Abstract
Nuclear cap binding complex (CBC) is recruited cotranscriptionally and stimulates spliceosome assembly on nascent mRNAs; however, its possible functions in regulating transcription elongation or termination were not well understood. We show that, while CBC appears to be dispensable for normal rates and processivity of elongation by RNA polymerase II (Pol II), it plays a direct role in preventing polyadenylation at weak termination sites. Similarly to Npl3p, with which it interacts, CBC suppresses the weak terminator of the gal10-Δ56 mutant allele by impeding recruitment of termination factors Pcf11p and Rna15p (subunits of cleavage factor IA [CF IA]) and does so without influencing Npl3p occupancy at the termination site. Importantly, deletion of CBC subunits or NPL3 also increases termination at a naturally occurring weak poly(A) site in the RNA14 coding sequences. We also show that CBC is most likely recruited directly to the cap of nascent transcripts rather than interacting first with transcriptional activators or the phosphorylated C-terminal domain of Pol II. Thus, our findings illuminate the mechanism of CBC recruitment and extend its function in Saccharomyces cerevisiae beyond mRNA splicing and degradation of aberrant nuclear mRNAs to include regulation of CF IA recruitment at poly(A) selection sites.
Prior to export of mRNA from the nucleus, the m7Gppp 5′ cap is bound by the nuclear cap binding complex (CBC), comprised in Saccharomyces cerevisiae of Cbp20p and Cbp80p (1). The CBC subunits are not essential in yeast, but deletion of CBP20 (also known as CBC2, MUD13, and SAE1) was shown to reduce the efficiency of pre-mRNA splicing (9). Consistent with this, both CBC subunits reside in the splicing commitment complex (50), and CBC stimulates spliceosome assembly in yeast (13). CBC is a limiting factor for export of U snRNAs in Xenopus laevis oocytes but is apparently dispensable for efficient nuclear export of mRNAs in yeast (20). CBC is required for degradation of mRNAs retained in the nucleus and was implicated in blocking utilization of defective transcription termination sites in yeast cells (10, 11).
Synthesis of the m7G cap is carried out in budding yeast by a complex of three enzymes: RNA triphosphatase Cet1p, RNA guanylyltransferase Ceg1p, and guanine 7-methyltransferase Abd1p. Capping occurs cotranscriptionally in yeast and other eukaryotes (1) and is facilitated by recruitment of capping enzymes to the heptad repeats in the C-terminal domain (CTD) of the Rpb1p subunit of polymerase II (Pol II) when phosphorylated on serine-5 by the Kin28p subunit of transcription factor IIH (TFIIH) (23, 29, 39). It is likely that binding of CBC to the cap also occurs cotranscriptionally, as Cbp80p was coimmunoprecipitated with Pol II in mammals (25) and Cbp80p and Cbp20p were found associated with certain constitutively transcribed genes in yeast (49) in an RNA-dependent manner (38). Cotranscriptional recruitment is also implied by the fact that CBC stimulates spliceosome assembly at transcribed genes in yeast (13). However, the molecular mechanism of CBC recruitment has not been investigated.
Transcriptional activators in yeast recruit an array of coactivators, such as histone acetyltransferase complex SAGA, directly to the upstream activation sequences (UASs) of target genes independently of a functional downstream promoter or recruitment of Pol II itself (4, 34). Recruitment of other factors that function during elongation, such as the Paf1 complex (32), occurs indirectly and is stimulated by Rpb1p-CTD phosphorylation, as observed for capping enzymes (42). We considered that binding of CBC to the nascent transcripts could be stimulated indirectly by recruitment of CBC to the UAS by activators, or to the promoter region by the phosphorylated Rpb1p-CTD, prior to its interaction with the mRNA cap. Our analysis of CBC recruitment by chromatin immunoprecipitation (ChIP) assays in various yeast mutants disfavors an important role for CBC interaction with activators or phosphorylated Rpb1p and provides evidence that the m7G cap is the key factor driving CBC recruitment to nascent transcripts in yeast cells.
