we measured the span of its synteny block excluding the region spanned by the gene itself (in order to control for differences in gene size). Each curve shows the cumulative distribution of synteny block span, measured in Dmel bp, around genes in a particular category. Categories were defined from GO biological process annotation and HCNE density. The category “any biological process” contains all genes annotated with a GO biological process term other than “biological process unknown.” Genes in HCNE-dense regions overlap a 40-kb region where at least 1% (400 bp) of the sequence is in HCNEs. Numbers within parentheses indicate the number of genes annotated to the indicated process and assigned to a single synteny block.
To investigate whether maintained fly-mosquito microsynteny at the ct locus could be explained by a selective pressure to keep the HCNE-cluster intact, we searched for HCNEs conserved between Agam and the yellow fever mosquito Aëdes aegypti (Aeg) at the ct locus in mosquitoes. Indeed, there is a distinct island of mosquito-specific HCNEs confined to the region of the fly-mosquito synteny block (Fig. 2A). The picture is similar at several other loci, including the locus of the homeodomain transcription factor gene tailup (tup, Fig. 2B; see also Supplemental Figs. S5–S7). Curiously, few noncoding elements are highly conserved between flies and mosquitoes (Glazov et al. 2005). Only 612/6779 (9%) of our Drosophila HCNEs are at least partially aligned to the Agam sequence in a precomputed whole-genome alignment and only 264 (4%) are aligned with at least 30 base identities. The examples presented here suggest that, while many elements may have diverged beyond our ability to align them, the selective pressure to maintain their clusters has resulted in microsynteny conservation, which is detectable because protein-coding sequences align between Drosophila and Anopheles.
To quantitatively assess whether genes regulated by HCNE arrays are more likely to be in large regions of microsynteny between Dmel and Agam, we constructed synteny blocks between the two genomes, using a more relaxed approach than among the Drosophila because of the large evolutionary distance between flies and mosquitoes (see Methods). We then measured the span of Dmel–Agam synteny blocks around Dmel genes from several categories, including genes in HCNE-dense regions and genes annotated with Gene Ontology (GO) biological process terms that have been found to be associated with genes spanned by HCNE arrays (GO terms “multicellular organismal development” and “regulation of transcription, DNA-dependent;” see Supplemental Table S1 and Glazov et al. 2005). There was a tendency for genes in the HCNE-related categories to be within more extensive blocks of synteny than other types of genes (Fig. 2C). To better pinpoint the genes that are targets of HCNE arrays, we intersected the HCNE-related categories. Genes that were located in HCNE-dense regions, annotated to be involved in development, and annotated as transcriptional regulators were within significantly larger synteny blocks than genes from any of the non-HCNE related categories (P < 10−7 in pairwise one-tailed Kolmogorov-Smirnov tests against each of the categories “cellular protein metabolic process,” “cell organization and biogenesis,” “transport,” “signal transduction,” and “any biological process”).
HCNE-associated genes have specific types of core promoters
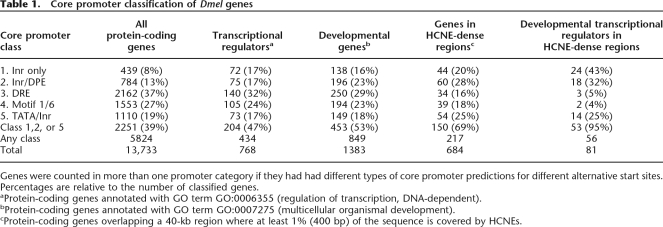
Data from this and earlier work suggests a model where insect and vertebrate HCNE arrays represent clusters of enhancers that specify expression programs for only a small subset of the genes that they span. How enhancer activity is specifically directed toward certain genes at HCNE-spanned loci is unknown. It has been demonstrated that enhancers can selectively target certain promoters (Li and Noll 1994; Merli et al. 1996) and that this selectivity may be facilitated by the occurrence of different core promoter types (Ohtsuki et al. 1998; Butler and Kadonaga 2001). A recent investigation of core promoters in Dmel classified them into five major types based on motif-content: TATA box followed by initiator (TATA/Inr), initiator followed by downstream promoter element (Inr/DPE), Motif 6 followed by Motif 1 (Motif 1/6), DNA replication element (DRE), and promoters containing only initiator, but none of the other elements (Inr only) (Ohler 2006). Based on these observations, the author designed a program (McPromoter) that predicts core promoters in the Dmel genome with high accuracy and classifies them as one of the five types.
Hypothesizing that enhancers in HCNE arrays may target specific genes within “striking distance” on the basis of their core promoter architecture, we used the genome-wide McPromoter predictions to investigate core promoter properties of likely target genes. Of 81 developmental transcriptional regulators located in HCNE-dense regions, 56 have a promoter prediction close to one or more annotated transcription start sites. Of these 56 genes, 53 (95%) are associated with a prediction of a type containing an Inr-motif (Inr only, Inr/DPE, or TATA/Inr; see Table 1). For comparison, only 39% all 5824 genes assigned a promoter prediction have a prediction with an Inr-motif. The enrichment is strongest for genes with Inr-only core promoters (P = 0.005, compared to Inr/DPE enrichment, by Fisher’s exact test). For examples of genes with different core promoter types, see Figures 2, 4, and 5 (see below), where gene models are colored according to associated promoter predictions (see also Supplemental Figs. S5, S6, S8, S9).
Table 1.
Core promoter classification of Dmel genes
Genes were counted in more than one promoter category if they had had different types of core promoter predictions for different alternative start sites. Percentages are relative to the number of classified genes.
aProtein-coding genes annotated with GO term GO:0006355 (regulation of transcription, DNA-dependent).
bProtein-coding genes annotated with GO term GO:0007275 (multicellular organismal development).
cProtein-coding genes overlapping a 40-kb region where at least 1% (400 bp) of the sequence is covered by HCNEs.
Figure 4.
