Abstract
The beyond 12/23 (B12/23) rule ensures inclusion of a Dβ gene segment in the assembled T-cell receptor (TCR) β variable region exon and is manifest by a failure of direct Vβ-to-Jβ gene segment joining. The restriction is enforced during the DNA cleavage step of V(D)J recombination by the recombination-activating gene 1 and 2 (RAG1/2) proteins and the recombination signal sequences (RSSs) flanking the TCRβ gene segments. Nothing is known about the step(s) at which DNA cleavage is defective or how TCRβ locus sequences contribute to these defects. To address this, we examined the steps of DNA cleavage by the RAG proteins using TCRβ locus V, D, and J RSS oligonucleotide substrates. The results demonstrate that the B12/23 rule is enforced through slow nicking of Jβ substrates and to some extent through poor synapsis of Vβ and Jβ substrates. Nicking is controlled largely by the coding flank and, unexpectedly, the RSS spacer, while synapsis is controlled primarily by the RSS nonamer. The results demonstrate that different Jβ substrates are crippled at different steps of cleavage by distinct combinations of defects in the various DNA elements and strongly suggest that the DNA nicking step of V(D)J recombination can be rate limiting in vivo.
V(D)J recombination is a site-specific recombination reaction responsible for assembling the exons encoding the antigen binding portion of immunoglobulins and T-cell receptors (TCRs). The many combinations of possible joining events of variable (V), diversity (D), and joining (J) coding segments generate diverse antigen receptor repertoires. V(D)J recombination is initiated during B- and T-cell development by the recombination-activating gene 1 and 2 (RAG1/2) proteins (21, 26).
RAG1/2 are required for the introduction of double-strand breaks between V, D, and J coding segments and their flanking recombination signal sequences (RSSs). RAG-mediated cleavage occurs in two ordered steps: nicking and hairpin formation (19). A nick is first introduced in the top strand 5′ of the heptamer at the junction between the heptamer and the V, D, or J coding segment. This exposes a 3′ hydroxyl group at the end of the coding flank and a 5′ phosphate group attached to the heptamer end. The 3′ hydroxyl group attacks the antiparallel strand in a transesterification reaction (31), which cleaves the DNA and creates a hairpin coding end and a blunt 5′ phosphorylated signal end. After cleavage of the two participating DNA substrates, the RSS ends are precisely joined and the coding ends are modified and joined in a process involving the nonhomologous end-joining DNA repair pathway (reviewed in references 3 and 9).
The RSS is composed of relatively conserved heptamer (consensus, 5′-CACAGTG) and nonamer (consensus, 5′-ACAAAAACC) sequences separated by a less well conserved spacer sequence of either 12 or 23 bp (17). The nonamer provides a major binding surface for RAG1, and the heptamer functions in binding and the establishment of a DNA structure appropriate for cleavage (7, 9). V(D)J recombination occurs primarily between a gene segment flanked by a 12RSS and one flanked by a 23RSS, a restriction termed the 12/23 rule. The 12/23 rule is imposed at or before the cleavage step of V(D)J recombination with efficient hairpin formation requiring the assembly of a stable paired complex (PC) between the RAG proteins, HMGB1/2, and a 12RSS and 23RSS (7, 9). In contrast, nicking can occur in the signal complex (SC) containing the RAG/HMGB proteins and one RSS (36).
The RSS can impose significant constraints on variable region gene assembly beyond enforcing the 12/23 rule. A particularly clear example is provided by the TCRβ locus, in which Vβ RSSs and 3′Dβ RSSs contain 23-base-pair spacers and 5′Dβ RSSs and Jβ RSSs contain 12-base-pair spacers (see Fig. 1). Rearrangement occurs first between Dβ and Jβ, followed by Vβ-to-DJβ rearrangement, with little or no direct Vβ-to-Jβ rearrangement occurring even though it is permissible by the 12/23 rule (reviewed in reference 29). This phenomenon, which was first experimentally analyzed using mice with a modified TCRβ locus, was termed the beyond 12/23 (B12/23) rule (2). These and other experiments revealed that the 5′Dβ1 12RSS, and not the Jβ 12RSSs, support rearrangement to various Vβ gene segments in a position-independent manner and that Dβ-to-Jβ rearrangement is not required for Vβ-to-Dβ rearrangement (2, 27). Subsequent studies demonstrated that extrachromosomal V(D)J recombination substrates together with RAG1/2 are able to recapitulate the B12/23 rule in nonlymphoid cells, suggesting that RAG1 and RAG2 are the only lymph-specific factors necessary to establish this restriction (14, 23, 30). Importantly, purified core RAG1/2 proteins and HMGB1/2 were found to be able to perform B12/23-restricted cleavage with TCRβ RSSs in a cell-free system (14, 23). Hence, the B12/23 rule is established in large part by the RAG proteins and RSSs during the cleavage phase of V(D)J recombination. It is still unknown, however, at which step(s) leading to DNA cleavage the B12/23 rule is imposed.
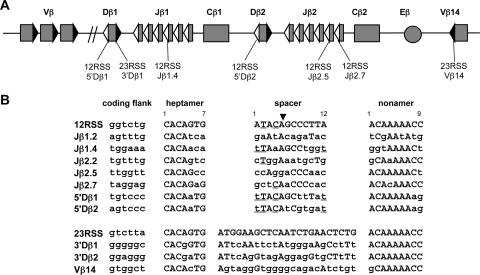
FIG. 1.
TCRβ locus. (A) Gene segments are depicted as rectangles, the enhancer as a circle, and 12RSSs and 23RSSs as white and black triangles, respectively. The RSSs examined in this study are indicated. The map is not drawn to scale. (B) Relevant TCRβ locus RSS and coding flank sequences. The TCRβ RSSs are displayed below the consensus 12RSS and 23RSS sequences with differences from the consensus sequence indicated by lowercase letters. All coding flank residues are indicated with lowercase letters. Four conserved positions in the 5′Dβ spacers are underlined, and identities at these positions in the Jβ spacers are also indicated by underlining (see Discussion). The well-conserved, functionally important fifth spacer position is indicated by a black arrowhead. The RSS elements are numbered as indicated. The DNA substrates used in our experiments extend both 5′ and 3′ of the sequences shown here (Table 1).
RSSs and their coding flanks have naturally occurring sequence variations in vivo that influence the efficiency of V(D)J recombination (reviewed in references 4 and 6). The heptamer and nonamer are the most important determinants of recombination efficiency, and the closer they are to their consensus sequences (derived from many functional endogenous RSSs), the more effectively an RSS will function (11). Coding flank sequences also influence RSS recombination potential, in large part by affecting the nicking step of the recombination reaction (10, 35). Naturally occurring variations in spacer sequence can also affect cleavage and recombination frequency (6).
The contributions of the coding flank, heptamer, nonamer, and spacer sequences to the B12/23 rule have been studied in vivo and in vitro (2, 13, 14, 22, 27, 32, 33). The integrity of the nonamer sequence and the highly conserved spacer nucleotides of the 5′Dβ1 RSS are important for efficient recombination with Vβ gene segments (13, 14). In addition, the Vβ coding flank and its 23-bp spacer influence the B12/23 restriction, while the heptamer and nonamer of the Vβ 23RSSs do not appear to strongly influence the B12/23 rule (14). The molecular mechanism by which the coding flank and components of the RSS exert these effects remains unknown.
