Abstract
H2A.Z is a histone H2A variant that is essential for viability in organisms such as Tetrahymena thermophila, Drosophila melanogaster, and mice. In Saccharomyces cerevisiae, loss of H2A.Z is tolerated, but proper regulation of gene expression is affected. Genetics and genome-wide localization studies show that yeast H2A.Z physically localizes to the promoters of genes and functions in part to protect active genes in euchromatin from being silenced by heterochromatin spreading. To date, the function of H2A.Z in mammalian cells is less clear, and evidence so far suggests that it has a role in chromatin compaction and heterochromatin silencing. In this study, we found that the bulk of H2A.Z is excluded from constitutive heterochromatin in differentiated human and mouse cells. Consistent with this observation, analyses of H2A.Z- or H2A-containing mononucleosomes show that the H3 associated with H2A.Z has lower levels of K9 methylation but higher levels of K4 methylation than those associated with H2A. We also found that a fraction of mammalian H2A.Z is monoubiquitylated and that, on the inactive X chromosomes of female cells, the majority of this histone variant is modified by ubiquitin. Finally, ubiquitylation of H2A.Z is mediated by the RING1b E3 ligase of the human polycomb complex, further supporting a silencing role of ubiquitylated H2A.Z. These new findings suggest that mammalian H2A.Z is associated with both euchromatin and facultative heterochromatin and that monoubiquitylation is a specific mark that distinguishes the H2A.Z associated with these different chromatin states.
While many studies have linked the function of H2A.Z to transcriptional regulation, its exact function in this process has remained enigmatic, since abundant evidence supports both positive and negative roles for this histone variant in transcription (reviewed in reference 14). Much of our current understanding comes from studies of the H2A.Z homolog Htz1 of Saccharomyces cerevisiae. Loss of Htz1 leads to defects in transcriptional activation of the PHO5 and GAL1 genes, as well as disruption of the transcriptional silencing of the HMR and telomeric loci of this organism (9, 30). Global gene expression profiling of Htz1 deletion mutants suggests that this variant functions to counteract the spreading of Sir2-mediated heterochromatin-type silencing (21). Studies combining chromatin immunoprecipitation and microarray analysis indicate that Htz1 is specifically localized to the 5′ end of genes, and furthermore, Htz1 is preferentially associated with inactive genes (13, 18, 26, 36). Interestingly, deletion of Htz1 does not affect transcriptional silencing of the associated genes but instead disrupts proper expression of those silenced genes under activating conditions. Taken together, these data suggest that Htz1 functions to maintain regulated promoters in a chromatin state that is poised and compatible with transcription.
In contrast to the yeast studies, in vitro and in vivo analyses of H2A.Z function in complex organisms have yielded confounding results. Structural and biophysical analyses have found both stabilizing and destabilizing effects of H2A.Z on the nucleosome structure as well as on the stability of oligonucleosome arrays (1, 10, 23, 33). In Tetrahymena thermophila, the H2A.Z equivalent, hv1, is found exclusively in the transcriptionally active macronucleus but not in the transcriptionally silent micronucleus (32). Drosophila melanogaster polytene chromosome staining shows a nonrandom distribution of the fly H2A.Z homolog, H2AvD, on both euchromatic and heterochromatic regions (17). In addition, chromatin immunoprecipitation assays show that H2AvD is localized to transcribing and nontranscribing genes. In trophoblasts and endoderm cells of mouse embryos, H2A.Z is enriched at the pericentric heterochromatin but is depleted on the transcriptionally silenced inactive X chromosome (27). In vivo cross-linking studies also show that H2A.Z associates with HP1α, a resident protein that marks constitutive heterochromatin in mammalian cells, and in vitro studies using reconstituted nucleosome arrays indicate that H2A.Z and HP1α can synergize to promote chromatin compaction (11). RNA interference (RNAi) studies have suggested that H2A.Z has a role in chromosome segregation (28), a function that may be specifically associated with the fraction of H2A.Z detected at centromeres (12). So far, these studies have focused on the structural functions of mammalian H2A.Z. In this study, we focused on the epigenetic aspects of mammalian H2A.Z and found that it is associated with both euchromatin and facultative heterochromatin. For example, in contrast to mouse embryonic cells, H2A.Z is depleted at pericentric heterochromatin in differentiated mouse and human cells. Consistent with that finding, H2A.Z-containing nucleosomes are enriched for K4-methylated H3 and are reduced for K9-methylated H3 compared to the methylation levels of nucleosomes containing H2A. We also found that a fraction of H2A.Z is monoubiquitylated at the C terminus and that H2A.Z located on the transcriptionally silent inactive X chromosome of female cells is mostly ubiquitylated. These data show a conservation of H2A.Z's function in transcriptional regulation in yeast and mammalian cells and that ubiquitylation of H2A.Z distinguishes the fraction of this variant that is associated with facultative heterochromatin from the euchromatin-associated H2A.Z.
MATERIALS AND METHODS
Cell culture, whole-cell lysis, and antibodies.
All cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Transfections were done using Lipofectamine 2000 (Invitrogen), and total cell lysates were obtained by directly lysing washed and pelleted cells in 2× sample buffer. The H2A.Z-specific antibodies were generated in rabbits against the two contiguous regions on H2A.Z most divergent from the canonical H2A: the H2A.Z-L1 antibody was raised against the peptide sequence KSSRTTSHGR, and the H2A.Z-C antibody was raised against the peptide sequence IPHIHKSLIGKKGQQKTV. Polyclonal antibodies against tri-MeK4 (trimethylation of lysine 4 of histone H3), di-MeK4, tri-MeK9, and di-MeK9 and monoclonal antibodies against HP1α and tri-MeK27 H3 were from Upstate Biotechnologies, Inc. The monoclonal antibody against di-MeK4/tri-MeK4 and the general H3 antibody were from Abcam. The Flag M2 antibody was from Sigma, the monoclonal hemagglutinin (HA) antibody was from Covance, and the Ring1b antibody was from MBL International.
RNAi studies.
The RNAi target sequences against H2A.Z and RING1b were based on previous publications that showed successful knockdown of the target proteins (10, 34). The RNAi target sequences, as well as the scrambled control sequences, were cloned in the pSuper vector (Oligoengine), and these constructs express short hairpin RNAs (shRNAs) when transfected into mammalian cells. For all RNAi knockdown experiments, the analyses were done 72 h after transfection of the pSuper constructs into 293T cells.
Immunofluorescence analyses.
Interphase and metaphase chromosome immunofluorescence microscopies were done as described previously (4). For metaphase spreads, cells were incubated with 0.1 to 0.2 μg/ml Colcemid (Invitrogen), and mitotic cells were harvested by mechanical shock. After cells were washed twice in phosphate-buffered saline (PBS), cells were swollen in 0.075 M KCl for 8 min at 37°C. Cells were applied to glass slides by centrifugation at 2,000 rpm for 5 min using a cytospin II (Shandon). Slides were incubated in KCM (120 mM KCl, 20 mM NaCl, 10 mM Tris, pH 8, 0.5 mM EDTA, and 0.1% Triton) for 15 min prior to the application of primary antibodies. Both primary and secondary antibodies were incubated for 1 h at 37°C, and following each incubation the slides were washed in KCM three times. The chromosomes were fixed in 3.7% formaldehyde for 10 min, counterstained with 4′,6′-diamidino-2-phenylindole (DAPI), and mounted using VectaShield (Vector Laboratories). Images were acquired using a Leica DM microscope and camera.