Given the role of CBC in stimulating cotranscriptional spliceosome assembly, we also wondered whether it enhances the rate or processivity of transcription elongation by Pol II or regulates 3′-end formation. The possibility of an elongation defect was prompted by the finding that cbp80Δ null mutants display increased sensitivity to 6-azauracil (6AU) and mycophenolic acid (MPA) (36). These drugs reduce the elongation rate and processivity of Pol II in vivo (26), and sensitivity to both is frequently displayed by mutants impaired for transcription elongation (36). Furthermore, cbp80 mutations interact genetically with mutations in HPR1 (47), encoding a subunit of the THO/TREX complex involved in transcription elongation and mRNA export (37, 44), and deletion of HPR1 (or other THO subunits) reduces processivity of Pol II elongation in vivo (26). cbp80 mutations also interact with mutations in DNA topoisomerase I and increase negative superhelicity of a plasmid in vivo, which could arise from longer RNA-DNA hybrids formed by nascent transcripts during elongation (47). Finally, Cbp80p copurified with certain factors implicated in transcription elongation, including Bur1p (Sgv1p), Bur2p, Nhp68p (yFACT complex), and Tho2p (THO complex) (12). However, using a sophisticated ChIP assay to measure the progression of Pol II molecules during transcription of an extended (∼8-kb) coding sequence (26), we show that inactivation of CBC has no detectable effect on the rate or processivity of elongating Pol II molecules in vivo.
The idea that CBC participates in termination was suggested previously by the finding that inactivation of CBP80 partially suppressed the effects of a mutation in the 3′ end of the CYC1 gene (cyc1-512) that leads to decreased CYC1 mRNA coupled with the appearance of longer, read-through transcripts. Although cbp80Δ decreased the degradation rate of certain read-through transcripts produced from cyc1-512, it also appeared to increase the level of properly terminated transcripts by enhancing termination at the correct poly(A) addition site (11). This finding suggested that CBC suppresses the recognition of weak or defective signals for 3′-end formation.
We have taken several approaches to obtaining direct evidence that CBC antagonizes transcription termination independently of its effects on turnover of read-through transcripts and to probe the molecular mechanism of this antitermination activity. First, we analyzed the effect of cbp80Δ on the gal10-Δ56 allele, which contains a defective terminator. The Gal− phenotype of this mutation results from interference with preinitiation complex (PIC) assembly downstream at the GAL7 promoter by elongating Pol II molecules that read through the defective poly(A) site at gal10-Δ56 (7). Our results indicate that cbp80Δ decreases the number of such “read-through” polymerases, directly implicating wild-type (WT) CBC in blocking recognition of the weak gal10-Δ56 terminator. Second, we conducted ChIP assays to demonstrate that cbp80Δ elevates recruitment of subunits of the termination complex cleavage factor IA (CF IA) to the gal10-Δ56 terminator, providing a molecular mechanism for CBC action in blocking termination. The same function was recently identified for the NPL3 product (7), and interestingly, Npl3p interacts with the CBC both genetically (synthetic lethal interactions) and physically (in the context of mRNP) (28, 41). Finding that recruitment of Npl3p to the termination region of gal10-Δ56 is unaffected by cbp80Δ, we conclude that CBC and Npl3p both function cotranscriptionally to impede CF IA recruitment to weak terminators. We further demonstrate that CBC and Npl3p cooperate to suppress termination at a naturally occurring weak poly(A) site within the native RNA14 coding sequences. Our finding that 6AU, like cbp80Δ and npl3Δ mutations, increases utilization of the internal RNA14 terminator helps to explain the 6AU sensitivity of these mutants.
MATERIALS AND METHODS
Yeast strains.
All yeast strains used in this work are listed in Table 1. The WT parent strain BY4741 and deletion derivatives were described previously (48) and purchased from Research Genetics. The presence of all reported deletion alleles was confirmed by PCR amplification or complementation of mutant phenotypes by plasmid-borne WT genes. Myc-tagged strains were constructed as described previously (32).
TABLE 1.