The B12/23 restriction has important implications for the regulation of variable region gene assembly and repertoire development. However, no studies have investigated the biochemical mechanisms by which endogenous RSSs impose the B12/23 rule and, in particular, at which step(s) of V(D)J recombination these restrictions are imposed. Here we show that preferential recombination of Vβ with 5′Dβ substrates, as opposed to Jβ substrates, is determined predominantly at the nicking and synapsis steps of DNA cleavage. The results reveal an important role of the spacer in controlling the nicking step of DNA cleavage and of the nonamer in modulating the stability of the PC. In addition, we find that different Jβ substrates have distinctive unfavorable DNA sequence elements which can confer strikingly different defects in cleavage. These findings reveal some of the forces that act to determine the choice of V, D, and J gene segments used in assembled antigen receptor genes and support the model that the B12/23 rule arose as a result of the accumulation of small sequence alterations in TCRβ locus gene segments during evolution.
MATERIALS AND METHODS
DNA substrates.
The sequences of the top-strand oligonucleotides of the substrates used in this study are shown in Table 1. All substrates contained 16 bp 5′ of the heptamer and 15 bp 3′ of the nonamer. Oligonucleotides were obtained from the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University and Invitrogen. Oligonucleotides were 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase, and substrates were prepared by annealing two (or three in the case of prenicked substrates) complementary oligonucleotides and gel purification as previously described (8). Prenicked substrates used in biotin pull-down experiments contained a top-strand dideoxy residue immediately 5′ of the heptamer, eliminating the 3′ OH normally used in hairpin formation (20).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Top-strand sequence (5′-3′) |
|---|---|
| 12RSS | GATCTGGCCTGGTCTGCACAGTGATACAGCCCTTAACAAAAACCTGCACTCGAGCGGAG |
| Jβ1.1 | GATCTGTCTGTGTTTGCACAGTGCCATAGGATGAGGAGAAAAATTGCACTCGAGCGGAG |
| Jβ1.4 | GATCTGTCGTTGGAAACACAACATTAAAGCCTGGTGGTAAAACTTGCACTCGAGCGGAG |
| Jβ2.5 | GATCTGTGTCTTGGTTCACAGCCCCAGGACCCAACACAAAAACTTGCACTCGAGCGGAG |
| Jβ2.7 | GATCTGTTCATAGGAGCACAGAGGCTCAACCCCACACACAAACCTGCACTCGAGCGGAG |
| 5′Dβ1 | GATCTGCCCCTGTCCCCACAATGTTACAGCTTTATACAAAAAAGTGCACTCGAGCGGAG |
| 5′Dβ2 | GATCTGCCCCAGTCCCCACAATGTTACATCGTGATACAAAAAAGTGCACTCGAGCGGAG |
| 5′DB1/JB1.4H | GATCTGCCCCTGTCCCCACAACATTACAGCTTTATACAAAAAAGTGCACTCGAGCGGAG |
| 5′DB1/JB1.4CF | GATCTGTCGTTGGAAACACAATGTTACAGCTTTATACAAAAAAGTGCACTCGAGCGGAG |
| 5′DB1/JB1.4Sp | GATCTGCCCCTGTCCCCACAATGTTAAAGCCTGGTACAAAAAAGTGCACTCGAGCGGAG |
| 5′DB1/JB1.4N | GATCTGCCCCTGTCCCCACAATGTTACAGCTTTATGGTAAAACTTGCACTCGAGCGGAG |
| Jβ2.7Sp | GATCTGTGTCTTGGTTCACAGCCGCTCAACCCCACACAAAAACTTGCACTCGAGCGGAG |
| Jβ2.7C/A | GATCTGTTCATAGGAGCACAGAGGCTCAACCCCACACAAAAACCTGCACTCGAGCGGAG |
| Jβ2.7CF | GATCTGTTCATAGGAGCACAGCCCCAGGACCCAACACAAAAACTTGCACTCGAGCGGAG |
| 23RSS | GATCTGGCCTGTCTTACACAGTGATGGAAGCTCAATCTGAACTCTGACAAAAACCTGCACTCGAGCGGAG |
| 3′Dβ1 | GATCTGGACAGGGGGCCACGGTGATTCAATTCTATGGGAAGCCTTTACAAAAACCTGCACTCGAGCGGAG |
| Vβ14 | GATCTGCATTGTGGCTCACACTGAGTAGGGTGGGGCAGACATCTGTGCAAAAACCTGCACTCGAGCGGAG |
Protein expression and purification.
Recombinant glutathione S-transferase-RAG2 core protein (amino acids [aa] 1 to 383) was expressed in 293T cells and purified by glutathione Sepharose affinity chromatography, and recombinant maltose binding protein-RAG1 core protein (aa 384 to 1008) was expressed in bacteria and purified by amylose affinity chromatography as previously described (8). Recombinant HMGB2 protein (aa 1 to 185) lacking the C-terminal acidic region was expressed in bacteria and purified as previously described (5). All protein preparations were at least 90% pure as judged by Coomassie blue staining. The activities of different RAG protein preparations were measured by cleavage of consensus oligonucleotide RSS substrates.
Binding, cleavage, and PC assays.
Binding and cleavage assays were performed essentially as previously described (8). Biotin pull-down assays to detect the PC contained 120 fmol of 5′ bottom-strand biotinylated oligonucleotide substrate which was incubated with 100 ng each of RAG1 and RAG2 and 30 ng HMGB2 at 37°C for 10 min in a total reaction volume of 20 μl of binding buffer (25 mM MOPS-KOH [pH 7.0], 5 mM Tris-HCl [pH 8.0], 2.4 mM dithiothreitol, 90 mM potassium acetate, 30 mM KCl, 10 mM CaCl2, 0.1 mg/ml bovine serum albumin, and 2% glycerol) as previously described (20). One hundred twenty femtomoles of 5′ end-labeled DNA (∼0.6 × 1018 to 1.2 × 1018 cpm/mol) was added, and the reaction mixture was incubated for another 10 min at 37°C. A sample was then removed to allow determination of the amount of “input” 32P. Streptavidin-coated magnetic beads (Dynabeads M-280; 10 μg/μl) were added, and the reaction mixture was incubated for 1 h at 37°C with occasional mixing, followed by separation of the magnetic beads and supernatant using a magnetic stand. Beads were washed six times at room temperature with 60 μl of binding buffer and resuspended in 10 μl of binding buffer and 10 μl formamide to extract DNA from the bead-bound complexes. The 32P released in this supernatant (“bound”) and in the “input” sample were quantified with a scintillation counter. Results are expressed as “bound” divided by “input” multiplied by 100, after subtraction of background binding (see Results).
RESULTS
Recapitulation of the B12/23 rule with oligonucleotide substrates.
The endogenous TCRβ locus contains 31 Vβ gene segments and two D-Jβ gene clusters, each with a single Dβ gene segment and six functional Jβ segments (Fig. 1A). To study the molecular mechanism of the B12/23 rule, we utilized purified HMGB2 and truncated “core” RAG proteins (minimal regions required for catalytic activity) together with oligonucleotide substrates whose sequences were derived from the Vβ14, Dβ1, Dβ2, Jβ1.4, Jβ2.5, and Jβ2.7 gene segments (Fig. 1B). Jβ2.5 and Jβ2.7 are the most frequently used segments in the second cluster, while Jβ1.4 is among the most infrequently used (18).