Expression of tagged H2A.Z.
For this study, H2A.Z was tagged by several different methods and was cloned into either pLNCX2 (Clontech) or pcDNA3.1 (Invitrogen). For green fluorescent protein (GFP) tagging, the GFP coding sequence was fused in frame to the 3′ end of the H2A.Z coding sequence, as described previously (15). For Flag or Avi tagging, the coding sequences for the tags were fused in frame to either the 5′ or 3′ end of the H2A.Z cDNA. The Avi tag refers to a 15-amino-acid tag that contains a biotinylation site for the Escherichia coli BirA enzyme. When cotransfected in mammalian cells, the BirA enzyme biotinylates the tagged H2A.Z in vivo, which facilitates detection and purification of the biotinylated H2A.Z based on the strong interaction between biotin and avidin (7).
Mononucleosome immunoprecipitation.
293T cells were seeded onto 150-mm-diameter plates and were transfected with pcDNA vector alone or with constructs that express Flag-tagged H2A or H2A.Z. Transfected cells were trypsinized and washed two times with PBS. Cellular pellets were washed once with buffer A (20 mM HEPES, pH 7.35, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 10 mM N-ethylmaleimide [NEM], 1 mM dithiothreitol, and 1 mM phenylmethanesulfonyl fluoride), resuspended in buffer A containing 0.2% Triton X-100, and then incubated on ice for 5 min. The resulting nuclear suspension was centrifuged at 600 × g, and the nuclei were resuspended in buffer A without Triton X-100 and without NEM. The content of chromatin was estimated by diluting a small amount of this nuclear suspension (1:40) into 0.1% sodium dodecyl sulfate (SDS) and reading the absorbance (1 A260 unit is 1 mg/ml). Nuclei were then resuspended in buffer A containing 2 mM CaCl2 (without Triton X-100 and NEM), and chromatin digestion was carried out with microccocal nuclease (MNase; Worthington) at 37°C for 30 min with 100 U/mg of chromatin in a volume of 400 μl. Digestion of chromatin by MNase was stopped by addition of EGTA to a final concentration of 1 mM. The resulting MNase-digested nuclei were centrifuged at 600 × g, resuspended in nuclear extraction buffer (20 mM HEPES, pH 7.35, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EGTA), left on ice for 1 h, and then centrifuged (1,000 × g for 5 min at 4°C) to remove nuclear debris. The salt concentration of the supernatant was then adjusted to 150 mM NaCl by addition of dilution and equilibration buffer (20 mM HEPES, pH 7.35, 1.5 mM MgCl2, 0.2 mM EGTA, and 25% glycerol) drop-wise while the supernatant was vortexed. The resulting H1-chromatin-enriched precipitate was cleared by centrifugation, and the resulting suspension containing mononucleosomes was used for immunoprecipitation.
For immunoprecipitation, M2-agarose beads (Sigma A2220; 5 μl bed volume/immunoprecipitation) were added to mononucleosome preparations, and the solution was incubated overnight at 4°C. Beads were washed eight times in buffer D (20 mM HEPES, pH 7.35, 150 mM NaCl, 1.5 mM MgCl2, 0.2 mM EGTA, 0.2% Triton X-100, and 10% glycerol) and then were eluted by being boiled in SDS-polyacrylamide gel electrophoresis sample buffer without reducing agent (100 mM Tris, pH 6.8, 4% SDS, and 20% glycerol). Eluted proteins were then removed with a Hamilton syringe fitted with a 27-gauge needle. Prior to boiling the samples, 1 mM dithiothreitol was added, and the samples were run on SDS-polyacrylamide gel electrophoresis gels for Western blot transfer according to standard practices.
RESULTS
Generation and characterization of H2A.Z-specific antibodies.
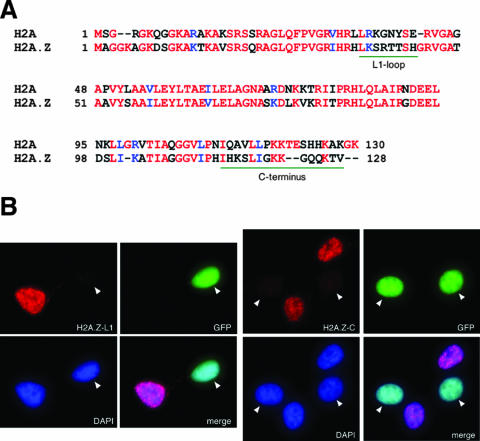
To investigate H2A.Z functions in mouse and human cells, we have generated and affinity purified two separate antibodies directed against the two regions most divergent between mammalian H2A.Z and H2A (Fig. 1A). The H2A.Z-L1 antibody is directed against the L1 loop of H2A.Z, whereas the H2A.Z-C antibody is directed against the C terminus of the protein. Since the amino acid sequences of human and mouse H2A.Z are identical, these antibodies recognize H2A.Z from both organisms. To confirm the specificities of the antibodies, we transfected constructs that express shRNAs against H2A.Z into 293T cells and examined the detection of the transfected and untransfected cells by these antibodies using immunofluorescence staining. To mark the cells that were transfected with the shRNA constructs, we also cotransfected an H2B-GFP expression construct so that the transfected (H2A.Z knocked down) cells would fluoresce green upon UV excitation. As shown in Fig. 1B, the H2A.Z-L1 and H2A.Z-C antibodies detected only the untransfected 293T cells but not the transfected cells, indicating that they specifically recognize epitopes on the endogenous H2A.Z. Furthermore, Western blot analyses showed that both H2A.Z-L1 and H2A.Z-C antibodies specifically detected H2A.Z-GFP, but not H2A-GFP, fusion proteins (see Fig. 4E). Together, these studies confirm that the H2A.Z antibodies are highly specific for H2A.Z and do not cross-react with the canonical H2A core histone by Western blot analysis or immunofluorescence assays.
FIG. 1.
The H2A.Z-L1 and H2A.Z-C antibodies are specific for H2A.Z. (A) Amino acid sequence alignment of mammalian H2A and H2A.Z. The two most divergent regions, to which the H2A.Z antibodies were raised, are underlined in green. (B) An shRNA construct that targets H2A.Z mRNA is cotransfected with a construct that expresses H2B-GFP into 293T cells. Transfected (green fluorescing) and nontransfected (nongreen) cells were examined by immunofluorescence using the H2A.Z-L1 and H2A.Z-C antibodies. Both antibodies stain the untransfected but not the transfected cells, indicating that both antibodies recognize the endogenous H2A.Z but not H2A.
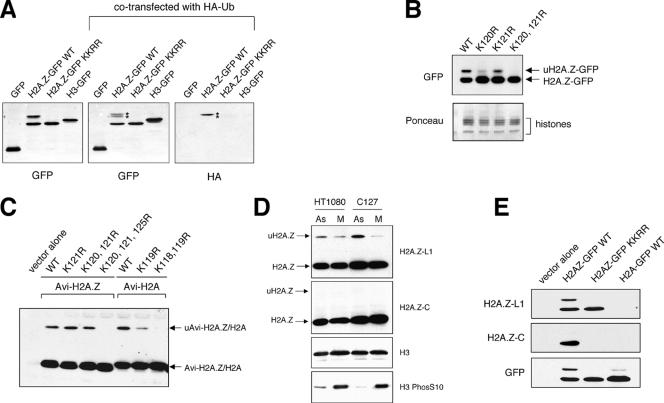
FIG. 4.