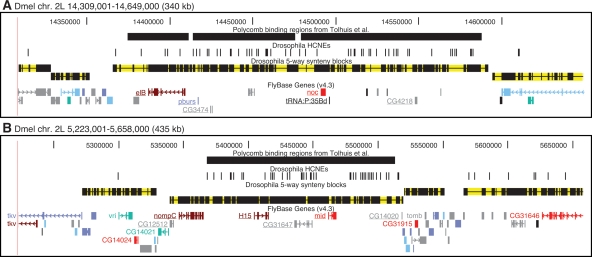
HCNE-clusters spanning coregulated genes and boundary agreement among synteny blocks, HCNE clusters, and Polycomb binding regions. Gene models are colored by predicted core promoter type as in Fig. 2. Only selected genes are labeled. (A) The paralogous zinc finger genes elB and noc, implicated in tracheal and appendage development, have different, but partially overlapping, spatial expression patterns during embryonic development (Dorfman et al. 2002) and are coexpressed in larval leg and wing discs (Weihe et al. 2004). Among the five flies, elB and noc are in conserved microsynteny with a tRNA gene and at least three protein-coding genes (underlined), which have no evidence of being functionally related to elB or noc: pburs encodes a subunit of the hormone bursicon required for wing expansion and associated cuticle changes after flies emerge from pupae (Luo et al. 2005); CG3474 is predicted to encode a cuticle component; CG4218 is predicted to encode a protein of unknown function. (B) The paralogous T-box genes H15 and mid are involved in regulation of heart development and have similar spatial expression patterns during embryonic development (Miskolczi-McCallum et al. 2005; Reim et al. 2005). They are in conserved microsynteny with four other genes (underlined) among the five flies: CG12512, predicted to encode a protein with an AMP-dependent synthetase and ligase domain; nompC, encoding a mechanosensory transduction channel (Walker et al. 2000); and two genes of unknown function. The developmental regulators vri (George and Terracol 1997) and tomb (Jiang et al. 2007) are centrally positioned in neighboring synteny blocks. Two transcript isoforms are shown for tkv because it has two major transcription start sites with different types of core promoter predictions.
Figure 5.
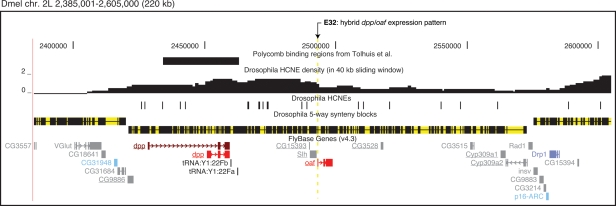
The E32 enhancer trap insertion at the Dmel decapentaplegic (dpp) locus. Gene models are colored by predicted core promoter type as in Fig. 2. Two transcript isoforms are shown for dpp because this gene has different core promoter predictions for two of its major transcription start sites (other dpp start sites are not shown, see St Johnston et al. 1990). The HCNE-spanned gene desert downstream from dpp contains several conserved enhancers that specify dpp expression in imaginal discs (Merli et al. 1996). Although the neighboring, divergently transcribed genes SLY1 homologous (Slh) and out at first (oaf) are insensitive to the array of dpp enhancers and have different expression patterns, the enhancer trap insertion E32, inserted into the 5′-untranslated region of oaf (arrow), reproduces part of the dpp expression pattern in imaginal discs (Merli et al. 1996). Five other genes (underlined) are in conserved microsynteny with dpp, Slh, and oaf among the investigated flies.
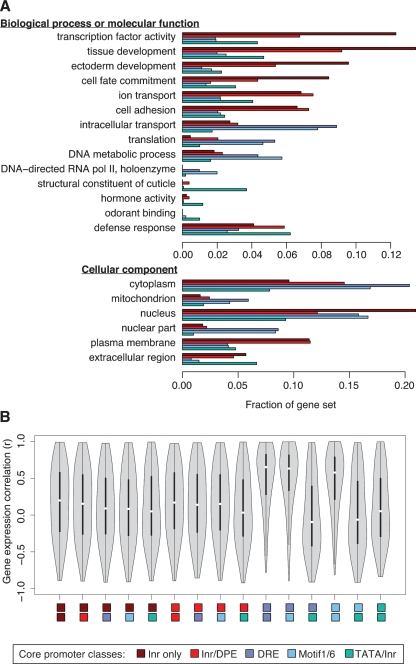
To further explore the association between core promoter types and gene functions, we performed a systematic search for enrichment of different GO annotations within each of the five core promoter classes (Fig. 3A; Supplemental Table S3). Consistent with the above results, we found enrichment for transcription factors among genes with Inr-only core promoters (P < 10−10) and enrichment for genes involved in developmental processes among genes with Inr-only core promoters (P < 10−13) and genes with Inr/DPE core promoters (P < 10−9). The latter gene set is also enriched for genes involved in ion transport (P < 10−4) and cell adhesion (P < 10−5). On the other hand, the set of genes with DRE core promoters is enriched for housekeeping functions (translation, P < 0.001) and mitochondrial proteins (P < 10−4). Genes with Motif 1/6 promoters showed a particular enrichment for RNA polymerase II components (P < 0.01), which also perform a housekeeping function. Although the set of genes with TATA/Inr core promoters appears to share some of the trends observed for Inr-only and Inr/DPE promoters, these trends are not statistically significant for the TATA/Inr-regulated genes, which instead are enriched for genes encoding proteins with more specialized, tissue-specific functions, such as cuticle constituents (P < 10−23), odorant binding proteins (P < 0.01), and defense-related proteins (P < 0.01). This finding is in agreement with results from mammals, where genes with TATA box core promoters tend to be expressed in tissue-specific contexts (Carninci et al. 2006). All P-values were adjusted for multiple testing with the Bonferroni method (see also Supplemental Table S3).
Figure 3.
Associations between core promoter types and gene functions. (A) Bars show the fraction of genes in each core promoter category that are annotated with indicated GO terms. All GO terms shown are significantly associated with a core promoter category at Bonferroni-adjusted P < 0.01 (see also Supplemental Table S3). (B) Violin plots (boxplots with added kernel density curves) show distributions of Pearson correlation coefficients for expression correlations between randomly selected gene pairs taken from pairs of core promoter categories indicated by colored rectangles below the plots. High correlations are frequent between genes with DRE core promoters and genes with Motif 1/6 core promoters, as well as among genes within each of those categories. Each distribution is based on a sample of 1000 randomly selected gene pairs. Genes were not compared against themselves.
To explore gene expression correlations among genes with different core promoter types, we used a published tiling array data set consisting of gene expression measurements across the Dmel genome at 12 time points during the 24 h of embryonic development (Manak et al. 2006). Consistent with a housekeeping nature of genes with DRE or Motif 1/6 core promoters, we found that randomly selected gene pairs from these sets often have highly correlated expression profiles, unlike gene pairs from the other sets (Fig. 3B). Genes in these two promoter categories also have the highest detection rates: 1423 (66%) of 2162 genes with DRE promoters and 1031 (66%) of 1553 genes with Motif 1/6 promoters were detected as expressed at some time point. Genes with TATA/Inr promoters have the lowest detection rate (46%; significantly different from genes with DRE or Motif 1/6 promoters: P < 10−15, χ2 test), consistent with more specialized roles for genes with TATA/Inr promoters.