Previous DNA cleavage analyses of the B12/23 rule utilized DNA substrates hundreds of base pairs long with the RSSs located on the same DNA molecule (14, 22, 23). To determine whether the B12/23 restriction could be recapitulated with oligonucleotide substrates, we performed DNA cleavage assays in the presence of Mg2+ with a Vβ14 23RSS substrate together with a 5′Dβ1, 5′Dβ2, Jβ1.4, Jβ2.5, or Jβ2.7 12RSS substrate. In the absence of a Vβ14 partner, the 12RSS substrates did not undergo hairpin formation, while addition of the Vβ14 substrate allowed hairpin formation with the 5′Dβ1 and 5′Dβ2, but not Jβ, substrates (Fig. 2A). These results indicate cleavage of the 5′D substrates in the PC with Vβ14 and are consistent with in vivo and in vitro observations of efficient Vβ-to-Dβ, but not Vβ-to-Jβ, rearrangement. We conclude that neither lymphoid cell-specific trans-acting factors nor endogenous chromosomal sequences, except the RSS and a short coding flank, are required to recreate the B12/23 restriction. In addition, as reported previously (14, 23), the purified core RAG proteins and HMGB2 support B12/23 rule-restricted cleavage in vitro.
FIG. 2.
DNA binding and cleavage. (A) DNA cleavage reactions (Mg2+ buffer) were performed for 60 min with the proteins and DNA substrates indicated above the lanes, with the 12RSS labeled at the 5′ end of the top strand. Products were separated on a 10% denaturing polyacrylamide gel, which was imaged with a PhosphorImager. The reaction mixtures for lanes 1 and 2 contained (+) consensus (Consen) 12RSS and 23RSS substrates, while other reactions contained the Vβ14 23RSS substrate and 12RSS substrates as indicated. The structure and position of the input substrate and the products of nicking and hairpin formation are indicated to the left of the gel. (B) Gel shift analysis of SC formation. The RAG1/2 and HMGB2 proteins were allowed to bind to labeled DNA substrates (Ca2+ buffer) as indicated above the lanes, and the reaction mixtures were analyzed on an 8% native acrylamide gel. The structure and position of the free DNA and shifted complex (SC) are shown to the left of the gel. The consensus 23RSS (lane 9) and 3′Dβ1 (lane 13) substrates exhibit two discrete shifted complexes, the upper of which might be generated by RSS synapsis.
SC formation.
One possible mechanism for selective Vβ-to-5′Dβ recombination would be preferential binding of the RAG proteins to the 12RSS of 5′Dβ as opposed to Jβ. Previous experiments (with Jβ1.1, Jβ2.2, Jβ2.5, and Jβ2.7) failed to observe consistently stronger binding to the 5′Dβ1 RSS than to Jβ RSSs (23). To confirm this with our RAG proteins, we performed electrophoretic mobility shift assay analyses with individual TCRβ locus oligonucleotide substrates under conditions leading to formation of the SC. Binding to the 12RSSs of Jβ1.1 and Jβ1.4 was substantially weaker than to the consensus 12RSS but comparable to binding to the 5′Dβ1 12RSS (Fig. 2B). These results were reproducible in four independent experiments with two different preparations of RAG1 and RAG2 proteins. While our experiments do not provide a quantitative assessment of binding, they, like previous results, fail to support the idea that the B12/23 restriction can be explained by preferential binding of the RAG proteins to the 12RSS of 5′Dβ compared to the 12RSS of Jβ.
Synapsis.
An appealing hypothesis is that preferential formation of a stable PC between Vβ and 5′Dβ RSSs as opposed to Jβ RSSs contributes to the establishment of the B12/23 rule (14, 23). The PC can be observed in mobility shift assays with consensus 12 and 23RSS oligonucleotide substrates, but it is difficult to detect using endogenous RSSs derived from the TCRβ locus (data not shown and reference 22). We considered the possibility that a previously described biotin pull-down assay (12) might provide a more sensitive method to assess synapsis between endogenous RSSs and to test the contribution of preferential synapsis to the B12/23 rule.
A biotinylated Vβ14 substrate was incubated in the presence of 32P-labeled Dβ or Jβ 12RSS substrates (Fig. 3A), RAG proteins, HMGB1/2, and Ca2+. Biotin-tagged Vβ14 was immobilized on streptavidin-coated magnetic beads, the beads were washed, and the input and final counts of 32P-labeled Dβ and Jβ substrates retained on the beads were measured. The 5′Dβ1 substrate was recovered in a complex with the biotin-tagged Vβ14 partner more efficiently than the three Jβ substrates tested, with Jβ1.4, Jβ2.5, and Jβ2.7 recovered at 8%, 50%, and 12% of 5′Dβ1, respectively (Fig. 3B). To assess the background of nonspecific DNA binding to the streptavidin beads, in every experiment the 32P-labeled substrates were incubated in a reaction mixture with the appropriate 23RSS substrate lacking a biotin tag and 32P recovery was measured. The results displayed reflect subtraction of this background level of nonspecific DNA binding (less than 3% of input in all cases). In addition, recovery of a 32P-labeled 12RSS substrate with scrambled heptamer and nonamer sequences with biotin-tagged Vβ14 substrate was similar to background (data not shown). We conclude that these Jβ gene segments synapse poorly with the Vβ14 gene segment and that this could contribute to the establishment of B12/23 restriction.
FIG. 3.
Assessment of paired complex formation. (A) The biotin pull-down assay to assess synapsis was performed with 23RSS substrates labeled with biotin (Bio) at the 5′ end of the bottom strand and 32P-labeled 12RSS (asterisk) substrates and the RAG and HMGB2 proteins (shaded ovals) in a Ca2+ buffer. In heminicked assays, 12RSS substrates contained a top-strand nick immediately 5′ of the heptamer with the labeled top-strand oligonucleotide terminating in a 3′ dideoxy residue to prevent hairpin formation. (B) The 12RSS and 23RSS substrates used are indicated below the graph. The mean of at least three independent measurements for each substrate pair (see Materials and Methods) is plotted (error bars indicate standard deviations). Statistical significance was assessed using the two-tailed Student t test (*, P < 0.02). Synapsis with Vβ14 of the three intact Jβ substrates was significantly lower than for the 5′Dβ1 substrate, as indicated by asterisks above the relevant gray bars. All three Jβ substrates formed the PC significantly more efficiently with 3′Dβ1 than with Vβ14 (Jβ1.4, P = 0.014; Jβ2.5, P = 0.022; Jβ2.7, P = 0.008).
Nicking.
The nicking step of the cleavage reaction does not require synapsis with a partner RSS. Despite the fact that Jβ and 5′Dβ substrates form the SC with approximately equal efficiency, it appeared that Jβ substrates were nicked substantially less efficiently (Fig. 1A). This was of interest because while hairpin formation is typically the rate-limiting step in cleavage by the RAG proteins (34), nicking can become the rate-limiting step under some circumstances (35). On this basis, we hypothesized that slow or inefficient nicking of Jβ gene segments could interfere with their ability to undergo coupled cleavage with Vβ gene segments by slowing hairpin formation at both the J and V partner (35). Hence, differential nicking could play a previously unsuspected role in helping to establish the B12/23 rule.