Yeast strains used in this study
| Name | Parent | Genotypeb | Reference or source |
|---|---|---|---|
| Myc-tagged strains | |||
| HQY691 | BY4741a | SUA7-myc13::HIS3* | 32 |
| CMY005 | BY4741a | CBP20-myc13::HIS3* | This work |
| CMY006 | BY4741a | CBP80-myc13::HIS3* | This work |
| CMY007 | 249a | CBP20-myc13::HIS3* gcn4Δ::kanMX4 | This work |
| CMY008 | HQY700 | CBP20-myc13::HIS3* arg1-ΔTATA | This work |
| CMY009 | 6566a | CBP20-myc13::HIS3* cbp80Δ::kanMX4 | This work |
| CMY010 | 4268a | CBP20-myc13::HIS3* npl3Δ::kanMX4 | This work |
| CMY011 | 2074a | CBP80-myc13::HIS3* cbp20Δ::kanMX4 | This work |
| CMY012 | 4268a | CBP80-myc13::HIS3* npl3Δ::kanMX4 | This work |
| CMY013 | HQY957 | CBP80-myc13::HIS3* kin28Δ::kanMX4[LEU2 KIN28-HA] | This work |
| CMY014 | HQY958 | CBP80-myc13::HIS3* kin28Δ::kanMX4[LEU2 kin28-HA-ts16] | This work |
| CMY015 | ABD1WT | CBP80-myc13::HIS3* abd1Δ::LEU2[TRP1 ABD1] | This work |
| CMY016 | abd1-5 | CBP80-myc13::HIS3* abd1Δ::LEU2[TRP1 abd1-5] | This work |
| CMY017 | CMY125 | RPB3-myc13::HIS3* PGAL1-YLR454w | This work |
| CMY018 | CMY126 | RPB3-myc13::HIS3* cbp80Δ::kanMX4 PGAL1-YLR454w | This work |
| CMY019 | BY4741 | PCF11-myc13::HIS3* | This work |
| CMY020 | CMY127 | PCF11-myc13::HIS3* gal10-Δ56 | This work |
| CMY021 | CMY128 | PCF11-myc13::HIS3* cbp80Δ::kanMX4 gal10-Δ56 | This work |
| CMY022 | CMY129 | PCF11-myc13::HIS3* npl3Δ::kanMX4 gal10-Δ56 | This work |
| CMY023 | BY4741 | RNA15-myc13::HIS3* | This work |
| CMY024 | CMY127 | RNA15-myc13::HIS3* gal10-Δ56 | This work |
| CMY025 | CMY128 | RNA15-myc13::HIS3* cbp80Δ::kanMX4 gal10-Δ56 | This work |
| CMY026 | CMY129 | RNA15-myc13::HIS3* npl3Δ::kanMX4 gal10-Δ56 | This work |
| CMY027 | BY4741 | NPL3-myc13::HIS3 | This work |
| CMY028 | 2074a | NPL3-myc13::HIS3* cbp20Δ::kanMX4 | This work |
| CMY029 | 6566a | NPL3-myc13::HIS3* cbp80Δ::kanMX4 | This work |
| Untagged strains | |||
| BY4741 | matahis3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 | Research Genetics | |
| 249a | BY4741a | gcn4Δ::kanMX4 | Research Genetics |
| 2074a | BY4741a | cbp20Δ::kanMX4 | Research Genetics |
| 6566a | BY4741a | cbp80Δ::kanMX4 | Research Genetics |
| 4268a | BY4741a | npl3Δ::kanMX4 | Research Genetics |
| HQY700 | BY4741a | arg1-ΔTATA | 32 |
| HQY957 | BY4741a | kin28Δ::kanMX4[LEU2 KIN28-HA] | 32 |
| HQY958 | BY4741a | kin28Δ::kanMX4[LEU2 kin28-HA-ts16] | 32 |
| CMY125 | BY4741a | PGAL1-YLR454w | This work |
| CMY126 | 6566a | cbp80Δ::kanMX4 PGAL1-YLR454w | This work |
| CMY127 | BY4741a | gal10-Δ56 | This work |
| CMY128 | 6566a | cbp80Δ::kanMX4 gal10-Δ56 | This work |
| CMY129 | 4268a | npl3Δ::kanMX4 gal10-Δ56 | This work |
| W303 | matα leu2 ura3 lys2 trp1 his3 | ||
| ABD1 WT | W303 | abd1Δ::LEU2[TRP1 ABD1] | 40 |
| abd1-5 | W303 | abd1Δ::LEU2[TRP1 abd1-5] | 40 |
Purchased from Research Genetics (48).
HIS3* designates the HIS3 allele from Saccharomyces kluyveri.