HCNE arrays mark regulatory domains maintained in evolution
While the data presented here suggest that the need to maintain HCNE clusters is a major reason for microsynteny conservation in insects, other reasons for microsynteny conservation exist. A genome-wide comparison of Dmel–Dpse synteny blocks to changes in gene expression throughout the Dmel life cycle suggested that microsynteny is preserved at some loci in order to maintain coregulation of neighboring genes (Stolc et al. 2004; see also erratum at http://bussemaker.bio.columbia.edu/papers/Science2004/). Figure 4 shows two loci that are likely to be under dual pressures to maintain HCNE arrays and coregulated genes. Each of these loci contains two coexpressed and paralogous developmental regulatory genes (elB/noc, H15/mid), spanned by a HCNE cluster that delimits roughly the same genomic region as its surrounding synteny block, suggesting that these loci constitute genomic regulatory blocks with dual targets for some of the enhancer activity likely contained in their HCNEs.
Further evidence for the existence of large regulatory domains in Drosophila genomes comes from genome-wide mapping of Polycomb binding sites in embryonic cell lines, where Polycomb was found to bind large regions, preferentially around developmental regulators (Schwartz et al. 2006; Tolhuis et al. 2006). Similar findings have been reported for human embryonic stem cells, where the Polycomb repressive complex 2 subunit SUZ12 shows a strong tendency to bind across developmental transcription factor genes and around HCNEs (Lee et al. 2006). We inspected the Dmel Polycomb binding regions determined by Tolhuis et al. (2006) and noted an association with HCNEs, as expected. Tolhuis and colleagues interrogated ∼30% of the Dmel genome and found that 10% of the interrogated sequence corresponds to large Polycomb binding regions (Pc domains). HCNE sequence is more than twofold enriched in these Pc domains: 114 kb of the sequence interrogated by Tolhuis and colleagues corresponds to HCNEs, and 23% of this HCNE sequence is within Pc domains. The association of HCNEs with Pc domains is significant (P < 10−5; Wilcoxon test) when one compares the density of HCNEs in Pc domains to the density of HCNEs in regions randomly sampled from the part of the genome interrogated by Tolhuis and colleagues and with similar size distribution as the Pc domains. Interestingly, we also found a very good agreement between the boundaries of synteny blocks, HCNE clusters and Pc domains at a number of loci, including the three shown in Figures 2B and 4 (see also Supplemental Figs. S8, S9). These examples indicate that synteny blocks, HCNE clusters, and Polycomb binding regions can independently pinpoint the same large regulatory domains in insect genomes, suggesting that they reveal different aspects of the same evolutionarily conserved regulatory mechanism.
Discussion
Experimental evidence for long-range regulation and GRBs in Drosophila
Genomic regulatory blocks (GRBs) are regions containing long-range regulatory elements that have been interlocked in cis with their target genes as well as unrelated genes (Kikuta et al. 2007). We show here that this concept also applies to insect genomes. In the zebrafish genome, GRBs were discovered through enhancer detection events where the reporter insertion was close to or in a bystander gene, yet recapitulated the expression pattern of the target gene further away (Kikuta et al. 2007). Since enhancer detection has been performed extensively in Drosophila, we searched for examples of such insertions near bystander genes in the literature. Such insertions can be used to support the notion that regulatory elements form GRBs and thereby conserve microsynteny. The most striking example we found is the E32 enhancer detection line, which represents an insertion in the 5′ untranslated region of out at first (Merli et al. 1996). The insertion replicates part of the expression pattern of decapentaplegic (dpp), a developmental regulatory gene located 33 kb away. The region between dpp and the insertion contains a gene desert with HCNEs (Fig. 5) corresponding to known enhancers and a nonessential gene (Slh) with an unrelated expression pattern (Merli et al. 1996). We consider this a typical example of a GRB in Drosophila. At the loci of developmental regulators teashirt, engrailed, and u-shaped, insertions confirm that regulatory information is present at large distances from target genes, but do not directly show that this information is present inside bystander genes (Hayashi et al. 2002). Notably, dpp, teashirt, engrailed, and a gene desert with insertions upstream of u-shaped all coincide with Polycomb binding regions determined at high resolution (Tolhuis et al. 2006) (Fig. 5; Supplemental Figs. S8, S9). We conclude that, although it is commonly accepted that enhancer detection insertions in Drosophila reproduce the expression pattern of the nearest gene, these examples show that there are exceptions, in agreement with our enhancer detection results in zebrafish and in agreement with the notion that GRBs occur in both insects and vertebrates (Kikuta et al. 2007; this work).
Regulatory HCNE arrays are a fundamental feature of metazoan genomes
Most target genes in Drosophila GRBs appear to be developmental regulatory genes that have well-conserved vertebrate orthologs spanned by equivalent arrays of HCNEs (Sandelin et al. 2004). In addition to noncoding conservation and the types of genes they contain, other parallels between GRBs in insects and vertebrates are evident. They often harbor relatively long regions devoid of genes (gene deserts; Ovcharenko et al. 2005) and are characterized by microsynteny conserved deep in evolution (Kikuta et al. 2007; this work). Our demonstration of similarly organized HCNE arrays at orthologous Drosophila and Anopheles loci (where gene order has been partially preserved) reveals that microsynteny conservation, while constrained by regulatory elements, can outlive the sequence conservation of those elements.
The match between synteny blocks, HCNE arrays, and experimentally determined Polycomb binding regions in Drosophila is striking and supports the notion that these features are signatures of GRBs. In vertebrates, Polycomb group proteins are also preferentially found at the loci of developmental regulatory genes (Boyer et al. 2006; Lee et al. 2006), were shown to bind to evolutionarily conserved CpG islands that overlap large portions of developmental regulatory genes (Tanay et al. 2007), and directly control CpG methylation (Vire et al. 2006). Even though insects do not have genome methylation or CpG islands, one can speculate that Polycomb binding regions in Drosophila are functionally equivalent to conserved CpG islands in mammals. At present, it is unknown whether those regions in insects have any specific sequence properties analogous to CpG islands.
Together with a recent demonstration of the presence of HCNE clusters in nematode genomes of the genus Caenorhabditis (Vavouri et al. 2007), our findings indicate that arrays of HCNEs are central to developmental regulation of most, if not all, Metazoa. The association of HCNEs with orthologous genes among nematodes, insects, and vertebrates (Kikuta et al. 2007; Vavouri et al. 2007) suggests that long-range regulation and clusters/arrays of HCNEs are an ancient property of metazoan genomes. The role of HCNEs in constraining microsynteny has not yet been explored beyond vertebrates and insects, however.