Endogenous H2A.Z and transfected H2A.Z are monoubiquitylated in vivo. (A) 293T cells were transfected with constructs that express GFP, H3-GFP, wild-type (WT) H2A.Z-GFP, or mutant (KKRR) H2A.Z-GFP, in which K120 and 121 were mutated to arginines. For the samples shown at the center and right panels, the HA-ubiquitin (HA-Ub) expression construct was cotransfected with the GFP or histone-GFP construct. Western blotting with the GFP antibody shows that a fraction of H2A.Z-GFP migrates slower than the bulk of H2A.Z-GFP (marked by asterisks). Blotting with the HA antibody indicates that this slower-migrating band represents the ubiquitylated form of H2A.Z-GFP (uH2A.Z-GFP). In contrast, neither H3-GFP nor the mutant H2A.Z-GFP is monoubiquitylated. (B) Mapping of the sites of monoubiquitylation by mutagenesis. Expression constructs for the wild type or K-to-R mutants of H2A.Z-GFP were transfected into 293T cells and detected by Western blotting using a GFP antibody. Ponceau-stained histones were used as loading controls. (C) Transfection studies using various point mutants of N-terminally tagged H2A.Z show that K120, K121, and K125 all can be used as monoubiquitylation sites in vivo. In contrast, only K118 and K119 of H2A are sites of monoubiquitylation. (D) Total cell extracts were harvested from asynchronously growing (As) or mitotically arrested (M) human HT1080 or mouse C127 cells, and the endogenous histone levels were examined by Western blot analyses. While the H2A.Z-L1 antibody detected both the unmodified H2A.Z and uH2A.Z, the H2A.Z-C antibody detected only the unmodified H2A.Z. Total H3 was used as a loading control, and the increased levels of phosphorylated H3 were used as an indication of cells arrested at mitosis. (E) 293T cells were transfected with the vector control or plasmids that express wild-type H2A.Z-GFP, the K-to-R mutant of H2A.Z-GFP, or wild-type H2A-GFP, and whole-cell extracts were examined by Western blotting using H2A.Z-L1, H2A.Z-C, or GFP antibody.
H2A.Z is excluded from constitutive heterochromatin in differentiated mouse and human cells.
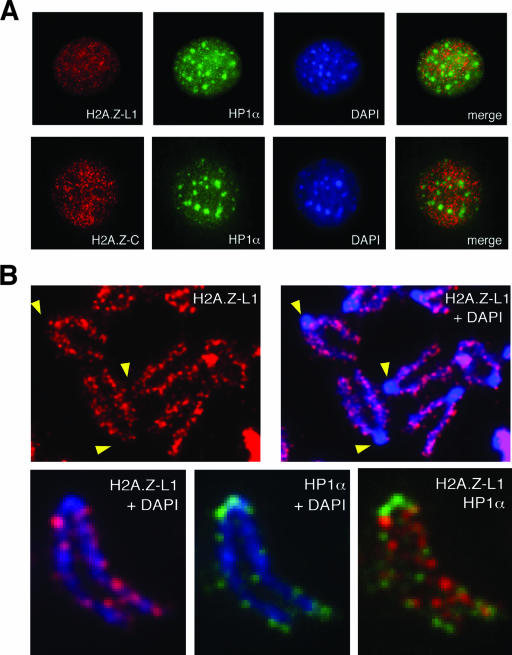
Immunofluorescence staining of interphase mouse fibroblasts using either the L1- or C-terminal antibody shows that H2A.Z is distributed across the nucleus in a punctate pattern but is excluded from the DAPI-dense pericentric heterochromatin (Fig. 2A). This exclusion is also seen when H2A.Z is costained with HP1α protein (a major component of heterochromatin) and is especially evident in metaphase chromosome spreads (Fig. 2B). In human cells, the clustering of heterochromatin is not as distinct as in mouse cells. Nevertheless, the mostly mutually exclusive distribution of H2A.Z and HP1α is also seen in the IMR90 human fibroblasts (data not shown). These findings are the opposite of those of the previously observed enrichment of H2A.Z at the pericentric heterochromatin in mouse embryonic trophoblast cells (27), but they are consistent with the reported exclusion of H2A.Z in tri-MeK9 H3-enriched regions in differentiated mouse cells (5), since trimethylation of K9 on H3 is a hallmark of constitutive pericentric heterochromatin (2, 16).
FIG. 2.
H2A.Z is excluded from pericentric heterochromatin in differentiated mouse fibroblasts. (A) Mouse 10T1/2 fibroblasts were stained using antibodies against H2A.Z and HP1α and were examined by immunofluorescence microscopy. Both H2A.Z antibodies show punctate nuclear staining but are excluded from the DAPI-dense and HP1α-enriched pericentric heterochromatin. (B) Metaphase chromosome spreads from mouse 10T1/2 cells show the exclusion of H2A.Z from the pericentric heterochromatin. The mutually exclusive localization of H2A.Z and HP1α is seen both at the pericentric heterochromatin and on the chromosome arms.
H2A.Z preferentially associates with di- and tri-MeK4 H3 in the nucleosome context.
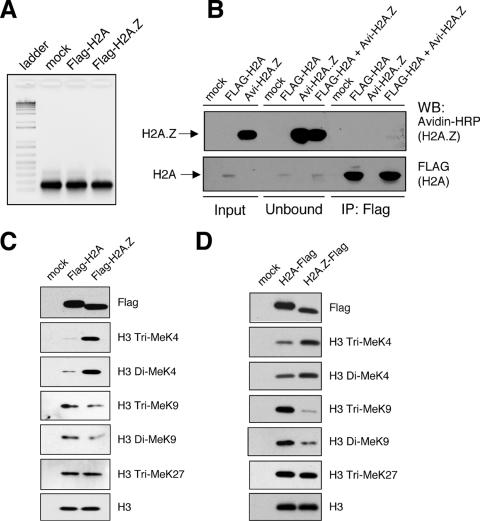
To further clarify whether mammalian H2A.Z functionally associates with euchromatin or heterochromatin, we sought to examine the methylation status of the histone H3 associated with H2A.Z in the nucleosome context. Methylation of histone H3 at lysines 4, 9, and 27 are well-characterized modification marks that determine different functional states of chromatin (reviewed in reference 19). K4-methylated H3 is associated with euchromatin and is enriched in regions of the genome poised for transcription. K9-methylated H3 binds HP1 proteins as part of the initiating steps in constitutive heterochromatin formation, and tri-MeK9 is enriched at the pericentric heterochromatin in mouse cells. K27-methylated H3 recruits polycomb complex-silencing proteins and is enriched in facultative heterochromatin, such as the inactive X chromosome in female cells. To elucidate the methylation status of H3 associated with H2A.Z, we used a modified version of the small-scale biochemical purification scheme described by Wysocka et al. (35). In brief, we expressed Flag-tagged H2A or H2A.Z in transiently transfected 293T cells, digested the chromatin to mononucleosomes by MNase treatment, and immunoprecipitated the Flag-H2A- or Flag-H2A.Z-containing nucleosomes by using Flag antibody-coupled M2 agarose beads. The transiently expressed histones are readily incorporated into chromatin, and we used the H2A.Z antibodies to confirm that expression levels of the tagged histones are well below the endogenous levels of H2A.Z (data not shown). Because of the low abundance of H2A.Z relative to that of the canonical H2A in mammalian cells, complete digestion of chromatin to single nucleosomes is essential to prevent inclusion of adjacent H2A-containing nucleosomes in the immunoprecipitation analyses. We therefore monitored each batch of nucleosomes used in the immunoprecipitation assays by DNA extraction and agarose gel analyses to ensure that only mononucleosomes were used as input (Fig. 3A). Furthermore, to test for potential nonspecific association of nucleosomes during the immunoprecipitation process, we mixed nucleosomes harvested from cells transfected with Flag-tagged H2A or Avi-tagged H2A.Z (see Materials and Methods for details) and found that the M2 beads specifically immunoprecipitated the Flag-tagged H2A, whereas none of the Avi-tagged H2A.Z nucleosomes was pulled down nonspecifically (Fig. 3B). With these conditions met, we immunoprecipitated the H2A- or H2A.Z-containing mononucleosomes, normalized the amount of immunoprecipitated nucleosomes based on the amount of total H3 (detected by the unmodified H3 antibody), and examined the levels of the various histone H3 modifications by Western blot analyses (Fig. 3C).