To test this hypothesis, we incubated 5′Dβ1 and various Jβ substrates with RAG1/2 and HMGB1/2 in a buffer containing Mg2+ and assessed the kinetics of production of nicked products (Fig. 4). The initial rate of nicking (estimated from the 5-min time point) was at least ninefold faster for 5′Dβ1 than for the two fastest nicking Jβ substrates (Jβ2.7 and Jβ1.4). For the Jβ2.5 substrate, the difference was at least 100-fold, with very little nicked product detected at any time point (Fig. 4). Therefore, it is plausible that slow nicking of Jβ gene segments contributes to the B12/23 rule. We also noted that nicking and synapsis efficiencies were inversely correlated: the more rapidly nicking Jβ1.4 and Jβ2.7 substrates formed the PC with the lowest efficiency, and Jβ2.5 nicked very poorly but was the most efficient substrate in the synapsis assay (Fig. 3 and 4). This suggested that both the nicking and synapsis steps of the cleavage reaction contribute to the B12/23 rule and that deficiencies in these two steps combine in different ways to cripple the ability of different Jβ gene segments to recombine with Vβ gene segments.
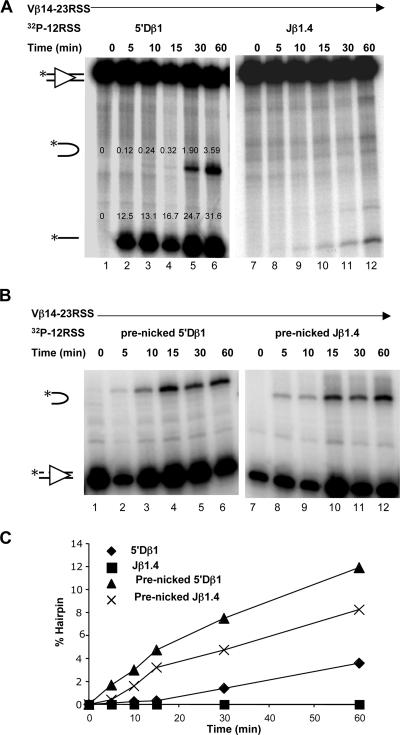
FIG. 4.
Assessment of DNA nicking. (A) Nicking reactions were performed with individual 32P-labeled 12RSS substrates in Mg2+ buffer for the times shown above the lanes. Analysis and symbols are as in the legend to Fig. 2A. (B) Quantitation of nicking of 12RSS substrates. The mean of at least three independent measurements of nicking is plotted (error bars indicate standard deviations).
If nicking contributes to the B212/23 rule, we would expect that a prenicked Jβ substrate would be able to undergo hairpin formation when paired with a Vβ substrate. To investigate this, we performed cleavage reactions using intact or prenicked 5′Dβ1 and Jβ1.4 substrates coupled with the intact Vβ14 substrate. Endogenous RSSs from the TCRβ locus generally cleave poorly in vitro, and hairpin formation with the intact 5′Dβ1 substrate was inefficient while that with the intact Jβ1.4 substrate was undetectable (Fig. 5A and C). Prenicking of 5′Dβ1 substantially increased hairpin formation, and importantly, prenicking of Jβ1.4 also resulted in a striking enhancement of hairpin formation to levels that were 50 to 75% of those observed with the prenicked 5′Dβ1 substrate (Fig. 5B and C). In the absence of a 23RSS partner, prenicked Jβ14 did not undergo cleavage (data not shown). These results were reproducible in multiple experiments. Therefore, prenicking substantially equalizes the ability of 5′Dβ1 and Jβ1.4 substrates to undergo cleavage, supporting the model that poor nicking of Jβ contributes substantially to the B12/23 rule.
FIG. 5.
Prenicking enables hairpin formation with the Jβ1.4 substrate. (A and B) DNA cleavage analyses were performed as described in the legend to Fig. 2A with intact (A) or prenicked (B) 5′Dβ1 and Jβ1.4 12RSS substrates together with the Vβ14 23RSS substrate. The percentages of input substrate converted to the nicked and hairpin products are indicated for the 5′Dβ1 substrate for comparison to the results shown in Fig. 6B. (C) Quantitation of hairpin formation for the intact and prenicked substrates. Results are representative of at least three (A) or two (B) independent experiments.
Nicking is a prerequisite for hairpin formation and has also been implicated in stabilizing the PC with consensus RSS substrates (20). Therefore, we hypothesized that poor nicking of Jβ substrates could exacerbate inefficient synapsis in vivo. To test this, we compared PC formation of intact and prenicked Jβ substrates with the Vβ14 substrate. Interestingly, prenicking only modestly increased synapsis for Jβ1.4 and Jβ2.7 (not statistically significant), while it substantially enhanced PC formation for Jβ2.5 (2.6-fold; P < 0.01) (Fig. 3B). Prenicking of 5′Dβ1 also significantly increased PC formation (1.7-fold; P < 0.02), but this was not observed for the prenicked consensus 12RSS, perhaps because synapsis was already quite efficient with the intact substrate (Fig. 3B). Synapsis of the nicked Jβ2.5 substrate was more efficient than the intact 5′Dβ1 substrate and 75% as efficient as with the nicked 5′Dβ1 substrate. Together, these results indicate that nicking does not enhance synapsis equally for all Jβ substrates and that some Jβ substrates—notably, those that exhibit clearly detectable nicking—are intrinsically poor at forming the PC. We conclude that while Jβ substrates uniformly synapse with Vβ14 less well than does 5′Dβ1, poor nicking of Jβ substrates is also critical for establishment of the B12/23 rule.
Roles of the heptamer, spacer, nonamer, and coding flank.
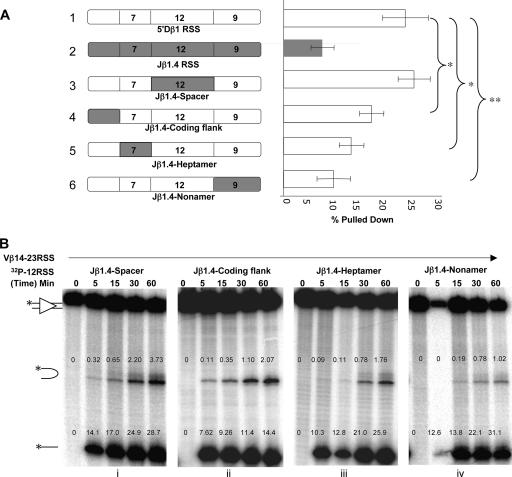
While the nonamer and spacer elements of 5′Dβ1 have been shown to play an important role in preferential recombination of Vβ with Dβ as opposed to Jβ (13, 14), it is unknown how these components or the heptamer and coding flank contribute mechanistically to the B12/23 restriction. To examine this, we measured synapsis and DNA cleavage with a panel of chimeric 5′Dβ1-Jβ1.4 RSSs (Fig. 6A). Substitution of the spacer sequence of 5′Dβ1 with that of Jβ1.4 had no significant effect on synapsis with Vβ14 compared to unmodified 5′Dβ1 (Fig. 6A, substrates 1 and 3). In contrast, replacement of the nonamer sequence of 5′Dβ1 with that of Jβ1.4 resulted in a substantial (60%) decrease in synapsis to levels almost equal to those observed with the Jβ1.4 substrate in this series of experiments. Substitution of the coding flank or heptamer of 5′Dβ1 with those of Jβ1.4 resulted in intermediate decreases of ≈25 or 40%, respectively, in PC formation. These results indicate that the coding flank, heptamer, and nonamer can all influence synapsis, with the nonamer playing the most significant role. However, the spacer does not appear to contribute to the preferential synapsis of Vβ14 with 5′Dβ1 as opposed to Jβ1.4. This might be because the spacer makes little contribution to the stability of the PC or because Jβ1.4 has a “good” spacer that makes it unable to interfere with PC formation by the 5′Dβ1 RSS (see Discussion).