Responsiveness of genes to long-range enhancers
The apparent unresponsiveness of bystander genes to long-range enhancers in GRBs remains mysterious. Distance does not seem to be crucial for enhancer action (Nobrega et al. 2003; Ellingsen et al. 2005). In the study mentioned above (Merli et al. 1996), the Drosophila gene out at first does not normally react to dpp enhancers but did so after exchanging its promoter with a dpp promoter. Thus, one explanation for enhancer specificity could be differential responsiveness of core promoters to enhancers (Smale 2001). In mammals, different types of core promoters have been clearly shown to be related to different modes of regulation (Carninci et al. 2006). In Drosophila, a recent study classified many known promoter regions into a number of different subtypes according to the principal motif (or combinations thereof) they contain (Ohler 2006). In this work we have shown that this classification discriminates between developmental genes (Inr with or without DPE), housekeeping genes (DRE or Motif 1/6), and tissue-specific genes (TATA). Based on these results, we speculate that it is the Inr-type of promoters without TATA boxes that are most likely to respond to long-range regulation. Indeed, inspection of dozens of Drosophila GRBs strongly supports the hypothesis that nonresponsive bystander genes, with expression patterns unrelated to the target gene in the same region, have core promoters of the DRE or Motif 1/6 types. In this way, Ohler’s classification of Drosophila core promoters is more powerful than that for vertebrate promoters made by Carninci et al. (2006); in vertebrates, we still do not know the fundamental difference between core promoters for housekeeping and developmental regulatory genes, which both seem to have CpG island core promoters, most without TATA boxes and with “broad”-type transcription start regions.
While borders of GRBs can be identified as synteny block boundaries by comparative genomics, it is still unclear how the cellular machinery recognizes those borders. Some regulatory domains are known to be delimited by insulator elements, which bind proteins that block the reach of enhancers or inhibit the spread of repressed chromatin (Valenzuela and Kamakaka 2006). Recent studies have revealed an abundance of putative insulator elements bound by the enhancer-blocking protein CTCF in mammalian genomes, and predicted a similar number of binding sites in Tetraodon (Kim et al. 2007; Xie et al. 2007). Human CTCF is functionally conserved in Drosophila, where several other enhancer-blocking proteins also are known (Moon et al. 2005). It will be interesting to see whether insulator elements are present at the borders of vertebrate and Drosophila GRBs.
Conclusions
The evidence presented in this paper establishes GRBs as a fundamental property of metazoan genomes. The long distances of regulatory elements from their developmental regulatory target genes will have to be taken into account in future studies of these genes and their regulatory networks. Additionally, these findings provide guidelines for designing enhancer trap experiments and their interpretation, including an informed choice of core promoter type for enhancer trap constructs.
Methods
Sequences and annotations
We used the following genome assemblies: Dmel release 4 (Berkeley Drosophila Genome Project); Dpse release 1.03 (Baylor HGSC); Dana, Dvir, and Dmoj Aug. 2005 (Agencourt); Agam MOZ2 (The International Anopheles Genome Project); Aaeg AaegL1 (The Broad Institute and TIGR), and A. mellifera Amel_2.0 (Baylor HGSC). We obtained Aaeg sequences from Ensembl (Hubbard et al. 2007; http://www.ensembl.org), and the other genome sequences, pairwise chained BLASTZ alignments between them, and annotations from the UCSC Genome Browser Database (Kuhn et al. 2007; http://genome.ucsc.edu). We used FlyBase v. 4.3 gene and CDS annotations (Crosby et al. 2007; http://flybase.org) and Dmel GO annotations (rev. 1.93) from http://www.geneontology.org.
HCNE detection
We identified elements highly conserved among flies by scanning pairwise BLASTZ net whole-genome alignments (Kent et al. 2003) between Dmel and each of the other four Drosophila species for regions with at least 98% identity over 50 alignment columns. Highly conserved elements were merged if they overlapped on the Dmel assembly. We discarded elements whose Dmel coordinates overlapped with any exon in FlyBase 4.3 genes, RefSeq genes, Dmel cDNA sequences from GenBank, or GENSCAN predictions. Remaining elements from each pairwise comparison were intersected based on their Dmel coordinates, to obtain elements conserved among all five species. Such elements spanning at least 50 bp of Dmel sequence were considered fly HCNEs. To detect mosquito HCNEs at selected Agam loci, we identified homologous Aaeg contigs by inspecting translated BLAT alignments in Ensembl v. 42–43 (Hubbard et al. 2007). We aligned Agam and Aaeg sequences with Shuffle-LAGAN v. 2.0 (Brudno et al. 2003) with default settings and used the resulting alignments to detect HCNEs as described for flies above, but using a lower identity threshold (80%) and removing elements that overlapped exons by comparing with the following UCSC Genome Browser database annotations on the Agam assembly: Ensembl genes, Agam cDNAs from GenBank, aligned Dmel proteins and GENSCAN predictions. To assess conservation of Drosophila HCNEs in Agam, we used a BLASTZ net alignment from Dmel to Agam.
Computation of feature densities and density peak detection
For images of loci, we computed HCNE densities by a sliding-window approach (Fig. 2) to provide easily interpreted density values. For genome-wide detection of density peaks, we required smoothed curves and therefore used the density function in R (http://www.R-project.org) with a Gaussian kernel and the indicated bandwidths (30,000 unless stated) to compute one density value every kilobase. We detected peaks by searching for density values that were higher than a threshold value (Fig. 1B) and their five preceding and five following values along the chromosome arm.
Identification of synteny blocks and RA sequence among flies
To identify synteny blocks, we made use of the utilities and C functions in the UCSC Genome Browser source package (http://genome.ucsc.edu/FAQ/FAQlicense).
Starting from pairwise chained BLASTZ alignments (chains) between the Dmel genome and each of the four other genomes, we constructed pairwise net alignments (nets) by running the program chainNet with option –minSpace = 1. chainNet filters a set of chains to retain only the best alignment for each position in one of the genomes (Kent et al. 2003). The chainNet algorithm tends to prioritize large chains and therefore its output is suitable for identifying synteny blocks. For each of the four pairwise genome comparisons, we constructed two sets of nets (one from the perspective of each genome), and used them to filter the chains into a set of reciprocal-best chains (rb-chains) that only contain alignment columns included in the nets for both genomes. To find the bases in the Dmel sequence that were aligned in a reciprocal-best manner in all four parwise genome comparisons (RA sequence), we identified the Dmel bases that were in ungapped blocks (i.e., were aligned to some base) in all four sets of rb-chains. We constructed pairwise synteny blocks from rb-chains in three steps: (1) Rb-chains were split at gaps that spanned nets if, within the gap, nets for either genome contained at least 10 kb in ungapped blocks. We used nets to split rb-chains because they include alignments that are not reciprocal-best, thus allowing us to capture synteny breaks caused, for example, by species-specific duplications. Only rb-chains that contained ≥10 kb in ungapped blocks after this step were retained. (2) We classified regions spanned by multiple (nested) rb-chains as being outside synteny blocks, and truncated nested rb-chains accordingly. Again, rb-chains containing <10 kb in ungapped blocks were discarded. (3) To avoid artificial synteny breaks due to failure to link scaffolds together in any of the non-Dmel assemblies, we joined rb-chains that were nearest neighbors along the same Dmel chromosome arm, but on different scaffolds in the non-Dmel assembly, unless the gap between the rb-chains in either genome contained nets with at least 10 kb of sequence in ungapped blocks (i.e., the same criterion as used to split chains in step 2 above). The set of rb-chains after this third step constituted our pairwise synteny blocks. Although joining of chains may overestimate synteny in pairwise comparisons, any such effects should be minimal after pairwise synteny blocks are intersected into five-way synteny blocks. We created five-way synteny blocks by intersecting the pairwise synteny blocks based on their coordinates on the Dmel assembly: Any two Dmel bases were assigned to the same five-way synteny block if, and only if, they were part of the same synteny block in each of the pairwise comparisons. We discarded five-way synteny blocks that did not contain at least 10 kb in ungapped alignments across all pairwise synteny blocks.