FIG. 3.
H2A.Z-containing mononucleosomes have higher levels of di-MeK4/tri-MeK4 H3 and lower levels of tri-MeK9 H3 than H2A-containing nucleosomes. (A) Representative agarose gel analysis showing complete MNase digestion of chromatin into single nucleosomes before being used in the immunoprecipitation procedure. The image is a negative image of the original ethidium bromide-stained gel, so the DNA band is black and the gel background is white. (B) 293T cells were either mock transfected, transfected with Flag-H2A, or transfected with Avi-H2A.Z, and total cell lysates from the two transfected cell samples were mixed before being used in the Flag immunoprecipitation assay. Flag-H2A, but not Avi-H2A.Z, was brought down by the M2 beads, indicating that the immunoprecipitation procedure is highly specific. (C) Flag-H2A.Z- or Flag-H2A-containing mononucleosomes are immunoprecipitated using the Flag antibody-coupled M2 beads. The amounts of immunoprecipitated H3 were normalized relative to each other, and the relative levels of various H3 modifications were examined using the indicated antibodies. (D) The same mononucleosome immunoprecipitation was repeated using C-terminally Flag-tagged H2A and H2A.Z. For both experiments (C and D), H2A.Z-containing nucleosomes are enriched for K4-methylated H3, whereas H2A-containing nucleosomes are enriched for K9-methylated H3. WB, Western blot; IP, immunoprecipitation.
Comparing the H3 methylation status of the two nucleosome fractions, the most striking difference is the enrichment of di- and tri-MeK4 H3 on the H2A.Z-containing nucleosomes compared to the methylation status of H2A nucleosomes. In addition, in contrast to the K4 methylation status, the di- and tri-MeK9 H3 levels are consistently lower in the H2A.Z nucleosomes than in H2A nucleosomes. To further validate these observations, we also repeated the experiment using H2A and H2A.Z that were Flag tagged at the C terminus and saw the exact same trend, in that nucleosomes that contain H2A.Z are preferentially enriched for K4-methylated H3, whereas those that contain H2A are enriched for K9-methylated H3 (Fig. 3D). We note that the differences in the absolute levels of K4- and K9-methylated H3 between the H2A- and H2A.Z-containing nucleosomes may vary from experiment to experiment; however, the trends are consistent over multiple repeats of the experiment. In yeast, H2A.Z is localized to the transcription start site of genes (13, 18, 26, 36). Given that the 5′ ends of genes generally are enriched for tri-MeK4 H3 (25), the higher levels of K4-methylated H3 associated with H2A.Z nucleosomes in 293T cells is consistent with the euchromatin-associated functions of H2A.Z in yeast and suggests that this function is conserved between yeast and human cells. Since tri-MeK9 H3 is enriched at constitutive heterochromatin and since this motif physically binds to HP1α, the reduced K9-methylated H3 on H2A.Z nucleosomes corroborates our immunofluorescence data showing depletion of H2A.Z at constitutive heterochromatin (Fig. 2). The low levels of K9-methylated H3 associated with H2A.Z likely represent the recently reported minor amounts of H2A.Z at centromeres (12) that may not be apparent at the resolution of our immunofluorescence studies. Overall, our mammalian immunofluorescence and biochemical analyses are consistent with previous findings with yeast and suggest that H2A.Z preferentially associates with euchromatin but is depleted at constitutive heterochromatin in mouse and human cells. Moreover, given the equal amount of K27-methylated H3 found in H2A and H2A.Z nucleosomes, they may have a similar propensity to associate with facultative heterochromatin.
H2A.Z is monoubiquitylated at its C terminus.
To further our studies of H2A.Z, we generated an H2A.Z-GFP expression construct, as done in other histone-GFP studies (15). Expression of H2A.Z-GFP and the control H3-GFP in transiently transfected mammalian cells shows nuclear localization of the GFP signals (data not shown), and Western blot analyses of transfected cell lysates show abundant expression of the respective GFP-tagged histones with the expected molecular sizes (Fig. 4A). Interestingly, we consistently saw a slower-migrating form of H2A.Z-GFP that was detected by the antibodies against GFP (Fig. 4A, B, and E) and H2A.Z (Fig. 4E). Given that H2A is known to be monoubiquitylated, we tested whether the slower-migrating band is due to ubiquitylation by cotransfecting the H2A.Z-GFP construct with an HA-ubiquitin plasmid. Whole-cell extracts were harvested from the transfected cells, and proteins conjugated to the HA-ubiquitin in the total extracts were detected by Western blotting using an antibody against the HA epitope. Short exposures of Western blots show that the ubiquitin epitope is specifically present on the higher-molecular-weight band but not on the unmodified form of H2A.Z-GFP (Fig. 4A). In this assay, the HA antibody seemed to preferentially detect the ubiquitylated form of H2A.Z-GFP. This is likely due to the timing of the coexpression of the HA-ubiquitin and H2A.Z-GFP molecules, as well as the fact that H2A.Z-GFP is one of very few cellular proteins that are monoubiquitylated (and thus detected as a discrete band by the HA antibody), whereas most other cellular proteins are polyubiquitylated. Indeed, upon longer exposures, the HA signal is found as a high-molecular-weight smear, which reflects the fact that polyubiquitylated proteins contain multiple and branched ubiquitin chains (data not shown). Based on the incorporation of the HA-ubiquitin epitope and the calculated molecular weight shift, we conclude that the slower-migrating band represents the monoubiquitylated form of H2A.Z-GFP (uH2A.Z-GFP).