FIG. 6.
Analyses of Jβ1.4-5′Dβ1 chimeric substrates. (A) Synapsis was measured for the chimeric substrates shown to the left of the graph. The shaded DNA regions were derived from Jβ1.4. The data in this panel are derived from a different set of experiments than the experiments shown in Fig. 3B. Analysis and data presentation are described as for Fig. 3B. Statistically significant differences are indicated (*, P < 0.05; **, P < 0.01). (B) DNA cleavage analyses were performed as described in the legend to Fig. 2A with chimeric 12RSS substrates (named as in panel A) together with the Vβ14 23RSS substrate. The percentages of input substrate converted to the nicked and hairpin products are indicated. The comparable data for the 5′Dβ1 substrate are presented in Fig. 5A, left panel.
Next, we examined the effects of these substitutions on nicking and hairpin formation in coupled cleavage reactions with the Vβ14 23RSS. Consistent with the synapsis results, the spacer substitution had little effect on either step of DNA cleavage, with results comparable to those for unmodified 5′Dβ1 (Fig. 5A and 6B, panel i), and the nonamer substitution had the strongest deleterious effect on hairpin formation (Fig. 6B, panel iv). Importantly, the nonamer substitution had no effect on nicking, indicating that binding of the RAG proteins to the RSS to form the SC was not substantially altered by the nonamer swap. Instead, the effects of the nonamer substitution on hairpin formation can be explained largely by its effects on synapsis. Also consistent with the results of the synapsis assay, the coding flank and heptamer substitutions yielded intermediate effects on hairpin formation (Fig. 6B, panels ii and iii). Interestingly, the chimeric substrate containing the Jβ1.4 coding flank exhibited a ≈50% decrease in nicking compared to the other three substitution substrates (compare panel ii to panels i, iii, and iv), although nicking remained much more efficient than for Jβ1.4 (e.g., Fig. 5A). These data suggest that nicking is not rate limiting for hairpin formation with these chimeric substrates and that the poor nicking observed with the Jβ1.4 substrate is due to the influence of the coding flank together with elements of the RSS. We conclude that in these substrates, the nonamer plays the most important role in controlling hairpin formation by virtue of its effect on synapsis and that several elements of the Jβ1.4 RSS act in concert to cause its poor nicking/poor synapsis properties.
Jβ2.5 and Jβ2.7 are two of the most frequently used J gene segments in the Jβ2 gene cluster (18). They exhibit distinct weaknesses in our biochemical assays: Jβ2.5 nicks very poorly but pairs well with Vβ14, while Jβ2.7 supports detectable nicking but relatively poor synapsis (Fig. 3 and 4). It was therefore of interest to determine how the different components of the RSS and the coding flank contribute to these distinct behaviors. To this end, we created hybrid substrates in which the coding flank or spacer motifs of Jβ2.5 were replaced with the corresponding motifs from Jβ2.7, or the nonconsensus cytosine residue in the fourth position of the nonamer of Jβ2.7 was replaced with the consensus adenine residue found in Jβ2.5 (Fig. 1B). Mutations in the fourth position of the nonamer interrupt the A tract and have been shown to reduce recombination in artificial recombination substrates in vivo (1, 11). The heptamer was not investigated because previous results indicated that the heptamers of these two Jβ gene segments function equivalently (22).
Substitution of the spacer of Jβ2.5 with that of Jβ2.7 decreased synapsis ≈20% (Fig. 7A) but increased nicking dramatically (36-fold at 60 min [Fig. 7B]; note that it is difficult to estimate the increase in the initial rate of nicking because no product was detected at early time points for Jβ2.5). Similar results were obtained when the Jβ2.5 coding flank was replaced with that from Jβ2.7: little if any change in PC formation but a dramatic enhancement of nicking (42-fold at 60 min [Fig. 7B]). Each of these chimeric substrates nicks more efficiently than Jβ2.7 itself (Fig. 7B). We conclude that the spacer and coding flank of Jβ2.5 contribute to its poor nicking ability, although it is clear from examination of initial rates of nicking that the coding flank plays a dominant role (the coding flank-substituted substrate nicks nearly fivefold faster in the first 5 minutes than the spacer-substituted substrate; Fig. 7B).
FIG. 7.
Analyses of Jβ2.5-Jβ2.7 chimeric substrates. (A) Synapsis was measured for the chimeric substrates shown to the left of the graph. The shaded DNA regions were derived from Jβ2.7. The Jβ2.7 (C/A) substrate contains a C→A substitution in the fourth position of the nonamer. The data in this panel are derive from a different set of experiments than the experiments shown in Fig. 3B. Analysis and data presentation are as described in the legend to Fig. 3B. Statistically significant differences are indicated (*, P < 0.01; **, P < 0.001). (B) Quantitation of nicking of chimeric 12RSS substrates (named as in panel A). Experiments were performed as described in the legend to Fig. 4. Data analysis and presentation are as described in the legend to Fig. 4B. (C) DNA cleavage analyses were performed as described in the legend to Fig. 2A with consensus (lanes 1 and 2) or chimeric (lanes 3 to 11) 12RSS substrates together with the consensus (lanes 1 and 2) or Vβ14 23RSS (lanes 3 to 11) substrates as indicated above the lanes. Faint bands corresponding to hairpin products detected in lanes 5 and 8 are indicated with black arrows.
The C→A mutation of the fourth position of the Jβ2.7 nonamer creates a consensus nonamer and causes a nearly fivefold increase in synapsis with Vβ14 (Fig. 7A) and a sevenfold increase in nicking efficiency (Fig. 7B). The Jβ2.7 (C→A) substrate nicks comparably to 5′Dβ1 (≈30% nicking at 60 min [Fig. 5A]) and forms the PC with Vβ14 at least as efficiently as Jβ2.5 (Fig. 7A). Given this, we predicted that this substrate would be competent for hairpin formation in cleavage reactions with a Vβ14 partner. Indeed, hairpin formation was readily detected with the Jβ2.7 (C→A) substrate but only when the reaction mixture included a Vβ14 23RSS partner (Fig. 7C, lanes 10 and 11). We also examined the two chimeric Jβ2.5 substrates, containing either the spacer or coding flank of Jβ2.7 in this assay. In both cases, a low level of hairpin formation was detected specifically in reaction mixtures containing a Vβ14 partner (Fig. 7C, lanes 4 and 5 and lanes 7 and 8). Together, our results indicate that the B12/23 rule is established at different steps of the reaction by different DNA elements for Jβ2.5 and Jβ2.7: for Jβ2.5, the primary deficit is in nicking and is enforced by the coding flank and spacer, whereas for Jβ2.7, the primary deficit is in synapsis and is imposed by a single nucleotide alteration in the nonamer.