Analysis of Dmel–Agam synteny
To identify Dmel–Agam synteny blocks, we first computed reciprocal-best BLASTZ net alignments between Dmel and Agam as described for fly comparisons above. We then constructed a graph where two alignments (nodes) were connected if separated by ≤100 kb in both genomes (not considering strand, to allow local inversions within synteny blocks). We considered each connected component in the graph to be one synteny block. The threshold of 100 kb is arbitrary; we tested several values in the range 0–300 kb with similar results. Considering all protein-coding FlyBase genes, we assigned a gene to a synteny block if that gene had a transcript with at least 25% of its CDS aligned to the syntenic Agam locus. Genes that belonged to multiple blocks according to this rule were excluded.
Core promoter analysis
We assigned a McPromoter prediction (Ohler 2006) to a FlyBase transcript if it was within 250 bp upstream of the annotated start site of the transcript or within the noncoding part of its first exon. In rare cases where multiple promoter predictions satisfied these criteria, the prediction closest to the annotated start site was chosen. For illustrated loci, core promoter assignments to genes were reviewed and changed if available transcript data motivated modifications to FlyBase gene models.
Expression analysis
To assign expression values to genes, we processed FlyBase gene models as follows. Because the expression signals from the tiling array study (Manak et al. 2006) are not strand-specific, we masked parts of exons that overlapped exons on the other genomic strand. We disregarded any gene that had more than half of its total exon sequence masked. For each remaining gene i, we computed its maximum transfrag coverage cmaxi as maxj(cij), where cij is the number of unmasked exon bases covered by transfrags for gene i at time point j. Any gene i with cmaxi ≥70% of its unmasked exon sequence was considered expressed (a similar criterion was used in the original analysis of the data; Manak et al. 2006); other genes were assigned an expression value of 0 for all time points. If two expressed genes (annotated on the same strand) shared unmasked exon sequence, only the gene with highest cmax was considered further, because we were not interested in comparing expression profiles between genes that share the same transcriptional unit. Each retained gene was then, for each time point, assigned an expression value equal to the median signal over its unmasked exon sequence. Only genes that showed at least a twofold difference in expression values between some time points were used in comparisons of expression profiles.
Acknowledgments
This work was supported by the Functional Genomics Programme (FUGE) of the Research Council of Norway and a core grant from the Sars Centre. S.H.S. is supported by the National Sciences and Research Council of Canada (NSERC), and is a MSFHR senior trainee. We thank David Fredman, Fernando Casares, and Wyeth Wasserman for useful discussions, and Agencourt for permission to use the Dana, Dvir, and Dmoj genome assemblies.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6669607
References
- Bailey P.J., Klos J.M., Andersson E., Karlen M., Kallstrom M., Ponjavic J., Muhr J., Lenhard B., Sandelin A., Ericson J., Klos J.M., Andersson E., Karlen M., Kallstrom M., Ponjavic J., Muhr J., Lenhard B., Sandelin A., Ericson J., Andersson E., Karlen M., Kallstrom M., Ponjavic J., Muhr J., Lenhard B., Sandelin A., Ericson J., Karlen M., Kallstrom M., Ponjavic J., Muhr J., Lenhard B., Sandelin A., Ericson J., Kallstrom M., Ponjavic J., Muhr J., Lenhard B., Sandelin A., Ericson J., Ponjavic J., Muhr J., Lenhard B., Sandelin A., Ericson J., Muhr J., Lenhard B., Sandelin A., Ericson J., Lenhard B., Sandelin A., Ericson J., Sandelin A., Ericson J., Ericson J. A global genomic transcriptional code associated with CNS-expressed genes. Exp. Cell Res. 2006;312:3108–3119. doi: 10.1016/j.yexcr.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Stephen S., Kent W.J., Mattick J.S., Haussler D., Kent W.J., Mattick J.S., Haussler D., Mattick J.S., Haussler D., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., Levine S.S., Wernig M., Tajonar A., Ray M.K., Wernig M., Tajonar A., Ray M.K., Tajonar A., Ray M.K., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brudno M., Malde S., Poliakov A., Do C.B., Couronne O., Dubchak I., Batzoglou S., Malde S., Poliakov A., Do C.B., Couronne O., Dubchak I., Batzoglou S., Poliakov A., Do C.B., Couronne O., Dubchak I., Batzoglou S., Do C.B., Couronne O., Dubchak I., Batzoglou S., Couronne O., Dubchak I., Batzoglou S., Dubchak I., Batzoglou S., Batzoglou S. Glocal alignment: Finding rearrangements during alignment. Bioinformatics. 2003;19:i54–i62. doi: 10.1093/bioinformatics/btg1005. [DOI] [PubMed] [Google Scholar]
- Butler J.E., Kadonaga J.T., Kadonaga J.T. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes & Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., Taylor M.S., Engstrom P.G., Frith M.C., Engstrom P.G., Frith M.C., Frith M.C., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Carroll S.B. Endless forms most beautiful: The new science of Evo Devo and the making of the animal kingdom. W.W. Norton & Company; New York: 2005. [Google Scholar]
- Crosby M.A., Goodman J.L., Strelets V.B., Zhang P., Gelbart W.M., Goodman J.L., Strelets V.B., Zhang P., Gelbart W.M., Strelets V.B., Zhang P., Gelbart W.M., Zhang P., Gelbart W.M., Gelbart W.M. FlyBase: Genomes by the dozen. Nucleic Acids Res. 2007;35:D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Calle-Mustienes E., Feijoo C.G., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Feijoo C.G., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Manzanares M., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Tena J.J., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Rodriguez-Seguel E., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Letizia A., Allende M.L., Gomez-Skarmeta J.L., Allende M.L., Gomez-Skarmeta J.L., Gomez-Skarmeta J.L. A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res. 2005;15:1061–1072. doi: 10.1101/gr.4004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R., Glazer L., Weihe U., Wernet M.F., Shilo B.Z., Glazer L., Weihe U., Wernet M.F., Shilo B.Z., Weihe U., Wernet M.F., Shilo B.Z., Wernet M.F., Shilo B.Z., Shilo B.Z. Elbow and Noc define a family of zinc finger proteins controlling morphogenesis of specific tracheal branches. Development. 2002;129:3585–3596. doi: 10.1242/dev.129.15.3585. [DOI] [PubMed] [Google Scholar]