H2A is generally known to be monoubiquitylated at K119 at the C terminus, which is the second lysine of a double-lysine motif conserved between H2A and H2A.Z. To map the site of monoubiquitylation of H2A.Z-GFP, we mutated the corresponding lysine 121 of H2A.Z-GFP to a nonmodifiable arginine residue (K121R) and found that this single-point mutant still was abundantly monoubiquitylated (Fig. 4B). Mutating the adjacent K120 residue greatly diminished the levels of uH2A.Z-GFP, and mutating both K120 and K121 completely abolished monoubiquitylation of H2A.Z-GFP. These data suggest that both lysine residues can be monoubiquitylated. Furthermore, mutating K120 has a significantly greater effect on the monoubiquitylation levels than the K121 mutation, suggesting that K120 might be a preferred site of monoubiquitylation, whereas K121 might be a secondary backup site of modification. Mutating both lysine residues at positions 120 and 121 to arginine residues also abolished incorporation of the HA-ubiquitin onto the mutated H2A.Z-GFP proteins (Fig. 4A) in cotransfection experiments, further confirming these as sites of monoubiquitylation on this histone variant.
To test whether the C-terminal addition of GFP affects the site of monoubiquitylation, we also expressed H2A.Z that is tagged at the N terminus with an Avi tag (see Materials and Methods for details). With the N-terminally tagged H2A.Z, we found that the K120, 121R double mutant still retained residual amounts of monoubiquitylation, but mutating K125 in addition completely abolished monoubiquitylation of the tagged H2A.Z (Fig. 4C). In comparison, mutation of K119 of the N-terminally tagged H2A greatly reduced the uH2A levels, and mutating both K118 and K119 completely abolished monoubiquitylation of the tagged H2A (Fig. 4C). These studies suggest that although only one lysine per H2A or H2A.Z molecule generally is modified by ubiquitylation (since is it monoubiquitylated), several different lysine residues at the C termini of these histones can function as redundant acceptor sites for ubiquitylation.
Monoubiquitylation of H2A.Z blocks the recognition of this variant by antibodies directed to its C terminus.
To examine whether endogenous H2A.Z is also monoubiquitylated, we harvested total cell extracts from human and mouse cells and performed Western blot analyses using the H2A.Z antibodies. We found that the H2A.Z-L1 antibody readily detects the slower-migrating uH2A.Z band, whereas the H2A.Z C-terminal antibody detected only the nonubiquitylated H2A.Z (Fig. 4D). In both human and mouse cells, the levels of uH2A.Z are higher in asynchronous cells than in cells arrested at mitosis by nocodazole treatment, which is consistent with previous reports that showed a loss of uH2A in Chinese hamster cell lines at mitosis (20). The differential ability of the L1- and C-terminal H2A.Z antibodies to detect endogenous uH2A.Z could be due to an epitope-masking effect, since monoubiquitylation occurs at the lysines at the C terminus of H2A.Z. We tested the recognition of transiently expressed wild type-GFP or the K120, 121R mutant form of H2A.Z-GFP by these antibodies in Western blot analyses and found that, indeed, the H2A.Z-C antibody did not recognize the uH2A.Z-GFP band of the wild-type protein (Fig. 4E). Moreover, this antibody also did not detect the K120, 121R mutant form of H2A.Z-GFP, indicating that it specifically recognizes the KK motif at the C-terminal end of H2A.Z and that ubiquitylation of H2A.Z at that region prevented its recognition by this antibody. The unique characteristics of these antibodies were further exploited for examining the ubiquitylation of H2A.Z in vivo (see below).
Monoubiquitylated H2A.Z is enriched on the inactive X chromosome of female cells.
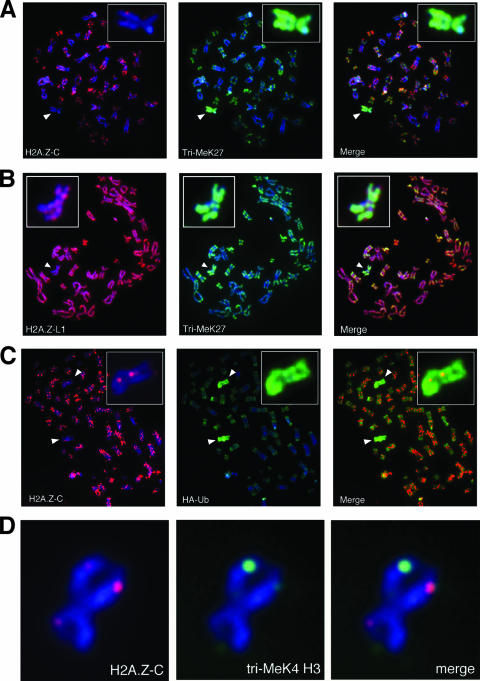
Using the antibodies discussed above, we looked at the distribution of H2A.Z on metaphase chromosome spreads. Staining of chromosomes from human female IMR90 cells using the H2A.Z-C antibody shows that one chromosome per spread consistently lacks staining by this antibody (Fig. 5A). Closer examination of this chromosome (insets in Fig. 5A and D) shows that, apart from a few bright foci, H2A.Z-C staining is completely lacking along the length of the chromosome. H2A.Z has been reported to be excluded from the inactive X chromosome (27), and we confirmed that the understained chromosome is indeed the inactive X chromosome by costaining the sample with a tri-MeK27 H3 antibody, a histone modification enriched on the inactive X chromosome as part of its silencing mechanism (24, 31) (Fig. 5A). Staining of the IMR90 metaphase chromosomes with the H2A.Z-L1 antibody also shows reduced staining on the tri-MeK27 H3-enriched inactive X chromosome (Fig. 5B); however, we still consistently see significant amounts of H2A.Z staining along the entire X chromosome (inset in Fig. 5B).
FIG. 5.
uH2A.Z is enriched on the inactive X chromosome in human cells. (A and B) Metaphase chromosomes were harvested from human female IMR90 cells and cytospun onto glass slides, and the indicated antibodies (H2A.Z-L1, H2A.Z-C, and tri-MeK27) were used for immunofluorescence staining of the chromosome spreads. The inactive X chromosomes from spreads (identified by enrichment of tri-MeK27 levels) were enlarged in the inset at the top left or right corners. (C) 293T cells were transfected with an HA-ubiquitin (HA-Ub) expression plasmid, and metaphase chromosomes were harvested and analyzed by immunofluorescence staining using the indicated antibodies. One of the inactive X chromosomes from the 293T cells is enlarged in the inset at the top right corner. (D) A representative inactive X chromosome from IMR90 cells costained with the H2A.Z-C and a monoclonal antibody that recognizes di- and tri-MeK4 H3.
