Abstract
Many Gram-negative bacteria regulate gene expression in response to their population size by sensing the level of acyl-homoserine lactone signal molecules which they produce and liberate to the environment. We have developed an assay for these signals that couples separation by thin-layer chromatography with detection using Agrobacterium tumefaciens harboring lacZ fused to a gene that is regulated by autoinduction. With the exception of N-butanoyl-l-homoserine lactone, the reporter detected acyl-homoserine lactones with 3-oxo-, 3-hydroxy-, and 3-unsubstituted side chains of all lengths tested. The intensity of the response was proportional to the amount of the signal molecule chromatographed. Each of the 3-oxo- and the 3-unsubstituted derivatives migrated with a unique mobility. Using the assay, we showed that some bacteria produce as many as five detectable signal molecules. Structures could be assigned tentatively on the basis of mobility and spot shape. The dominant species produced by Pseudomonas syringae pv. tabaci chromatographed with the properties of N-(3-oxohexanoyl)-l-homoserine lactone, a structure that was confirmed by mass spectrometry. An isolate of Pseudomonas fluorescens produced five detectable species, three of which had novel chromatographic properties. These were identified as the 3-hydroxy- forms of N-hexanoyl-, N-octanoyl-, and N-decanoyl-l-homoserine lactone. The assay can be used to screen cultures of bacteria for acyl-homoserine lactones, for quantifying the amounts of these molecules produced, and as an analytical and preparative aid in determining the structures of these signal molecules.
Keywords: autoinduction, signaling systems, quorum sensing, gene regulation/detection systems
Many Gram-negative bacteria regulate the expression of specialized gene sets in response to their population density by a mechanism called autoinduction (1). These bacteria measure their cell densities by sensing the level of a chemical signal, called the autoinducer (AI), that they themselves produce during growth. In the paradigm system Vibrio fischeri regulates expression of the lux operon by autoinduction through the transcriptional activator LuxR, and the specific autoinducer signal, VAI (2). This signal molecule, which diffuses out of, and back into, the cells, serves as a coinducer, apparently by interacting with LuxR (3, 4). Among other organisms, Pseudomonas aeruginosa regulates expression of secreted toxins and enzymes by means of the transcriptional activator LasR and the autoinducer PAI (5), and Agrobacterium tumefaciens uses TraR and the autoinducer AAI (6, 7) to regulate conjugal transfer of the Ti plasmid.
All AIs identified to date are N-acylated derivatives of l-homoserine lactone (acyl-HSLs). Specificity is conferred by the length and the nature of the substitution at carbon 3 of the acyl side chain. For example, VAI is N-(3-oxohexanoyl)-l-HSL (8), AAI is N-(3-oxooctanoyl)-l-HSL (9), and PAI is N-(3-oxododecanoyl)-l-HSL (10). In some systems carbon 3 is hydroxylated (11, 12), whereas in others, carbon 3 is fully reduced (13–16). Although a given R protein is activated most efficiently by its cognate autoinducer, analogs differing in chain length and the nature of the substitution at carbon 3 can show activity, albeit at much higher concentrations (17–19).
Analysis of compounds with AI activity usually couples fractionation by HPLC with biodetection using reporter bacteria in which a detectable phenotype—such as light emission, expression of β-galactosidase activity, or production of pigments—requires addition of an exogenous active AI (8, 10, 20–22). Because of the specificity requirements of the R protein, most of the detection systems are limited in the range of acyl-HSLs to which they respond. This can be problematic, since bacteria often produce more than one acyl-HSL species (9, 12–16). Thus, a thorough screening for production of these compounds can require the use of several reporter systems, each responding to signal molecules with different structural features.
We report here a simple, rapid, and sensitive thin-layer chromatography (TLC) assay useful for analyzing bacterial cultures for the presence of AIs. The assay is particularly well suited for screening many samples simultaneously. Using this assay, we show that several organisms, some not previously examined, produce one or more distinguishable compounds with AI activity. Moreover, in previously untested bacteria, the assay predicted in some cases the production of known acyl-HSLs and in others, the existence of AIs with novel structures. We confirmed the predicted structures and characterized the novel forms by using mass spectrometry (MS).
MATERIALS AND METHODS
Bacterial Strains and Culture Conditions.
A. tumefaciens strain NT1(pDCI41E33) was used as the tra reporter. Plasmid pDCI41E33 contains a traG::lacZ fusion and traR cloned into pDSK519 (23). P. aeruginosa PAO1 and Pseudomonas syringae pv. tabaci 2024 were from our collections. V. fischeri MJ1 was from E. P. Greenberg (University of Iowa, Iowa City). Ralstonia solanacearum K60 was from L. Sequeira (University of Wisconsin, Madison), and Pseudomonas fluorescens 2-79 was from L. Thomashow (Washington State University, Pullman). Rhizobium meliloti L5–30 was from S. Long (Stanford University, Stanford, CA), while R. meliloti YA2 was from W. Lepps (University of Wisconsin, Madison). Rhodobacter sphaeroides 2.4.1 was from S. Kaplan (University of Texas Health Science Center, Houston).