How does the 3′Dβ1 23RSS overcome these deficits so as to allow D-to-J recombination at biologically useful efficiencies? We hypothesized that the answer would lie at least in part in the synapsis step of the reaction. Indeed, the 3′Dβ1 23RSS supports PC formation with Jβ1.4, Jβ2.5, and Jβ2.7 2- to 3.5-fold more efficiently than does the Vβ14 23RSS (Fig. 3B; P < 0.025). Enhanced synapsis is therefore likely to provide part of the explanation for the ability of 3′Dβ RSSs to recombine with Jβ gene segments and for its demonstrated ability to support coupled cleavage with Jβ RSS substrates in vitro (22, 23).
DISCUSSION
The B12/23 rule is largely or entirely enforced during the DNA cleavage phase of V(D)J recombination by the RAG proteins, the TCRβ RSSs, and their flanking coding sequences (reviewed in reference 29). We sought to determine which step(s) of the DNA cleavage process represents the critical control points for the B12/23 restriction and how the coding flank and components of the RSS contribute to these control points. Our data support the conclusions that Vβ-to-Jβ recombination is short circuited at several steps through the combined action of several RSS elements and that each Jβ gene segment is defective for a unique constellation of reasons. Below we consider each step in the cleavage reaction (binding, nicking, synapsis, and hairpin formation) and examine the contribution of each DNA element (heptamer, spacer, nonamer, and coding flank).
DNA binding to form the SC.
Better binding of the RAG proteins to 5′Dβ RSSs than to Jβ RSSs was previously found not to be a general explanation for the B12/23 rule. This was based on the finding that two Jβ RSSs (Jβ1.1 and Jβ2.5) formed the SC more efficiently than the 5′ Dβ1 RSS did, while two other RSSs (Jβ2.2 and Jβ2.7) bound with ≈35% and 20% of the efficiency, respectively, of the 5′ Dβ1 RSS (23). We also did not observe obviously better SC formation with the 5′Dβ1 substrate than with Jβ substrates. Nonetheless, for certain Jβ RSSs, it is plausible that weak SC formation is one factor contributing to poor recombination with Vβ (see below).
Nicking.
The most surprising aspect of our study is the link between inefficient nicking of Jβ gene segments and the B12/23 rule. The three Jβ substrates examined here nick substantially more slowly than the 5′Dβ1 substrate (Fig. 4B), and it is clear that this is not due simply to poor SC formation. Indeed, the Jβ substrate (Jβ2.5) with the strongest binding (23) and which undergoes synapsis with Vβ14 most efficiently (Fig. 3B) also exhibits the poorest nicking (Fig. 4). The importance of poor nicking is dramatically illustrated by the finding that a prenicked Jβ1.4 substrate supports hairpin formation substantially more efficiently than an intact 5′Dβ1 substrate. Furthermore, alterations of the Jβ2.5 substrate that enhance nicking but not synapsis also allow detectable hairpin formation (Fig. 7 and see below). With optimal RSS substrates, nicking is ≈150-fold faster than hairpin formation (34). With certain coding flanks, however, nicking becomes rate limiting even with consensus heptamer/nonamer sequences (35), leading the authors of that study to predict that, depending on the substrate, either nicking or hairpin formation could be rate limiting for DNA cleavage in vivo. Our data strongly support this idea and the hypothesis that slow nicking of Jβ substrates is a major underlying component of the B12/23 rule. Our findings also predict the formation in vivo of synaptic complexes containing a nicked Vβ and intact Jβ substrate. We propose that dissociation of such complexes occurs much faster than nicking of Jβ. After its release, the nicked Vβ substrate can then participate in recombination with a 5′Dβ substrate, be repaired by a DNA ligase, or possibly engage in aberrant recombination reactions (16).
Synapsis.
Given that hairpin formation is quite slow (34), it is highly likely that the stability of the synaptic complex strongly influences cleavage efficiency. We find that Vβ14 forms the PC more efficiently with 5′Dβ1 than with any of the Jβ substrates tested (Fig. 3B). Jβ substrates pair more efficiently with the 3′Dβ1 substrate than with the Vβ14 substrate (Fig. 3B), and where substrate alterations perturb the efficiency of synapsis, the changes are closely mirrored by changes in cleavage efficiencies (Fig. 6 and 7). Particularly noteworthy in this regard are substitutions of the Jβ1.4 heptamer or nonamer for the corresponding element of the 5′Dβ1 substrate, both of which reduce synapsis and hairpin formation with a Vβ14 23RSS partner but not nicking of 12RSS itself (Fig. 6). Overall, our data strongly argue that synapsis is an important step at which the B12/23 rule is enforced. It is worth noting that synapsis was measured under conditions that do not support nicking (Ca2+ buffer) and hence that differences in nicking efficiencies cannot explain the differences with which the various 12RSS substrates synapse with Vβ14.
It was previously observed, using a biotin pull-down assay, that prenicking one or both of a consensus 12/23 RSS pair of substrates enhances PC stability (20). While prenicking the 12RSS partner of Vβ14 enhanced synapsis in most cases tested, the effect was minimal for some substrates and was not observed at all for the consensus 12RSS substrate (Fig. 3B). The ability of 12RSS substrate nicking to stabilize the PC might rely on nicking of the Vβ14 23RSS substrate, and it might depend heavily on as yet uncharacterized sequence features of the RSSs.
Hairpin formation.
The in gel method developed by Swanson to assess hairpin formation independent of binding and synapsis (28) could not be used for analysis of endogenous TCRβ substrates because of their poor formation of the PC in electrophoretic mobility shift assays. Our finding that prenicking of Jβ1.4 allows relatively efficient hairpin formation with Vβ14 indicates that hairpin formation is not a critical control point for this substrate pair. Nonetheless, we cannot rule out the possibility that hairpin formation is also a B12/23-regulated step.
Heptamer.
Insertion of the heptamer of Jβ1.2 into the 5′Dβ1 RSS reduced minilocus Vβ14-to-DJβ recombination by ≈50%, indicating a modest role for the heptamer in B12/23 restriction (13). The heptamer was also found to contribute only modestly to the relative strength of Jβ RSSs (22). Our results are consistent with these findings, with substitution of the Jβ1.4 heptamer into the 5′Dβ1 RSS reducing synapsis and hairpin formation by nearly twofold. The heptamer does not appear to be a major determinant of the enormous (up to 500-fold [14]) preference of Vβ to recombine with 5′Dβ as opposed to Jβ.
Spacer.