We wondered whether the different staining pattern of the inactive X chromosomes by the H2A.Z-C and H2A.Z-L1 antibodies is due to the inability of the H2A.Z-C antibody to detect monoubiquitylated H2A.Z, since previous studies have shown that uH2A is enriched on the inactive X chromosome (8, 34). We transfected an HA-ubiquitin expression construct into 293T cells (also of female origin and known to contain multiple inactive X chromosomes [24]), performed metaphase chromosome staining of the transfected cells, and found that the chromosomes understained by the H2A.Z C-terminal antibody are indeed enriched for the HA-ubiquitin epitope (Fig. 5C). In contrast, the HA-ubiquitin-enriched inactive X chromosomes are still stained by the H2A.Z-L1 antibody (data not shown). Together, these results suggest that H2A.Z is not excluded from the inactive X chromosome; rather, detection of this variant on this chromosome is masked due to ubiquitylation of the antibody recognition site. We also have tested a commercially available H2A.Z antibody directed against the C terminus of this variant and found that it also did not detect the ubiquitylated form of endogenous H2A.Z (Fig. 6). Given that most of the mammalian H2A.Z studies utilize antibodies against the C terminus of H2A.Z, the occlusion of the antibody recognition epitope due to monoubiquitylation or other potential posttranslational modifications could explain the previously reported lack of H2A.Z on the inactive X chromosome (27).
FIG. 6.
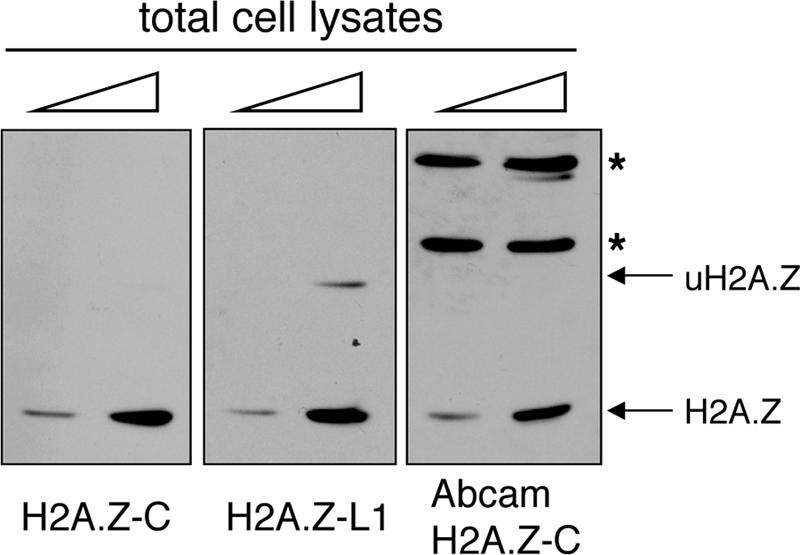
Antibodies directed against the C terminus of H2A.Z do not recognize the monoubiquitylated form of H2A.Z. Total cell lysates from 293T cells were examined by Western blot analyses using the indicated H2A.Z antibodies. Both the H2A.Z-C antibody and the commercially available antibody have difficulty detecting the endogenous monoubiquitylated H2A.Z. The bands marked by an asterisk represent nonspecific proteins within the total cell extracts that are recognized by the Abcam H2A.Z antibody.
Closer examination of the inactive X chromosome shows that there are distinct foci on this chromosome that are still stained by the H2A.Z-C antibody (Fig. 5A, inset, and D), presumably representing regions on the inactive X chromosome containing nonubiquitylated H2A.Z. This staining pattern is highly reminiscent of our previously reported hot spots of di-MeK4 H3 staining on the inactive X chromosome (4). Dual antibody labeling shows that the centers of the foci of H2A.Z-C and tri-MeK4 H3 staining do indeed colocalize positionally on the long arm of the inactive X chromosome (Fig. 5D). Given that our mononucleosome immunoprecipitation assays show that H2A.Z nucleosomes are enriched for K4-methylated H3, the simplest interpretation of our data is that the nonubiquitylated H2A.Z and K4-methylated H3 are preferentially associated with each other. Intriguingly, we often find that the staining intensities of H2A.Z foci are unequal between the two sister chromatids of the inactive X chromosome. Moreover, we consistently notice a reciprocal nature in the staining intensities of H2A.Z-C and MeK4 H3, inasmuch as the brighter H2A.Z-C foci often is paired with weaker MeK4 H3 staining on one chromatid, whereas the reverse trend is seen on the other sister chromatid. At present, we do not have an explanation for this observation, nor do we know whether the differential staining intensities by these antibodies are physiologically significant. Nevertheless, these results hint at a complex connection between nonubiquitylated H2A.Z and K4-methylated H3 on the inactive X chromosome that should be explored in future studies.
H2A.Z is partially depleted but is not excluded from the inactive X chromosome.
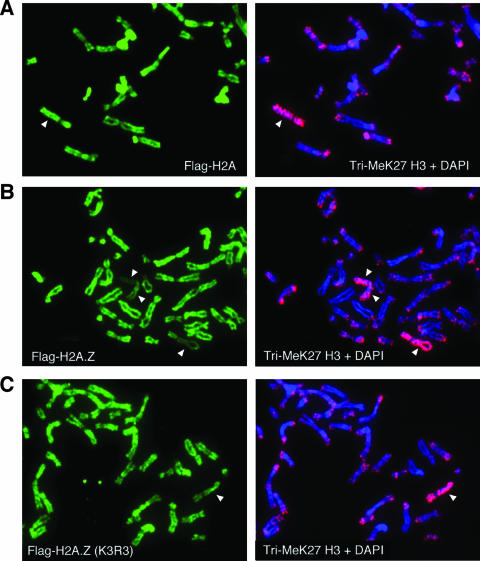
Given that previous studies have concluded that H2A.Z is mostly absent on the inactive X chromosome, we wanted to confirm our H2A.Z-L1 staining result that suggests that, albeit reduced, a significant amount of H2A.Z is still present along the length of the inactive X chromosome. To independently validate this observation, we transfected Flag-tagged versions of H2A and H2A.Z into 293T cells to examine the localization of these histones on the different chromosomes. As mentioned earlier, 293T cells are female in origin and contain multiple inactive X chromosomes per cell. In these studies, we used enriched tri-MeK27 H3 staining to locate the inactive X chromosome(s) in the metaphase spreads and compared the Flag staining on the autosomes to that on inactive X chromosomes (Fig. 7). Immunofluorescence staining shows that similar amounts of Flag-H2A are incorporated onto the autosomes and the inactive X chromosomes (Fig. 7A). Flag-tagged wild-type H2A.Z is also efficiently incorporated onto chromosomes in the transfected cells; however, compared to the incorporation of autosomes, its incorporation onto the inactive X chromosomes (a total of three shown in Fig. 7B) is noticeably and consistently reduced. Significant amounts of Flag-H2A.Z are still present along the entire length of the inactive X chromosomes, and this staining pattern is analogous to that seen on the inactive X chromosomes of IMR90 cells when stained with the H2A.Z-L1 antibody (Fig. 5B). Finally, using a mutant H2A.Z for which the three potentially ubiquitylated lysines are mutated to arginines (K3R3 mutant), we found that this mutant is still efficiently incorporated onto the inactive X chromosome (Fig. 7C), indicating that monoubiquitylation of H2A.Z is not a prerequisite for its deposition onto this chromosome. Interestingly, we consistently noticed that the reduced levels of the tagged H2A.Z on the inactive X chromosome are less apparent for the mutant H2A.Z than for the wild-type version. It is possible that monoubiquitylation or the C-terminal amino acid sequences of H2A.Z are involved in the retention or removal of this variant on the inactive X chromosome. Regardless, these transfection studies corroborate our previous conclusion that H2A.Z is not completely excluded from the inactive X chromosomes.
FIG. 7.