The A. tumefaciens indicator strain was grown in AB minimal mannitol liquid medium (ABM; ref. 21) at 28°C. Cultures used to prepare extracts for analysis were grown as follows: P. aeruginosa and P. fluorescens, AB minimal glucose medium, 37°C and 28°C, respectively; Ralstonia solanacearum, CPG medium (24), 28°C; P. syringae pv. tabaci, VB medium (25), 28°C; V. fischeri, autoinduction assay medium (26), 28°C; and Rhizobium meliloti strains, GTS medium containing 0.1% yeast extract (27), 28°C. Rhodobacter sphaeroides was grown in Luria–Bertani medium (LB) at room temperature under anaerobic conditions with constant illumination from a 60-W incandescent bulb. All other cultures were grown to late exponential or early stationary phase with shaking to ensure adequate aeration.
Extraction of Culture Supernatants.
Extracts for analytical TLC were prepared from 5-ml cultures, while those used to process samples for analysis by MS were prepared from 500-ml cultures. Bacteria were removed by centrifugation, the supernatants were extracted twice with equal volumes of ethyl acetate, and the combined extracts were dried over anhydrous magnesium sulfate, filtered, and evaporated to dryness. Residues from 5-ml cultures were dissolved in 50–100 μl of HPLC-grade ethyl acetate. Residues from 500-ml cultures were dissolved in the minimum volume of HPLC-grade ethyl acetate, usually 20–50 μl.
Analytical TLC.
Samples, in volumes of 1–4 μl, were applied to C18 reversed-phase TLC plates (200 μm layer; Baker) and the chromatograms were developed with methanol/water (60:40, vol/vol). After development, the solvent was evaporated, and the dried plates were overlaid with a culture of the indicator bacterium prepared as follows. For 20 × 20 cm plates, a 5-ml overnight culture of NT1(pDCI41E33) was used to inoculate 50 ml of ABM and the new culture was grown to late exponential phase. The entire 50 ml of culture was added to 100 ml of the same medium containing 1.12 g of melted agar and 60 μg/ml 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal) maintained at 45°C. The culture was mixed thoroughly and immediately spread over the surface of the developed plate held in a Plexiglas jig designed to produce a uniform layer of agar about 3 mm thick. After the agar solidified, the coated plates were incubated at 28°C for 12–18 hr in a closed plastic container.
Preparative TLC.
Samples from 500-ml cultures were partially purified by preparative TLC. For a given sample, a portion of the concentrated extract was diluted in ethyl acetate and 1 μl was spotted onto a reporter TLC plate along with a set of standards. The remainder of the concentrated sample was applied to a second TLC plate as a series of two to four spots along the baseline, and the two plates were chromatographed in the same chamber. Following development of the reporter TLC plate with the A. tumefaciens strain, the C18 matrix in the regions of the preparative plate corresponding to the compound to be analyzed were scraped off, combined, and extracted three times with 1–2 ml volumes of 1:1 (vol/vol) chloroform/dichloromethane. The combined extracts were clarified by centrifugation and passed through a fine sintered glass filter. The filtrates were taken to dryness and the residue was redissolved in the appropriate solvent for mass spectrometric analysis.
MS.
Electrospray ionization mass spectrometry (ESIMS) and MS/collisionally induced dissociation (CID)/MS were performed using a VG Quattro triple-quadrupole mass spectrometer. Samples for direct analysis were dissolved in 30 μl of a solvent of 1:1 (vol/vol) acetonitrile/water, made 0.3% with respect to formic acid (ACNF). Ten-microliter volumes were introduced into the mass spectrometer by loop injection using as carrier 50% acetonitrile in water at a flow rate of 10 μl/min. Alternatively, samples dissolved in 30 μl of 1:1 (vol/vol) methanol/water were analyzed by coupled, online microbore HPLC-ESIMS. Ten-microliter volumes were injected onto a 100 × 1 mm C18 column (Alltech) and fractionated with a gradient of 20–100% methanol made 0.1% (vol/vol) in formic acid at a flow rate of 30 μl/min. Tandem mass spectrometric analysis was performed using ultra-high-purity argon as the collision gas. High-resolution chemical ionization mass spectrometry (CIMS) was performed using a two-sector VG 70-VSE spectrometer at a resolution of 5000. One- or 2-μl volumes of sample, dissolved in ACNF, were introduced by direct insertion probe, vaporized by heating, and ionized using methane.
Acyl-HSL Standards.