- Ellingsen S., Laplante M.A., Konig M., Kikuta H., Furmanek T., Hoivik E.A., Becker T.S., Laplante M.A., Konig M., Kikuta H., Furmanek T., Hoivik E.A., Becker T.S., Konig M., Kikuta H., Furmanek T., Hoivik E.A., Becker T.S., Kikuta H., Furmanek T., Hoivik E.A., Becker T.S., Furmanek T., Hoivik E.A., Becker T.S., Hoivik E.A., Becker T.S., Becker T.S. Large-scale enhancer detection in the zebrafish genome. Development. 2005;132:3799–3811. doi: 10.1242/dev.01951. [DOI] [PubMed] [Google Scholar]
- George H., Terracol R., Terracol R. The vrille gene of Drosophila is a maternal enhancer of decapentaplegic and encodes a new member of the bZIP family of transcription factors. Genetics. 1997;146:1345–1363. doi: 10.1093/genetics/146.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazov E.A., Pheasant M., McGraw E.A., Bejerano G., Mattick J.S., Pheasant M., McGraw E.A., Bejerano G., Mattick J.S., McGraw E.A., Bejerano G., Mattick J.S., Bejerano G., Mattick J.S., Mattick J.S. Ultraconserved elements in insect genomes: A highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res. 2005;15:800–808. doi: 10.1101/gr.3545105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottgens B., Barton L.M., Gilbert J.G., Bench A.J., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Barton L.M., Gilbert J.G., Bench A.J., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Gilbert J.G., Bench A.J., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Bench A.J., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Sanchez M.J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., Mistry S., Grafham D., McMurray A., Vaudin M., Grafham D., McMurray A., Vaudin M., McMurray A., Vaudin M., Vaudin M., et al. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Ito K., Sado Y., Taniguchi M., Akimoto A., Takeuchi H., Aigaki T., Matsuzaki F., Nakagoshi H., Tanimura T., Ito K., Sado Y., Taniguchi M., Akimoto A., Takeuchi H., Aigaki T., Matsuzaki F., Nakagoshi H., Tanimura T., Sado Y., Taniguchi M., Akimoto A., Takeuchi H., Aigaki T., Matsuzaki F., Nakagoshi H., Tanimura T., Taniguchi M., Akimoto A., Takeuchi H., Aigaki T., Matsuzaki F., Nakagoshi H., Tanimura T., Akimoto A., Takeuchi H., Aigaki T., Matsuzaki F., Nakagoshi H., Tanimura T., Takeuchi H., Aigaki T., Matsuzaki F., Nakagoshi H., Tanimura T., Aigaki T., Matsuzaki F., Nakagoshi H., Tanimura T., Matsuzaki F., Nakagoshi H., Tanimura T., Nakagoshi H., Tanimura T., Tanimura T., et al. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 2002;34:58–61. doi: 10.1002/gene.10137. [DOI] [PubMed] [Google Scholar]
- Hubbard T.J., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Aken B.L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Clarke L., Coates G., Cunningham F., Cutts T., Coates G., Cunningham F., Cutts T., Cunningham F., Cutts T., Cutts T., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Benson E., Bausek N., Doggett K., White-Cooper H., Benson E., Bausek N., Doggett K., White-Cooper H., Bausek N., Doggett K., White-Cooper H., Doggett K., White-Cooper H., White-Cooper H. Tombola, a tesmin/TSO1-family protein, regulates transcriptional activation in the Drosophila male germline and physically interacts with Always early. Development. 2007;134:1549–1559. doi: 10.1242/dev.000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D., Baertsch R., Hinrichs A., Miller W., Haussler D., Hinrichs A., Miller W., Haussler D., Miller W., Haussler D., Haussler D. Evolution's cauldron: Duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta H., Laplante M., Navratilova P., Komisarczuk A.Z., Engstrom P.G., Fredman D., Akalin A., Caccamo M., Sealy I., Howe K., Laplante M., Navratilova P., Komisarczuk A.Z., Engstrom P.G., Fredman D., Akalin A., Caccamo M., Sealy I., Howe K., Navratilova P., Komisarczuk A.Z., Engstrom P.G., Fredman D., Akalin A., Caccamo M., Sealy I., Howe K., Komisarczuk A.Z., Engstrom P.G., Fredman D., Akalin A., Caccamo M., Sealy I., Howe K., Engstrom P.G., Fredman D., Akalin A., Caccamo M., Sealy I., Howe K., Fredman D., Akalin A., Caccamo M., Sealy I., Howe K., Akalin A., Caccamo M., Sealy I., Howe K., Caccamo M., Sealy I., Howe K., Sealy I., Howe K., Howe K., et al. Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res. 2007;17:545–555. doi: 10.1101/gr.6086307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B., Zhang M.Q., Lobanenkov V.V., Ren B., Lobanenkov V.V., Ren B., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura-Yoshida C., Kitajima K., Oda-Ishii I., Tian E., Suzuki M., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Kitajima K., Oda-Ishii I., Tian E., Suzuki M., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Oda-Ishii I., Tian E., Suzuki M., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Tian E., Suzuki M., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Suzuki M., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Yamamoto M., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Suzuki T., Kobayashi M., Aizawa S., Matsuo I., Kobayashi M., Aizawa S., Matsuo I., Aizawa S., Matsuo I., Matsuo I. Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development. 2004;131:57–71. doi: 10.1242/dev.00877. [DOI] [PubMed] [Google Scholar]