Ubiquitylation of H2A.Z is not required for its incorporation onto the inactive X chromosomes. 293T cells were transfected with expression constructs for wild-type Flag-H2A (A), wild-type Flag-H2A.Z (B), or the nonubiquitylatable mutant (K3R3) form of Flag-H2A.Z (C). Metaphase chromosomes were isolated from the transfected cells and were immunostained using the Flag and tri-MeK27 H3 antibodies. The chromosomes enriched for tri-meK27 H3 (marked by white arrowheads) represent the inactive X chromosomes. Since the panels show only part of the chromosome spreads, not all of the inactive X chromosomes present per cell are shown.
Monoubiquitylation of H2A.Z is mediated by the RING1b E3 ligase.
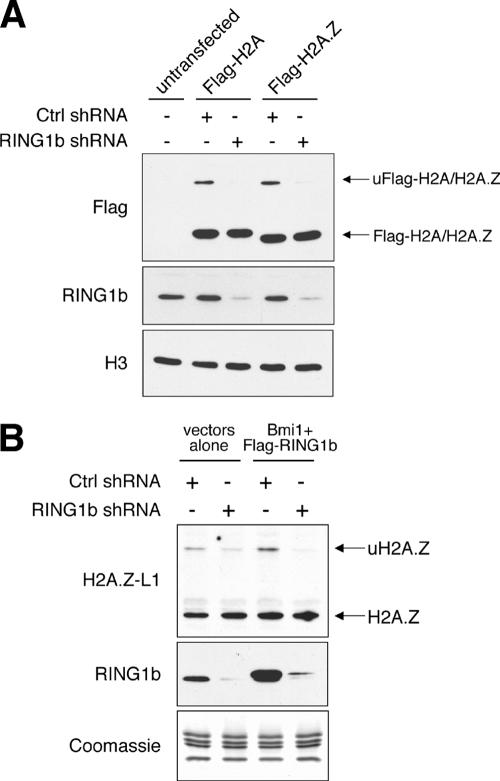
In mammalian cells, the RING1b E3 ligase is part of the polycomb complex responsible for monoubiquitylation of H2A (8, 34). This polycomb complex mediates silencing of many developmentally regulated genes as well as the transcriptional silencing of the inactive X chromosome in mammalian female cells. Given that our metaphase chromosome spread data suggest that a significant amount of monoubiquitylated H2A.Z, much like ubiquitylated H2A, is present on the inactive X chromosome, one prediction is that both H2A and H2A.Z are monoubiquitylated by the same E3 ligase. To test this hypothesis, we examined the effects of RNAi knockdown of RING1b on the ubiquitylation of H2A.Z. We first generated shRNA constructs based on previously published RING1b-RNAi target sequences and identified the construct most effective in knocking down expression of the cotransfected Flag-RING1b construct (data not shown). For a control, we also generated an shRNA construct with a scrambled sequence that does not target any known genes. Unlike the control, expression of this RING1b-shRNA construct effectively knocked down expression of endogenous RING1b as well as the ubiquitylation levels of the coexpressed Flag-H2A (Fig. 8A). Similarly, the levels of ubiquitylated Flag-H2A.Z are also greatly reduced in the RING1b-shRNA-transfected cells compared to levels in wild-type cells (Fig. 8A). Analogous to its effect on exogenously introduced H2A.Z, expression of the RING1b-shRNA construct also reduced the accumulation of the monoubiquitylated form of the endogenous H2A.Z (Fig. 8B). Previous publications have shown that an additional polycomb protein, Bmi1, enhances the ability of RING1b to ubiquitylate H2A in vitro (6). In our hands, coexpression of RING1b and Bmi1 in 293T cells modestly increased the endogenous levels of uH2A.Z but showed a more obvious knockdown of the uH2A.Z levels when the RING1b shRNA construct was coexpressed with these two enzymes (Fig. 8B). Therefore, additional components of the polycomb complex, besides RING1b and Bmi1, may be involved in regulating the overall levels of uH2A.Z. These results together show that ubiquitylation of H2A and H2A.Z is mediated by the same enzyme complexes and suggest that ubiquitylation of both H2A and H2A.Z is part of the polycomb-silencing pathway.
FIG. 8.
Expression of shRNA targeting RING1b reduces the levels of ubiquitylated H2A.Z. (A) 293T cells were transfected with the control scrambled shRNA construct (Ctrl shRNA) or with the shRNA construct targeting human RING1b along with either the empty vectors (untransfected) or the Flag-H2A or Flag-H2A.Z construct. The total cell lysate was harvested, and the expression of Flag-H2A and Flag-H2A.Z was detected by Western blotting using the Flag-M2 antibody. As shown, the RING1B shRNA, but not the control shRNA, specifically knocked down expression of the endogenous RING1b protein as well as the monoubiquitylated forms of Flag-H2A and Flag-H2A.Z. The Western blot using the H3 antibody is shown as a loading control. (B) 293T cells were transfected with the control or the RING1b shRNA constructs, along with empty vectors or constructs expressing Bmi1 and Flag-RING1b. The effects of these constructs on the ubiquitylation levels of endogenous H2A.Z were examined by Western blotting using the H2A.Z-L1 antibody. Note that the lysate from cells transfected with the Flag-RING1b and Bmi1 contains both the endogenous and overexpressed Flag-tagged RING1b, and these two versions of RING1b were not well resolved on the gel due to the high percentage (15%) of acrylamide used.
DISCUSSION
The function of H2A.Z has so far been best characterized for S. cerevisiae, in which H2A.Z is localized to gene promoters and protects the euchromatic regions from the spreading of heterochromatin silencing (13, 18, 26, 36). However, whether these functions of H2A.Z in yeast are conserved in mammalian cells is still not clear (for example, see reference 3). In this report, we show that in differentiated mouse cells, H2A.Z is localized to chromosome arms and is depleted at pericentric heterochromatin. Moreover, H2A.Z-containing nucleosomes are enriched for di- and tri-MeK4 H3 and have lower levels of K9-methylated H3. These findings are consistent with results of yeast Htz1 studies and support the conclusion that the majority of this variant is associated with euchromatin in both yeast and mammalian cells. Through our studies, we also found that a small fraction of H2A.Z is monoubiquitylated in mammalian cells and that monoubiquitylation can occur on any one of several lysine residues at the C terminus of H2A.Z. Using a pair of antibodies that differ in their abilities to detect the monoubiquitylated form of H2A.Z, we surmised that the majority of H2A.Z on the inactive X chromosomes of female cells is monoubiquitylated and propose that such modified H2A.Z is associated with the transcriptionally silent facultative heterochromatin. This conclusion is based on indirect inference; direct proof of the enrichment of uH2A.Z on the inactive X chromosome will have to wait until someone is able to generate an antibody that recognizes only the ubiquitylated form of H2A.Z. Nevertheless, consistent with the idea that uH2A.Z is associated with transcriptional silencing, RNAi studies show that the RING1b E3 ligase, which is part of the polycomb-silencing complex associated with the inactive X chromosome, regulates the overall level of uH2A.Z in human cells. Taken together, these studies suggest that the majority of H2A.Z in mouse and human cells is nonubiquitylated and is associated with euchromatin. On the other hand, monoubiquitylated H2A.Z is associated with facultative heterochromatin, and this modification marks the fraction of H2A.Z associated with gene silencing.