Acyl-HSLs were synthesized as described by Zhang et al. (9) or were the gifts of D. Lynn (University of Chicago) and A. Eberhard (Ithaca College, Ithaca, NY). The preparation of N-butanoyl-HSL contained in addition a less polar bioactive compound of unknown structure. The synthetic standards were stored dry under nitrogen at −20°C. Standard stock solutions were prepared in HPLC-grade ethyl acetate.
Quantitation.
One-microliter volumes of sets of 2-fold dilutions of a given acyl-HSL standard along with samples from extracts prepared from a culture of the organism in question were chromatographed on the same plate. After development using the Agrobacterium reporter each blue spot resulting from the standard and test compounds was scanned using a Molecular Dynamics laser densitometer. Standard curves were constructed by plotting the measured parameter for each spot of the dilution series of the acyl-HSL standard against the log10 of the amount of standard chromatographed. The amount of the particular acyl-HSL present on the plate was determined from the standard curve for that compound, and the amount present in the culture was calculated from this value adjusted for the loading volume, and the dilution and concentration factors deriving from the culture extraction procedure described above. Concentrations are expressed as pmol/ml of culture at the time of harvest.
RESULTS
Separation and Detection of Acyl-HSLs by TLC.
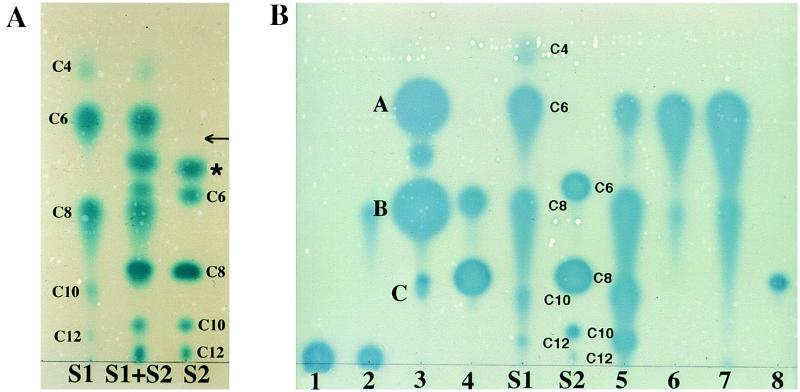
In preliminary experiments, we tested the usefulness of TLC to separate a mixture of synthetic acyl-HSL standards. The compounds were detected by overlaying the developed plate with a culture of an A. tumefaciens strain harboring a plasmid with a lacZ fusion to traG, a gene that requires the transcriptional activator TraR and an acyl-HSL for expression (6). This strain does not produce its own signal molecule, but it can induce the traG::lacZ reporter when supplied with an exogenous active acyl-HSL. Locations to which an active compound migrated on the plate yielded a blue zone resulting from hydrolysis of the X-Gal in the medium by the β-galactosidase expressed from the traG::lacZ reporter fusion. On C18 reversed-phase plates, of the solvent systems evaluated, methanol/water (60:40, vol/vol) separated each of the 10 standard compounds tested, even when all were present in a single sample (Fig. 1A). The A. tumefaciens reporter detected the 3-oxoacyl-HSLs with side chains ranging from C4 to C12 (Fig. 1A). The reporter also detected 3-unsubstituted-acyl-HSLs with side chains of 6 to 12 carbons, but it did not detect N-butanoyl-HSL at the amount tested (Fig. 1A). We did not have a full set of 3-hydroxy-substituted acyl-HSL standards. However, the A. tumefaciens reporter did respond to synthetic samples of N-(3-hydroxyoctanoyl)-l-HSL and N-(3-hydroxydecanoyl)-l-HSL (data not shown).
Figure 1.
TLC of acyl-HSLs. Samples were chromatographed on C18 reversed-phase thin-layer plates, developed with methanol/water (60:40, vol/vol), and the spots were visualized with the A. tumefaciens reporter strain. (A) Separation and detection of acyl-HSL standards. S1, 3-oxo-acyl-HSL standards; S2, 3-unsubstituted-acyl-HSL standards; S1 + S2, a mixture of the S1 and S2 standards. The acyl chain lengths are indicated for each compound. The asterisk marks an unknown active component. The arrow shows the position at which authentic N-butanoyl-l-HSL migrates. (B) Detection of acyl-HSLs produced by selected bacteria. Samples are from culture extracts of the following: 1, Rhizobium meliloti L5–30; 2, R. meliloti, YA2; 3, P. fluorescens 2-79; 4, Ralstonia solanacearum K60; 5, P. aeruginosa PAO1; 6, V. fischeri MJ1; 7, P. syringae, pv. tabaci 2024; and 8, Rhodobacter sphaeroides 2.4.1. Spots labeled A–C in lane 3 correspond to those compounds subjected to MS analysis as described in the text and in Fig. 3.