Previous studies have indicated an important role for the spacers of the 23RSS (14) and 12RSS (13, 22) in establishing the B12/23 rule. Substitution of the Jβ1.2 spacer into the 5′Dβ1 12RSS resulted in a dramatic reduction in minilocus V-to-DJβ recombination, an effect that was narrowed down to spacer residues 1, 2, 4, and 12 (underlined in Fig. 1B) that are highly conserved in 5′Dβ RSSs (13). Our finding that insertion of the Jβ1.4 spacer into the 5′Dβ1 RSS had no effect on synapsis or cleavage stands in apparent contradiction to this. However, whereas the spacer of Jβ1.2 differs from that of 5′Dβ1 at positions 1, 2, 4, and 12, the spacer of Jβ1.4 matches that of 5′Dβ1 at three of these four positions (Fig. 1B). This strongly supports the hypothesis that the “deterioration” (23) of different Jβ substrates is due to the accumulation of mutations in different DNA elements: for Jβ1.2, crippling sequence changes have occurred in the spacer and nonamer (13), whereas for Jβ1.4, the spacer is “good” and deterioration focuses on the other DNA elements (Table 2).
TABLE 2.
Jβ gene segment defects that contribute to the B12/23 rule
| Jβ segment | Defective DNA element(s)a | Defective cleavage step(s)b | Source and/or reference(s) |
|---|---|---|---|
| 1.2 | Nonamer = spacer ≫ heptamer | ND | 13 |
| 1.4 | Nonamer > heptamer > coding flank | Nicking, synapsis | This work; 14 |
| 2.2 | Nonamer (pos. 4) > spacer | ND (SC formation?) | 22, 23 |
| 2.5 | Coding flank > spacer | Nicking | This work; 22, 23 |
| 2.7 | Nonamer (pos. 4) | Nicking, synapsis (SC formation?) | This work; 22, 23 |
DNA elements are ranked according to the significance of their contribution to the B12/23 rule (from most to least) where possible. pos., position.
Defects in SC formation might be important for certain Jβ gene segments. ND, not determined.
Jβ2.5 provides a second example of a suboptimal spacer. When it was replaced with the spacer of Jβ2.7, nicking was substantially enhanced and hairpin formation became detectable despite the fact that synapsis with Vβ14 was, if anything, reduced (Fig. 7). An identical spacer swap in a previous study found that the spacer of Jβ2.7 supported better recombination and cleavage than that of Jβ2.5, leading to the prediction that the spacer of Jβ2.7 functions by enhancing PC formation (22). Our data indicate that this is incorrect and that instead the Jβ2.7 spacer acts at the nicking step of the reaction. The Jβ2.7 RSS matches that of 5′Dβ1 at only one of the four spacer positions noted above (position 4), whereas Jβ2.5, like Jβ1.2, differs at all four (Fig. 1B). We suggest that the consensus C at spacer position 4 (found in virtually all 5′Dβ 12RSSs) together with the consensus A at position 5 (the most highly conserved [24] and functionally important [15] 12RSS spacer residue) are important in explaining the stronger function of the Jβ2.7 spacer compared to that of Jβ2.5. While it is not known how the spacer influences nicking, we speculate that favorable protein interactions with the spacer support DNA distortion/bending near the site of cleavage proposed to be important for nicking (25).
Nonamer.
Previous findings strongly implicated the nonamer in the B12/23 rule, most notably the observation that replacement of the nonamer of 5′Dβ1 with that of Jβ1.2 drastically reduced minilocus V-to-DJβ recombination (13). We now demonstrate that the nonamer acts to a significant extent at the synapsis step of the reaction (Fig. 6 and 7). Interestingly, substitution of the Jβ1.4 nonamer for that of 5′Dβ1 substantially reduced synapsis (and hairpin formation) without affecting nicking, arguing that this particular swap did not radically alter SC formation. A single C-to-A mutation in the fourth position of the nonamer of Jβ2.7 dramatically enhanced synapsis, hairpin formation, and nicking (Fig. 7). In this case, it is reasonable to think that the mutation enhanced SC formation, which could at least partially explain the other effects observed. We conclude that the nonamer exerts much of its influence on the B12/23 rule at the synapsis step and that at least one Jβ gene segment (Jβ2.7) is crippled for direct V-to-J recombination by virtue of a single deleterious change in the nonamer. Consistent with this, an A-to-C mutation of the fourth position of the Jβ2.5 nonamer was previously demonstrated to substantially compromise RSS function (22).
Coding flank.
We propose that the coding flank can strongly influence the B12/23 restriction through its effects on nicking. The Jβ2.5 substrate nicks extremely poorly, but substitution of a Jβ2.7 coding flank strongly enhanced nicking (more than 40-fold) and allowed hairpin formation (Fig. 7). This is consistent with the finding that a coding flank terminating with the sequence TT immediately adjoining the heptamer, as in Jβ2.5 (Fig. 1B), confers a slow nicking phenotype on a consensus RSS (35). The Jβ2.7 coding flank (terminating in AG) was not previously tested (35), but our data indicate that it supports efficient nicking. While a poor coding flank helps prevent V-to-Jβ2.5 recombination, the coding flank of Jβ1.4 appears not to play as large a role. Appending the Jβ1.4 coding flank onto the 5′Dβ1 RSS reduces recombination in vivo (14) and nicking and hairpin formation in vitro (Fig. 6) by only ≈50%.
Multiple elements and multiple steps enforce the B12/23 rule.
We propose that direct V-to-Jβ recombination is prevented by distinct mechanisms for each Jβ gene segment, with each Jβ gene segment having acquired its own idiosyncratic deleterious mutations during evolution (summarized in Table 2). In contrast, given the high level of conservation observed within 5′Dβ 12RSSs and within 3′Dβ 23RSSs, it is plausible that Dβ RSSs resisted most evolutionary changes to ensure the inclusion of Dβ gene segments within assembled TCRβ genes. For Jβ1.4, a poor nonamer together with relatively weak heptamer and coding flank sequences creates a substrate with significant defects in both synapsis and nicking. For Jβ2.5, a nearly consensus nonamer that supports strong synapsis is overwhelmed by a dramatic nicking defect caused by deleterious spacer and coding flank sequences. Jβ2.7 is crippled in large part by a single deleterious nucleotide in the nonamer that undermines both nicking and synapsis. The 3′Dβ1 RSS overcomes these deficits in part through enhanced synapsis (Fig. 3B), with the spacer implicated as a particularly important element (14). We predict that subtle sequence variations in the RSS and coding flank influence gene segment usage by the mechanisms identified here in antigen receptor loci other than the TCRβ locus.
Acknowledgments
We thank Mihai Ciubotaru for advice and technical assistance throughout this work and other members of the Schatz lab for helpful suggestions.