- Kuhn R.M., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., Stanke M., Smith K.E., Siepel A., Smith K.E., Siepel A., Siepel A., et al. The UCSC genome browser database: Update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Chevalier B., Johnstone S.E., Cole M.F., Isono K., Johnstone S.E., Cole M.F., Isono K., Cole M.F., Isono K., Isono K., et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Noll M., Noll M. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 1994;13:400–406. doi: 10.1002/j.1460-2075.1994.tb06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., Chang J.L., Kulbokas E.J., Zody M.C., Kulbokas E.J., Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Luo C.W., Dewey E.M., Sudo S., Ewer J., Hsu S.Y., Honegger H.W., Hsueh A.J., Dewey E.M., Sudo S., Ewer J., Hsu S.Y., Honegger H.W., Hsueh A.J., Sudo S., Ewer J., Hsu S.Y., Honegger H.W., Hsueh A.J., Ewer J., Hsu S.Y., Honegger H.W., Hsueh A.J., Hsu S.Y., Honegger H.W., Hsueh A.J., Honegger H.W., Hsueh A.J., Hsueh A.J. Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proc. Natl. Acad. Sci. 2005;102:2820–2825. doi: 10.1073/pnas.0409916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak J.R., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Cheng J., Bell I., Ghosh S., Piccolboni A., Bell I., Ghosh S., Piccolboni A., Ghosh S., Piccolboni A., Piccolboni A., et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- McGovern V.L., Pacak C.A., Sewell S.T., Turski M.L., Seeger M.A., Pacak C.A., Sewell S.T., Turski M.L., Seeger M.A., Sewell S.T., Turski M.L., Seeger M.A., Turski M.L., Seeger M.A., Seeger M.A. A targeted gain of function screen in the embryonic CNS of Drosophila. Mech. Dev. 2003;120:1193–1207. doi: 10.1016/s0925-4773(03)00159-x. [DOI] [PubMed] [Google Scholar]
- Merli C., Bergstrom D.E., Cygan J.A., Blackman R.K., Bergstrom D.E., Cygan J.A., Blackman R.K., Cygan J.A., Blackman R.K., Blackman R.K. Promoter specificity mediates the independent regulation of neighboring genes. Genes & Dev. 1996;10:1260–1270. doi: 10.1101/gad.10.10.1260. [DOI] [PubMed] [Google Scholar]
- Milewski R.C., Chi N.C., Li J., Brown C., Lu M.M., Epstein J.A., Chi N.C., Li J., Brown C., Lu M.M., Epstein J.A., Li J., Brown C., Lu M.M., Epstein J.A., Brown C., Lu M.M., Epstein J.A., Lu M.M., Epstein J.A., Epstein J.A. Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development. 2004;131:829–837. doi: 10.1242/dev.00975. [DOI] [PubMed] [Google Scholar]
- Miskolczi-McCallum C.M., Scavetta R.J., Svendsen P.C., Soanes K.H., Brook W.J., Scavetta R.J., Svendsen P.C., Soanes K.H., Brook W.J., Svendsen P.C., Soanes K.H., Brook W.J., Soanes K.H., Brook W.J., Brook W.J. The Drosophila melanogaster T-box genes midline and H15 are conserved regulators of heart development. Dev. Biol. 2005;278:459–472. doi: 10.1016/j.ydbio.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Moon H., Filippova G., Loukinov D., Pugacheva E., Chen Q., Smith S.T., Munhall A., Grewe B., Bartkuhn M., Arnold R., Filippova G., Loukinov D., Pugacheva E., Chen Q., Smith S.T., Munhall A., Grewe B., Bartkuhn M., Arnold R., Loukinov D., Pugacheva E., Chen Q., Smith S.T., Munhall A., Grewe B., Bartkuhn M., Arnold R., Pugacheva E., Chen Q., Smith S.T., Munhall A., Grewe B., Bartkuhn M., Arnold R., Chen Q., Smith S.T., Munhall A., Grewe B., Bartkuhn M., Arnold R., Smith S.T., Munhall A., Grewe B., Bartkuhn M., Arnold R., Munhall A., Grewe B., Bartkuhn M., Arnold R., Grewe B., Bartkuhn M., Arnold R., Bartkuhn M., Arnold R., Arnold R., et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M., Campbell G., Campbell G. Generating and interpreting the Brinker gradient in the Drosophila wing. Dev. Biol. 2005;286:647–658. doi: 10.1016/j.ydbio.2005.08.036. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Larkin D.M., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Everts-van der Wind A., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Bourque G., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Tesler G., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Auvil L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Beever J.E., Chowdhary B.P., Galibert F., Gatzke L., Chowdhary B.P., Galibert F., Gatzke L., Galibert F., Gatzke L., Gatzke L., et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Nepveu A. Role of the multifunctional CDP/Cut/Cux homeodomain transcription factor in regulating differentiation, cell growth and development. Gene. 2001;270:1–15. doi: 10.1016/s0378-1119(01)00485-1. [DOI] [PubMed] [Google Scholar]
- Nobrega M.A., Ovcharenko I., Afzal V., Rubin E.M., Ovcharenko I., Afzal V., Rubin E.M., Afzal V., Rubin E.M., Rubin E.M. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Ohler U. Identification of core promoter modules in Drosophila and their application in accurate transcription start site prediction. Nucleic Acids Res. 2006;34:5943–5950. doi: 10.1093/nar/gkl608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S., Levine M., Cai H.N., Levine M., Cai H.N., Cai H.N. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes & Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I., Loots G.G., Nobrega M.A., Hardison R.C., Miller W., Stubbs L., Loots G.G., Nobrega M.A., Hardison R.C., Miller W., Stubbs L., Nobrega M.A., Hardison R.C., Miller W., Stubbs L., Hardison R.C., Miller W., Stubbs L., Miller W., Stubbs L., Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D., Kislyuk A., Levine M., Dubchak I., Kislyuk A., Levine M., Dubchak I., Levine M., Dubchak I., Dubchak I. Conservation patterns in different functional sequence categories of divergent Drosophila species. Genomics. 2006;88:431–442. doi: 10.1016/j.ygeno.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Minovitsky S., Dubchak I., Holt A., Lewis K.D., Dubchak I., Holt A., Lewis K.D., Holt A., Lewis K.D., Lewis K.D., et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Loots G.G., Nobrega M.A., Ovcharenko I., Loots G.G., Nobrega M.A., Ovcharenko I., Nobrega M.A., Ovcharenko I., Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevzner P., Tesler G., Tesler G. Genome rearrangements in mammalian evolution: Lessons from human and mouse genomes. Genome Res. 2003;13:37–45. doi: 10.1101/gr.757503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim I., Mohler J.P., Frasch M., Mohler J.P., Frasch M., Frasch M. Tbx20-related genes, mid and H15, are required for tinman expression, proper patterning, and normal differentiation of cardioblasts in Drosophila. Mech. Dev. 2005;122:1056–1069. doi: 10.1016/j.mod.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Sandelin A., Bailey P., Bruce S., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Bailey P., Bruce S., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Bruce S., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Engstrom P.G., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Klos J.M., Wasserman W.W., Ericson J., Lenhard B., Wasserman W.W., Ericson J., Lenhard B., Ericson J., Lenhard B., Lenhard B. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y.B., Kahn T.G., Nix D.A., Li X.Y., Bourgon R., Biggin M., Pirrotta V., Kahn T.G., Nix D.A., Li X.Y., Bourgon R., Biggin M., Pirrotta V., Nix D.A., Li X.Y., Bourgon R., Biggin M., Pirrotta V., Li X.Y., Bourgon R., Biggin M., Pirrotta V., Bourgon R., Biggin M., Pirrotta V., Biggin M., Pirrotta V., Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Shin J.T., Priest J.R., Ovcharenko I., Ronco A., Moore R.K., Burns C.G., MacRae C.A., Priest J.R., Ovcharenko I., Ronco A., Moore R.K., Burns C.G., MacRae C.A., Ovcharenko I., Ronco A., Moore R.K., Burns C.G., MacRae C.A., Ronco A., Moore R.K., Burns C.G., MacRae C.A., Moore R.K., Burns C.G., MacRae C.A., Burns C.G., MacRae C.A., MacRae C.A. Human-zebrafish non-coding conserved elements act in vivo to regulate transcription. Nucleic Acids Res. 2005;33:5437–5445. doi: 10.1093/nar/gki853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., Clawson H., Spieth J., Hillier L.W., Richards S., Spieth J., Hillier L.W., Richards S., Hillier L.W., Richards S., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale S.T. Core promoters: Active contributors to combinatorial gene regulation. Genes & Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- St Johnston R.D., Hoffmann F.M., Blackman R.K., Segal D., Grimaila R., Padgett R.W., Irick H.A., Gelbart W.M., Hoffmann F.M., Blackman R.K., Segal D., Grimaila R., Padgett R.W., Irick H.A., Gelbart W.M., Blackman R.K., Segal D., Grimaila R., Padgett R.W., Irick H.A., Gelbart W.M., Segal D., Grimaila R., Padgett R.W., Irick H.A., Gelbart W.M., Grimaila R., Padgett R.W., Irick H.A., Gelbart W.M., Padgett R.W., Irick H.A., Gelbart W.M., Irick H.A., Gelbart W.M., Gelbart W.M. Molecular organization of the decapentaplegic gene in Drosophila melanogaster. Genes & Dev. 1990;4:1114–1127. doi: 10.1101/gad.4.7.1114. [DOI] [PubMed] [Google Scholar]
- Stolc V., Gauhar Z., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Gauhar Z., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Mason C., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Halasz G., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., van Batenburg M.F., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Rifkin S.A., Hua S., Herreman T., Tongprasit W., Barbano P.E., Hua S., Herreman T., Tongprasit W., Barbano P.E., Herreman T., Tongprasit W., Barbano P.E., Tongprasit W., Barbano P.E., Barbano P.E., et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Sumiyama K., Ruddle F.H., Ruddle F.H. Regulation of Dlx3 gene expression in visceral arches by evolutionarily conserved enhancer elements. Proc. Natl. Acad. Sci. 2003;100:4030–4034. doi: 10.1073/pnas.0530119100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A., O'Donnell A.H., Damelin M., Bestor T.H., O'Donnell A.H., Damelin M., Bestor T.H., Damelin M., Bestor T.H., Bestor T.H. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc. Natl. Acad. Sci. 2007;104:5521–5526. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor S., Thomas J.B., Thomas J.B. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron. 1997;18:397–409. doi: 10.1016/s0896-6273(00)81241-6. [DOI] [PubMed] [Google Scholar]
- Tolhuis B., de Wit E., Muijrers I., Teunissen H., Talhout W., van Steensel B., van Lohuizen M., de Wit E., Muijrers I., Teunissen H., Talhout W., van Steensel B., van Lohuizen M., Muijrers I., Teunissen H., Talhout W., van Steensel B., van Lohuizen M., Teunissen H., Talhout W., van Steensel B., van Lohuizen M., Talhout W., van Steensel B., van Lohuizen M., van Steensel B., van Lohuizen M., van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Valenzuela L., Kamakaka R.T., Kamakaka R.T. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- Vavouri T., Walter K., Gilks W.R., Lehner B., Elgar G., Walter K., Gilks W.R., Lehner B., Elgar G., Gilks W.R., Lehner B., Elgar G., Lehner B., Elgar G., Elgar G. Parallel evolution of conserved non-coding elements that target a common set of developmental regulatory genes from worms to humans. Genome Biol. 2007;8:R15. doi: 10.1186/gb-2007-8-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Van Eynde A., Bernard D., Vanderwinden J.M., Bernard D., Vanderwinden J.M., Vanderwinden J.M., et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Walker D.L., Wang D., Jin Y., Rath U., Wang Y., Johansen J., Johansen K.M., Wang D., Jin Y., Rath U., Wang Y., Johansen J., Johansen K.M., Jin Y., Rath U., Wang Y., Johansen J., Johansen K.M., Rath U., Wang Y., Johansen J., Johansen K.M., Wang Y., Johansen J., Johansen K.M., Johansen J., Johansen K.M., Johansen K.M. Skeletor, a novel chromosomal protein that redistributes during mitosis provides evidence for the formation of a spindle matrix. J. Cell Biol. 2000;151:1401–1412. doi: 10.1083/jcb.151.7.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe U., Dorfman R., Wernet M.F., Cohen S.M., Milan M., Dorfman R., Wernet M.F., Cohen S.M., Milan M., Wernet M.F., Cohen S.M., Milan M., Cohen S.M., Milan M., Milan M. Proximodistal subdivision of Drosophila legs and wings: The elbow-no ocelli gene complex. Development. 2004;131:767–774. doi: 10.1242/dev.00979. [DOI] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Smith S.F., North P., Callaway H., Kelly K., North P., Callaway H., Kelly K., Callaway H., Kelly K., Kelly K., et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Mikkelsen T.S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E.S., Mikkelsen T.S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E.S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E.S., Lindblad-Toh K., Kellis M., Lander E.S., Kellis M., Lander E.S., Lander E.S. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov E.M., Bork P., Bork P. Quantification of insect genome divergence. Trends Genet. 2007;23:16–20. doi: 10.1016/j.tig.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Zdobnov E.M., von Mering C., Letunic I., Torrents D., Suyama M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., von Mering C., Letunic I., Torrents D., Suyama M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Letunic I., Torrents D., Suyama M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Torrents D., Suyama M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Suyama M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., Thomasova D., Holt R.A., Subramanian G.M., Holt R.A., Subramanian G.M., Subramanian G.M., et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]