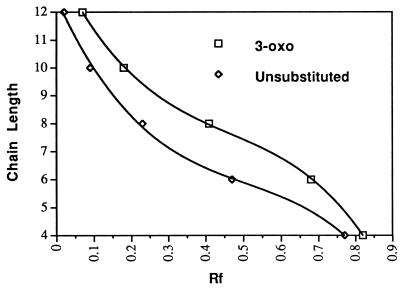
Each of the 3-oxo and 3-unsubstituted derivatives migrated with a characteristic Rf (Fig. 1A; Table 1) dependent on its acyl chain length and carbon 3 substitution (Fig. 2). In addition, the 3-oxo derivatives characteristically produced tailing spots with diffuse edges, whereas the 3-unsubstituted forms produced circular spots with sharp edges (Fig. 1). The 3-hydroxy-substituted compounds migrated with the same mobility as their 3-oxo analogs, but the spots did not tail.
Table 1.
Chromatographic properties of N-acyl-HSLs
| HSL | Rf* | Amount applied (in pmol) required
|
|
|---|---|---|---|
| For minimum detection | For routine detection† | ||
| 3-Oxo-C4 | 0.82 | NK | NK |
| 3-Oxo-C6 | 0.68 | 0.03 | 0.52 |
| 3-Oxo-C8 | 0.41 | 0.0005 | 0.03 |
| 3-Oxo-C10 | 0.18 | 0.19 | 3 |
| 3-Oxo-C12 | 0.07 | 2 | 16 |
| 3-Unsubstituted C4 | 0.77 | ND | ND |
| 3-Unsubstituted C6 | 0.47 | 300 | 1200 |
| 3-Unsubstituted C8 | 0.23 | 2.4 | 38 |
| 3-Unsubstituted C10 | 0.09 | 100 | 400 |
| 3-Unsubstituted C12 | 0.02 | NK | NK |
NK, not known. Amounts available were insufficient to determine weights. ND, not detected.
Rf values differ slightly from one experiment to the next. The values given represent migration rates taken from a single experiment in which all of the standards were chromatographed on the same plate.
Values represent amounts of the indicated N-acyl-HSL required to produce an easily detectable reaction with the A. tumefaciens indicator strain (see Fig. 1).
Figure 2.
Relationship between acyl side-chain length and chromatographic mobility. Rf values were determined following chromatography and detection.
Sensitivity of the Detection System.
The A. tumefaciens reporter strain responded to as little as 0.5 fmol of N-(3-oxooctanoyl)-l-HSL, the cognate signal molecule of the tra system, present on the chromatogram, while the other acyl-HSL species tested required substantially higher amounts for detection (Table 1). The limits of detection, as well as the amounts of each of the standard compounds required to give easily detectable spots with the A. tumefaciens reporter following chromatography, are presented in Table 1.
Analysis of Acyl-HSL Species Produced by Bacteria.
We tested the usefulness of the TLC assay by analyzing extracts prepared from culture supernatants of eight Gram-negative bacteria. Four of these, P. aeruginosa, V. fischeri, and the two Rhizobium meliloti isolates, have been reported to produce acyl-HSLs of known or undetermined structure (8, 12–15), while the others have not been characterized. The TLC assay detected at least one active compound in the supernatants of all eight cultures (Fig. 1B). Six of the organisms produced several active compounds, with P. aeruginosa PAO1 and P. fluorescens 2-79 yielding a minimum of four and five detectable compounds, respectively.
The four species detected in extracts from P. aeruginosa chromatographed with characteristics indistinguishable from those of 3-oxo derivatives with acyl chain lengths of 6, 8, 10, and 12 carbons (Fig. 1B, lane 5). The two Rhizobium meliloti strains produced active species that were strongly nonpolar, remaining close to the origin (Fig. 1B, lanes 1 and 2). Rhizobium meliloti YA2 produced a second active species that comigrated with, and had the tailing properties of, N-(3-oxooctanoyl)-l-HSL. Ralstonia solanacearum produced two active compounds, the more nonpolar of which could be N-octanoyl-l-HSL (Fig. 1B, lane 4). The more polar compound migrated with an Rf similar to that of N-(3-oxooctanoyl)-l-HSL. However the compound did not tail on the chromatogram, suggesting that it is not a 3-oxo derivative. Rhodobacter sphaeroides produced a single detectable species having the mobility and spot characteristics of N-octanoyl-l-HSL (Fig. 1B, lane 8).