This work was supported by Public Health Service grant AI32524 from the National Institute of Allergy and Immunology. S.D.F. was supported by the Irvington Institute for Immunological Research. D.G.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Akamatsu, Y., N. Tsurushita, F. Nagawa, M. Matsuoka, K. Okazaki, M. Imai, and H. Sakano. 1994. Essential residues in V(D)J recombination signals. J. Immunol. 153:4520-4529. [PubMed] [Google Scholar]
- 2.Bassing, C. H., F. W. Alt, M. M. Hughes, M. D'Auteuil, T. D. Wehrly, B. B. Woodman, F. Gartner, J. M. White, L. Davidson, and B. P. Sleckman. 2000. Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature 405:583-586. [DOI] [PubMed] [Google Scholar]
- 3.Bassing, C. H., W. Swat, and F. W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109:S45-S55. [DOI] [PubMed] [Google Scholar]
- 4.Cowell, L. G., M. Davila, D. Ramsden, and G. Kelsoe. 2004. Computational tools for understanding sequence variability in recombination signals. Immunol. Rev. 200:57-69. [DOI] [PubMed] [Google Scholar]
- 5.Eastman, Q. M., I. J. Villey, and D. G. Schatz. 1999. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol. Cell. Biol. 19:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feeney, A. J., P. Goebel, and C. R. Espinoza. 2004. Many levels of control of V gene rearrangement frequency. Immunol. Rev. 200:44-56. [DOI] [PubMed] [Google Scholar]
- 7.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 8.Fugmann, S. D., I. J. Villey, L. M. Ptaszek, and D. G. Schatz. 2000. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell 5:97-107. [DOI] [PubMed] [Google Scholar]
- 9.Gellert, M. 2002. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 10.Gerstein, R. M., and M. R. Lieber. 1993. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev. 7:1459-1469. [DOI] [PubMed] [Google Scholar]
- 11.Hesse, J. E., M. R. Lieber, K. Mizuuchi, and M. Gellert. 1989. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 3:1053-1061. [DOI] [PubMed] [Google Scholar]
- 12.Hiom, K., and M. Gellert. 1998. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell 1:1011-1019. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, M. M., R. E. Tillman, T. D. Wehrly, J. M. White, and B. P. Sleckman. 2003. The B12/23 restriction is critically dependent on recombination signal nonamer and spacer sequences. J. Immunol. 171:6604-6610. [DOI] [PubMed] [Google Scholar]
- 14.Jung, D., C. H. Bassing, S. D. Fugmann, H. L. Cheng, D. G. Schatz, and F. W. Alt. 2003. Extrachromosomal recombination substrates recapitulate beyond 12/23 restricted V(D)J recombination in nonlymphoid cells. Immunity 18:65-74. [DOI] [PubMed] [Google Scholar]
- 15.Lee, A. I., S. D. Fugmann, L. G. Cowell, L. M. Ptaszek, G. Kelsoe, and D. G. Schatz. 2003. A functional analysis of the spacer of V(D)J recombination signal sequences. PLoS Biol. 1:56-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, G. S., M. B. Neiditch, S. S. Salus, and D. B. Roth. 2004. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell 117:171-184. [DOI] [PubMed] [Google Scholar]
- 17.Lewis, S. M. 1994. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv. Immunol. 56:27-150. [DOI] [PubMed] [Google Scholar]
- 18.Livàk, F., D. B. Burtrum, L. Rowen, D. G. Schatz, and H. T. Petrie. 2000. Genetic modulation of T cell receptor gene segment usage during somatic recombination. J. Exp. Med. 192:1191-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 20.Nagawa, F., S. Hirose, H. Nishizumi, T. Nishihara, and H. Sakano. 2004. Joining mutants of RAG1 and RAG2 that demonstrate impaired interactions with the coding-end DNA. J. Biol. Chem. 279:38360-38368. [DOI] [PubMed] [Google Scholar]
- 21.Oettinger, M. A., D. G. Schatz, C. Gorka, and D. Baltimore. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248:1517-1523. [DOI] [PubMed] [Google Scholar]
- 22.Olaru, A., D. N. Patterson, H. R. Cai, and F. Livàk. 2004. Recombination signal sequence variations and the mechanism of patterned T-cell receptor-beta locus rearrangement. Mol. Immunol. 40:1189-1201. [DOI] [PubMed] [Google Scholar]
- 23.Olaru, A., D. N. Patterson, I. Villey, and F. Livàk. 2003. DNA-Rag protein interactions in the control of selective D gene utilization in the TCR beta locus. J. Immunol. 171:3605-3611. [DOI] [PubMed] [Google Scholar]
- 24.Ramsden, D. A., K. Baetz, and G. E. Wu. 1994. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 22:1785-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santagata, S., E. Besmer, A. Villa, F. Bozzi, J. S. Allingham, C. Sobacchi, D. B. Haniford, P. Vezzoni, M. C. Nussenzweig, Z.-Q. Pan, and P. Cortes. 1999. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol. Cell 4:935-947. [DOI] [PubMed] [Google Scholar]
- 26.Schatz, D. G., M. A. Oettinger, and D. Baltimore. 1989. The V(D)J recombination activating gene (RAG-1). Cell 59:1035-1048. [DOI] [PubMed] [Google Scholar]
- 27.Sleckman, B. P., C. H. Bassing, M. M. Hughes, A. Okada, M. D'Auteuil, T. D. Wehrly, B. B. Woodman, L. Davidson, J. Z. Chen, and F. W. Alt. 2000. Mechanisms that direct ordered assembly of T cell receptor beta locus V, D, and J gene segments. Proc. Natl. Acad. Sci. USA 97:7975-7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swanson, P. C. 2002. Fine structure and activity of discrete RAG-HMG complexes on V(D)J recombination signals. Mol. Cell. Biol. 22:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tillman, R. E., A. L. Wooley, M. M. Hughes, B. Khor, and B. P. Sleckman. 2004. Regulation of T-cell receptor beta-chain gene assembly by recombination signals: the beyond 12/23 restriction. Immunol. Rev. 200:36-43. [DOI] [PubMed] [Google Scholar]
- 30.Tillman, R. E., A. L. Wooley, B. Khor, T. D. Wehrly, C. A. Little, and B. P. Sleckman. 2003. Targeting of V beta to D beta rearrangement by RSSs can be mediated by the V(D)J recombinase in the absence of additional lymphoid-specific factors. J. Immunol. 170:5-9. [DOI] [PubMed] [Google Scholar]
- 31.van Gent, D. C., K. Mizuuchi, and M. Gellert. 1996. Similarities between initiation of V(D)J recombination and retroviral integration. Science 271:1592-1594. [DOI] [PubMed] [Google Scholar]
- 32.Wu, C., C. H. Bassing, D. Jung, B. B. Woodman, D. Foy, and F. W. Alt. 2003. Dramatically increased rearrangement and peripheral representation of V beta 14 driven by the 3′ D beta 1 recombination signal sequence. Immunity 18:75-85. [DOI] [PubMed] [Google Scholar]
- 33.Wu, C., S. Ranganath, M. Gleason, B. B. Woodman, T. M. Borjeson, F. W. Alt, and C. H. Bassing. 2007. Restriction of endogenous T cell antigen receptor beta rearrangements to Vbeta14 through selective recombination signal sequence modifications. Proc. Natl. Acad. Sci. USA 104:4002-4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, K., A. Taghva, Y. Ma, and M. R. Lieber. 2004. Kinetic analysis of the nicking and hairpin formation steps in V(D)J recombination. DNA Repair (Amsterdam) 3:67-75. [DOI] [PubMed] [Google Scholar]
- 35.Yu, K. F., and M. R. Lieber. 1999. Mechanistic basis for coding end sequence effects in the initiation of V(D)J recombination. Mol. Cell. Biol. 19:8094-8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, K. F., and M. R. Lieber. 2000. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol. 20:7914-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]