At present, ubiquitylated H2A.Z has not been found in S. cerevisiae, and a recent report using mass spectrometry suggested that the C terminus of H2A.Z in yeast is free of posttranslational modifications (22). It would not be surprising if the H2A.Z in S. cerevisiae were not ubiquitylated, since H2B is the predominant histone ubiquitylated in this organism (29). Also, the yeast genome is mostly maintained in a euchromatic state, and this organism lacks the H3 methylation marks (K9 and K27 methylation) associated with mammalian heterochromatin. In contrast to yeast cells, the majority of the mammalian genome is transcriptionally inactive, and the silenced regions are maintained by constitutive and facultative heterochromatin. Our studies of mammalian H2A.Z suggest that this variant is mostly absent within constitutive heterochromatin but is associated with facultative heterochromatin silencing when it is monoubiquitylated. It is likely that H2A.Z has evolved additional functions in mammalian cells beyond its function in yeast. In that regard, it is relevant to remember that, while the deletion of the H2A.Z gene is tolerated in S. cerevisiae, loss of H2A.Z expression in Tetrahymena, Drosophila, and mammalian cells is lethal in these organisms. Therefore, the essential function of H2A.Z in more complex organisms may be related to its silencing function, and the contribution of the monoubiquitylation of H2A.Z to this essential function in mammalian cells will need to be further examined.
Acknowledgments
We thank Scott Briggs and Edith Heard for critical reading of the manuscript. We also thank Upstate Biotechnology for their assistance in antibody generation.
This project was funded by a National Cancer Institute of Canada operating grant awarded to P.C. and by the Canada Research Chair program.
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Abbott, D. W., V. S. Ivanova, X. Wang, W. M. Bonner, and J. Ausio. 2001. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J. Biol. Chem. 276:41945-41949. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, E., and S. B. Hake. 2006. The nucleosome: a little variation goes a long way. Biochem. Cell Biol. 84:505-517. [DOI] [PubMed] [Google Scholar]
- 4.Boggs, B. A., P. Cheung, E. Heard, D. L. Spector, A. C. Chinault, and C. D. Allis. 2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 30:73-76. [DOI] [PubMed] [Google Scholar]
- 5.Bulynko, Y. A., L. C. Hsing, R. W. Mason, D. J. Tremethick, and S. A. Grigoryev. 2006. Cathepsin L stabilizes the histone modification landscape on the Y chromosome and pericentromeric heterochromatin. Mol. Cell. Biol. 26:4172-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, R., Y. Tsukada, and Y. Zhang. 2005. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20:845-854. [DOI] [PubMed] [Google Scholar]
- 7.de Boer, E., P. Rodriguez, E. Bonte, J. Krijgsveld, E. Katsantoni, A. Heck, F. Grosveld, and J. Strouboulis. 2003. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. USA 100:7480-7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Napoles, M., J. E. Mermoud, R. Wakao, Y. A. Tang, M. Endoh, R. Appanah, T. B. Nesterova, J. Silva, A. P. Otte, M. Vidal, H. Koseki, and N. Brockdorff. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7:663-676. [DOI] [PubMed] [Google Scholar]
- 9.Dhillon, N., and R. T. Kamakaka. 2000. A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell 6:769-780. [DOI] [PubMed] [Google Scholar]
- 10.Fan, J. Y., F. Gordon, K. Luger, J. C. Hansen, and D. J. Tremethick. 2002. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol. 9:172-176. [DOI] [PubMed] [Google Scholar]
- 11.Fan, J. Y., D. Rangasamy, K. Luger, and D. J. Tremethick. 2004. H2A.Z alters the nucleosome surface to promote HP1α-mediated chromatin fiber folding. Mol. Cell 16:655-661. [DOI] [PubMed] [Google Scholar]
- 12.Greaves, I. K., D. Rangasamy, P. Ridgway, and D. J. Tremethick. 2007. H2A.Z contributes to the unique 3D structure of the centromere. Proc. Natl. Acad. Sci. USA 104:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillemette, B., A. R. Bataille, N. Gevry, M. Adam, M. Blanchette, F. Robert, and L. Gaudreau. 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3:e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillemette, B., and L. Gaudreau. 2006. Reuniting the contrasting functions of H2A.Z. Biochem. Cell Biol. 84:528-535. [DOI] [PubMed] [Google Scholar]
- 15.Kimura, H., and P. R. Cook. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 17.Leach, T. J., M. Mazzeo, H. L. Chotkowski, J. P. Madigan, M. G. Wotring, and R. L. Glaser. 2000. Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J. Biol. Chem. 275:23267-23272. [DOI] [PubMed] [Google Scholar]
- 18.Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen, C. Seidel, J. Gerton, and J. L. Workman. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 102:18385-18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6:838-849. [DOI] [PubMed] [Google Scholar]
- 20.Matsui, S. I., B. K. Seon, and A. A. Sandberg. 1979. Disappearance of a structural chromatin protein A24 in mitosis: implications for molecular basis of chromatin condensation. Proc. Natl. Acad. Sci. USA 76:6386-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725-736. [DOI] [PubMed] [Google Scholar]
- 22.Millar, C. B., F. Xu, K. Zhang, and M. Grunstein. 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20:711-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, Y. J., P. N. Dyer, D. J. Tremethick, and K. Luger. 2004. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem. 279:24274-24282. [DOI] [PubMed] [Google Scholar]
- 24.Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer, H. Wang, C. C. de la Cruz, A. P. Otte, B. Panning, and Y. Zhang. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131-135. [DOI] [PubMed] [Google Scholar]
- 25.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517-527. [DOI] [PubMed] [Google Scholar]
- 26.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rangasamy, D., L. Berven, P. Ridgway, and D. J. Tremethick. 2003. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 22:1599-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rangasamy, D., I. Greaves, and D. J. Tremethick. 2004. RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 11:650-655. [DOI] [PubMed] [Google Scholar]
- 29.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 30.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411-422. [DOI] [PubMed] [Google Scholar]
- 31.Silva, J., W. Mak, I. Zvetkova, R. Appanah, T. B. Nesterova, Z. Webster, A. H. Peters, T. Jenuwein, A. P. Otte, and N. Brockdorff. 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell 4:481-495. [DOI] [PubMed] [Google Scholar]
- 32.Stargell, L. A., J. Bowen, C. A. Dadd, P. C. Dedon, M. Davis, R. G. Cook, C. D. Allis, and M. A. Gorovsky. 1993. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev. 7:2641-2651. [DOI] [PubMed] [Google Scholar]
- 33.Thambirajah, A. A., D. Dryhurst, T. Ishibashi, A. Li, A. H. Maffey, and J. Ausio. 2006. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 281:20036-20044. [DOI] [PubMed] [Google Scholar]
- 34.Wang, H., L. Wang, H. Erdjument-Bromage, M. Vidal, P. Tempst, R. S. Jones, and Y. Zhang. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431:873-878. [DOI] [PubMed] [Google Scholar]
- 35.Wysocka, J., P. T. Reilly, and W. Herr. 2001. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 21:3820-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, H., D. N. Roberts, and B. R. Cairns. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123:219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]