V. fischeri and P. syringae pv. tabaci each produced two detectable compounds which could be N-(3-oxohexanoyl)-l-HSL and N-(3-oxooctanoyl)-l-HSL (Fig. 1B, lanes 6 and 7). ESIMS analysis of the putative N-(3-oxohexanoyl)-l-HSL produced by P. syringae pv. tabaci, eluted from a preparative TLC plate as described in Materials and Methods, gave a parent ion signal at m/z 214, precisely that obtained with a sample of synthetic N-(3-oxohexanoyl)-l-HSL (data not shown). MS/MS analysis of this compound produced the HSL daughter ion with m/z 102, and additional ion fragments from the acyl side chain. The spectrum was indistinguishable from that of the N-(3-oxohexanoyl)-l-HSL standard (data not shown).
Analysis of Unknown Acyl-HSLs by Preparative TLC and MS.
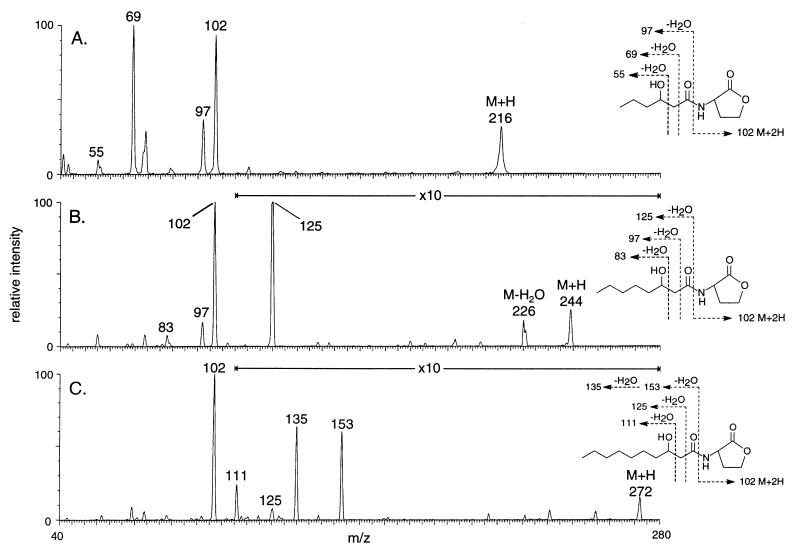
Only one of the five detectable species produced by P. fluorescens strain 2-79 showed chromatographic characteristics similar to those of our standards (Fig. 1B, lane 3). This low-activity species could be N-octanoyl-l-HSL. The two compounds with greatest activities migrated with N-(3-oxohexanoyl)-l-HSL and N-(3-oxooctanoyl)-l-HSL, but the spots did not tail. MS analysis of material recovered from the TLC plates yielded prominent ions at m/z 216 and m/z 244 for the more polar and the less polar compounds, respectively (Fig. 3 A and B). High-resolution CIMS analysis of these two compounds gave mass values of 216.1247, consistent with the formula C10H18NO4, and 244.1551, corresponding to the formula C12H22NO4. These match the formulae for N-(3-hydroxyhexanoyl)-l-HSL and N-(3-hydroxyoctanoyl)-l-HSL. MS/MS analysis of the two parent ions each generated a peak for the HSL at m/z 102 (Fig. 3 A and B) and a series of peaks from fragmentations in the acyl side chains (Fig. 3 A and B). The m/z values for each of these indicate a loss of water, which characteristically occurs during MS/MS analysis of compounds with hydroxyl groups (28). Moreover, the MS/MS spectrum of the putative N-(3-hydroxyoctanoyl)-l-HSL was indistinguishable from that of the synthetic standard (data not shown).
Figure 3.
Tandem MS analysis of active compounds from an extract of a culture of P. fluorescens 2-79. Samples recovered from a preparative TLC plate were analyzed by MS/CID/MS. The spectra are from samples corresponding to the three spots labeled A, B, and C in lane 3 of Fig. 1B.
The material recovered from the region of the preparative TLC plate corresponding to the low-activity doublet spot migrating below N-(3-hydroxyoctanoyl)-l-HSL (Fig. 1B, lane 3) gave a parent ion peak at m/z 272 (Fig. 3C). Tandem MS analysis of this peak produced the HSL ion at m/z 102 and additional peaks derived from the acyl side chain as shown in Fig. 3C. Although there was not enough material available for high-resolution CIMS analysis, the parent ion (M + H) at m/z of 272 and the fragmentation pattern produced by MS/MS is consistent with a molecular structure of N-(3-hydroxydecanoyl)-l-HSL (Fig. 3C).
Quantifying Acyl-HSLs in Culture Supernatants by TLC.
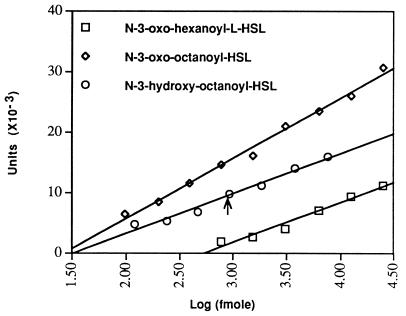
As measured by laser densitometry, the response of the A. tumefaciens indicator strain to decreasing amounts of a set of standards of N-(3-oxooctanoyl)-l-HSL, of N-(3-oxohexanoyl)-l-HSL, and of N-(3-hydroxyoctanoyl)-l-HSL present on the plates was linear over concentration ranges of at least two orders of magnitude (Fig. 4). Similar results were obtained by plotting the diameter of each spot versus the log10 of the concentration (data not shown).
Figure 4.
Quantitation of acyl-HSLs. Samples of a series of 2-fold dilutions of the indicated acyl-HSL, along with a sample of an extract from a culture of P. fluorescens 2-79 were chromatographed on a single plate. Following detection, the blue spots were scanned by densitometry and the values for each of the standards, expressed in arbitrary units, were plotted against the amount of the acyl-HSL in each spot. The arrow marks the location to which the densitometric value for the corresponding spot from the P. fluorescens extract intersects with the standard curve derived for authentic N-(3-hydroxyoctanoyl)-l-HSL.
We used this assay to quantify the amount of N-(3-hydroxyoctanoyl)-l-HSL present in culture supernatants of P. fluorescens 2-79 grown in AB minimal medium to late exponential phase. Fitting the densitometric value for the spot from the culture extract to the curve prepared from densitometric scans of a dilution series of the standard (Fig. 4) yielded a value of 18 pmol of the acyl-HSL per ml of culture.
DISCUSSION
Separation by TLC coupled with biodetection gives a direct, visual catalog of the acyl-HSL signal molecules, detectable by the A. tumefaciens indicator, present in supernatants of bacterial cultures. The assay is sensitive (Table 1), generally requiring extracts from cultures no larger than 5 ml. Moreover, it is simple, reasonably fast, amenable to processing many samples, and does not require elaborate or expensive instrumentation. Using this system, cultures of any cultivatable bacterium can be assayed for detectable acyl-HSL signal molecules in 2–3 days of work.
The assay must be used with caution. First, detection is limited to those acyl-HSLs to which the bioindicator will respond. In this regard, the A. tumefaciens traG::lacZ/traR reporter detects 3-oxo-substituted HSL derivatives with acyl chain lengths from 4 to 12 carbons as well as 3-hydroxy derivatives with acyl chain lengths of 6, 8, and 10 carbons. The reporter also can detect signal molecules with acyl chain lengths longer than 12. Extracts from cultures of the two Rhizobium meliloti strains tested contained an active compound that chromatographed with a mobility slower than that of either C12 standard. While the structures of these molecules are unknown, their presence is consistent with a report by Gray et al. (12) describing a highly nonpolar acyl-HSL activity in culture supernatants of R. meliloti strain Rm1021.
Second, the signal must be present at levels detectable by the reporter. The R proteins are highly specific for their cognate acyl-HSL signals but will respond to analogs if present at sufficiently high concentrations (9, 13, 17–19). Consistent with this, members of each carbon 3 substitution series were detected with lessening sensitivity the farther they diverged from the C8 chain length of the cognate Agrobacterium signal (Table 1). While it is extremely sensitive to the 3-oxo derivatives, and to the one 3-hydroxy derivative for which we have quantitative data (Table 1, Fig. 4), the A. tumefaciens reporter responds less well to most of the 3-unsubstituted acyl-HSLs, and it does not respond at all to N-butanoyl-l-HSL. The use of reporters that respond optimally to other acyl-HSL signals could extend the range as well as improve the sensitivity of the assay to certain of these signal molecules. However, in our hands, the A. tumefaciens strain is the most versatile single reporter available.
Third, although the shapes of the spots differ, the 3-oxo- and the 3-hydroxy derivatives of the same chain length migrate with indistinguishable mobilities in the methanol/water solvent system. In an extract containing both substitution types having the same acyl chain length, the presence of one, especially at amounts that result in high reporter activity, will mask that of the other. Moreover, a strongly active compound of one type may obscure the presence of a weakly active species that migrates with a similar but nonidentical Rf.
Finally, one cannot conclude from the absence of a signal that the tested bacterium does not produce one or more acyl-HSLs. Such organisms may produce signals that are not detectable by the reporter, or they may produce detectable molecules at levels below the threshold of the reporter. Modulation of acyl-HSL production by other regulatory circuits, as well as by the medium and culture conditions used to grow the bacteria being tested, also may influence whether these signal molecules are produced in amounts sufficient for detection by the reporter. For these reasons, the assay gives a minimum estimate of the complement of signal molecules that can be produced by a given microorganism.
Nevertheless, the TLC assay is well suited to screening even complex acyl-HSL complements produced by bacteria. As an example, the assay detected activities with chromatographic properties corresponding to N-(3-oxohexanoyl)-, N-(3-oxooctanoyl)-, N-(3-oxodecanoyl)-, and N-(3-oxododecanoyl)-l-HSL in extracts from P. aeruginosa (Fig. 1B, lane 5). These represent four of the six acyl-HSLs confirmed or inferred to be produced by this bacterium (10, 14–16). P. aeruginosa also produces N-butanoyl- and N-hexanoyl-l-HSL (15, 16). The A. tumefaciens reporter does not respond to the butanoyl derivative, and given its low responsiveness to the other 3-unsubstituted acyl-HSLs, the failure to detect N-hexanoyl-l-HSL in these extracts is not surprising. Most likely this signal is not produced at levels sufficient for detection by the A. tumefaciens reporter. Similarly, the assay detected activities with the chromatographic properties of two acyl-HSLs: N-(3-oxohexanoyl)-l-HSL and N-octanoyl-l-HSL, known to be produced by V. fischeri MJ1 (refs. 8 and 13; Fig. 1B, lane 6; and data not shown). We also detected an unreported activity which probably is N-(3-oxooctanoyl)-l-HSL. On the basis of the intensity of the spot, we estimate that the putative N-(3-oxooctanoyl)-l-HSL species is present in nanomolar amounts in culture supernatants of V. fischeri. Given that this molecule is the cognate for TraR, this level is easily detectable by the A. tumefaciens reporter (Table 1). However, it probably is below the level of detection by the lux reporter systems used in the previous studies (8, 13). We did not detect N-hexanoyl-l-HSL, the third known signal molecule of V. fischeri (13), in these preparations. Kuo, et al. (13) reported that, on an activity basis, this acyl-HSL is the least abundant signal in cultures of V. fischeri MJ1, and most likely it was present in the extracts at a concentration below the level of detection by the A. tumefaciens reporter.
Although the signal molecules cannot be identified unambiguously by TLC, their chromatographic properties can be used, in many instances, to assign tentative structures. Rf and spot shape predicted that the most active species produced by P. syringae pv. tabaci is N-(3-oxohexanoyl)-l-HSL. This was confirmed by MS (data not shown). Moreover, the TLC assay predicted that four of the five active compounds produced by P. fluorescens 2-79 were distinct from any of our standards. MS confirmed this, identifying three of the compounds as the previously unreported species, N-(3-hydroxyhexanoyl)-l-HSL, N-(3-hydroxyoctanoyl)-l-HSL, and N-(3-hydroxydecanoyl)-l-HSL (Fig. 3). These structural analyses also confirm that the traG::lacZ/traR reporter system responds to a range of 3-hydroxy-substituted acyl-HSLs. The N-(3-hydroxydecanoyl)-l-HSL forms a doublet with another active species (Fig. 1B, lane 3). While there was not enough of this material for analysis by tandem MS, the spot gave a peak at m/z 228 (M + H) which could correspond to N-octanoyl-l-HSL. This assignment is consistent with chromatographic behavior; the compound migrated with an Rf and spot shape indistinguishable from those of the N-octanoyl-l-HSL standard.
Like the TLC assay, current methods for quantifying acyl-HSLs use bioreporters, and they rely on comparing the inducer activity of a sample fractionated from a culture extract with that of a standard of the same compound at known concentration (29, 30). The TLC system simplifies and extends this assay by allowing the cochromatography of a dilution series of a set of standards, each at a known concentration, on the same assay plate as the unknown. This generates a series of standard curves, one for each of the known compounds. The amount of each of the compounds present in the culture sample can be determined by fitting the value of the intensity of the corresponding spot on the chromatogram to the appropriate curve. That the curves generated for three tested acyl-HSLs are linear over concentration ranges of two to three orders of magnitude provides considerable latitude for quantifying the corresponding signal molecules present in samples being tested.
In addition to analyzing acyl-HSLs produced by bacteria, the TLC assay can be used for monitoring the purification of these compounds. Moreover, preparative TLC can be used to fractionate these compounds prior to MS analysis. The assay also is well suited for assessing the patterns of acyl-HSL production during growth of bacteria, and for quantifying the levels of these molecules present in culture supernatants at different culture stages. This could be especially useful in monitoring such changes in cultures of bacteria that express more than one independent acyl-HSL signaling system.
Acknowledgments
We thank Ryan M. Farrand for expert graphics assistance. This work was supported by Grant R01 GM 52465 from the National Institutes of Health to S.K.F., and J.E.C. was supported by Grant R37 AI 15650 from the National Institues of Health. G.P. and S.L.D. were supported by a grant from the University of Illinois at Urbana–Champaign Research Board.
ABBREVIATIONS
- HSL
homoserine lactone
- AI
autoinducer
- ESIMS
electrospray ionization mass spectrometry
- CIMS
chemical ionization mass spectrometry
- CID
collisionally induced dissociation
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactopyranoside
Note Added in Proof
The Rf of authentic N-butanoyl-l-HSL (Fig. 1A, Table 1) was determined using Chromobacterium violaceum CV026 as the indicator. This strain responds strongly to 3-unsubstituted N-acyl-HSLs with chain lengths of 4, 6, and 8 carbons (ref. 31 and our results not shown).
References
- 1.Fuqua W C, Winans S C, Greenberg E P. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 2.Dunlap P V, Greenberg E P. In: Microbial Cell–Cell Interactions. Dworkin M, editor. Washington, DC: Am. Soc. Microbiol. Press; 1991. pp. 219–253. [Google Scholar]
- 3.Kaplan H B, Greenberg E P. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanzelka B L, Greenberg E P. J Bacteriol. 1995;177:815–817. doi: 10.1128/jb.177.3.815-817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 6.Piper K R, Beck von Bodman S, Farrand S K. Nature (London) 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 7.Fuqua W C, Winans S C. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberhard A, Burlingame A L, Eberhard C, Kenyon E L, Nealson K H, Oppenheimer N J. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Murphy P J, Kerr A, Tate M E. Nature (London) 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]
- 10.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao J-G, Meighan E A. J Biol Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- 12.Gray K M, Pearson J P, Downie J A, Boboye B E A, Greenberg E P. J Bacteriol. 1996;178:372–376. doi: 10.1128/jb.178.2.372-376.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo A, Callahan S M, Dunlap P V. J Bacteriol. 1996;178:971–976. doi: 10.1128/jb.178.4.971-976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson J P, Passador L, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winson M K, Camara M, Latifi A, Foglino M, Chhabra S R, Bally M, Chapon V, Salmond G P C, Bycroft B W, Lazdunski A, Stewart G S A B, Williams P. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Throup J P, Camara M, Briggs G S, Winson M K, Chhabra S R, Bycroft B, W, Williams P, Stewart G S A B. Mol Microbiol. 1995;17:345–356. doi: 10.1111/j.1365-2958.1995.mmi_17020345.x. [DOI] [PubMed] [Google Scholar]
- 17.Eberhard A, Widrig C A, McBath P, Schineller J B. Arch Microbiol. 1986;146:35–40. doi: 10.1007/BF00690155. [DOI] [PubMed] [Google Scholar]
- 18.Shaefer A L, Hanzelka B L, Eberhard A, Greenberg E P. J Bacteriol. 1996;178:2897–2901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Passador L, Tucker K D, Guertin K R, Journet M P, Kende A S, Iglewski B H. J Bacteriol. 1996;178:5995–6000. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engebrecht J, Nealson K, Silverman M. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 21.Hwang I, Li P-L, Zhang L, Piper K R, Cook D C, Tate M E, Farrand S K. Proc Natl Acad Sci USA. 1994;91:4639–4643. doi: 10.1073/pnas.91.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swift S, Winson M K, Chan P F, Bainton N J, Birdsall M, Reeves P J, Rees C E D, Chhabra S R, Hill P J, Throup J P, Bycroft B W, Salmond G P C, Williams P, Stewart G S A B. Mol Microbiol. 1993;10:511–520. doi: 10.1111/j.1365-2958.1993.tb00923.x. [DOI] [PubMed] [Google Scholar]
- 23.Cook D M, Li P-L, Hwang I, Ruchaud F, Padden S, Farrand S K. J Bacteriol. 1997;179:1291–1297. doi: 10.1128/jb.179.4.1291-1297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck von Bodman S, Farrand S K. J Bacteriol. 1995;177:5000–5008. doi: 10.1128/jb.177.17.5000-5008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel H J, Bonner D M. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 26.Greenberg E P, Hastings J W, Ulitzur S. Arch Microbiol. 1979;120:87–91. [Google Scholar]
- 27.Kiss G B, Vincze E, Kalmar Z, Forrai T, Kondorosi A. J Gen Microbiol. 1979;113:105–118. [Google Scholar]
- 28.McLafferty F W, Turecek F. Interpretation of Mass Spectra. 4th Ed. Mill Valley, CA: University Science Books; 1993. p. 77. [Google Scholar]
- 29.Eberhard A. J Bacteriol. 1972;109:1101–1105. doi: 10.1128/jb.109.3.1101-1105.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adar Y Y, Ulitzur S. J Biolumin Chemilumin. 1993;8:261–266. doi: 10.1002/bio.1170080506. [DOI] [PubMed] [Google Scholar]
- 31.Milton D L, Hardman A, Camara M, Chkabra S R, Bycroft B W, Stewart G S A B, Williams P. J Bacteriol. 1997;179:3004–3012. doi: 10.1128/jb.179.9.3004-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]