To construct gal10-Δ56 mutants, plasmid pCK200 (21) was linearized with NheI and used to transform the appropriate ura3Δ strains to Ura+, directing plasmid integration to the GAL10-GAL7 locus. Ura− derivatives were selected on medium containing 5-fluoroorotic acid and screened for recombination events that replaced WT GAL10-GAL7 with gal10-Δ56 by PCR analysis of genomic DNA using oligonucleotide primers complementary to sequences upstream or downstream of the GAL10 poly(A) site: 5′-CTTTTAGTTCTTAATTGCAACAC-3′ and 5′-CCGTCCATATCTTTCCATAG-3′. The PGAL1-YLR454w strains were prepared by single-step integration of a KpnI-linearized URA3 plasmid containing the GAL1 promoter fused to the first 300 bp of the YLR454w coding region, as described previously (26). All strains were verified by PCR analysis.
Biochemical methods.
The ChIP experiments were conducted as described previously (34) using the primers described elsewhere (6, 7, 22, 33) or provided on request. Northern analysis of total RNA was carried out as described previously (32). Poly(A)+ RNA was purified from total RNA on oligo(dT)-cellulose (Biolab) (3). Western analysis of whole-cell extracts was conducted using extracts prepared by trichloroacetic acid precipitation (35). Antibodies employed were the following: monoclonal anti-Myc (Roche), anti-Rpb3p (Neoclone), anti-Rpb1p-Ser5P (H14; Covance), anti-Rpb1p (8WG16; Covance), and polyclonal anti-Gcd6p (8).
RESULTS
Efficient cotranscriptional recruitment of CBC requires the m7G cap but is likely independent of CBC interaction with activator, Rpb1p-CTD, or Npl3p.
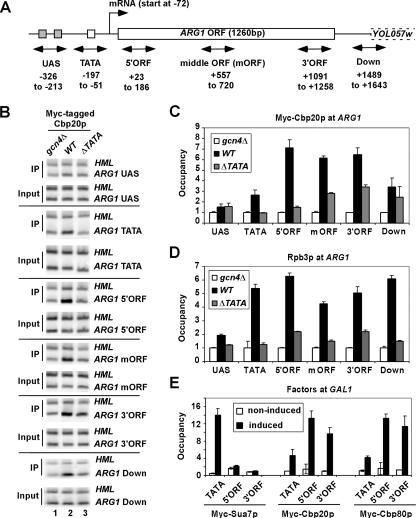
Although the cotranscriptional recruitment of CBC is well established, its mechanism of recruitment has not been examined in detail. We asked whether CBC might be recruited directly by a transcriptional activator to the UAS prior to its interaction with the cap. Induction of Gcn4p synthesis by starvation for isoleucine and valine (by treating cells with the inhibitor sulfometuron) leads to a strong increase in binding of Gcn4p and recruitment of coactivators to the UAS and of general transcription factors and Pol II to the promoter of ARG1 (14, 33, 34, 46). Importantly, deletion of the TATA element at ARG1 (arg1-ΔTATA) impairs recruitment of Pol II to the promoter but not the recruitment of coactivators to the UAS (34). We reasoned that if Gcn4p recruits CBC directly to the UAS, in the manner observed for coactivators, then we should observe high-level CBC occupancy at the UAS that would be unaffected by the ΔTATA mutation.
To test this prediction, we conducted ChIP analysis of strains containing Myc-tagged CBC subunits. The Myc-tagged strains grew identically to the parental strains on yeast extract-peptone-dextrose (data not shown), indicating that the tagged proteins were functional in vivo. At odds with the hypothesis that CBC is recruited directly by Gcn4p, the UAS element displayed the lowest level of Cbp20p occupancy of all locations tested at the ARG1 gene in WT cells treated with sulfometuron (Fig. 1A to C). Cbp20p occupancy was also much lower in the nontranscribed sequences in the promoter (TATA) or downstream of the polyadenylation site (“Down” primers) than in transcribed sequences throughout the ARG1 open reading frame (ORF). As expected, Cbp20p occupancy was reduced to background levels at all locations in the isogenic gcn4Δ mutant, confirming that appreciable CBC recruitment to ARG1 depends on transcriptional activation by Gcn4p (Fig. 1A to C).
FIG. 1.
Induction of Gcn4p increases Cbp20p occupancy in the transcribed, but not UAS, region of ARG1, dependent on the TATA element. (A) ARG1 locus with names and positions of fragments (relative to the ATG) that were PCR amplified for ChIP analyses given below. Dashed lines depict the adjacent gene YOL057w. (B to D) ChIP analysis of Myc-Cbp20p or Rpb3p occupancy at ARG1 following induction of Gcn4p. CBP20-myc strains CMY007 (gcn4Δ), CMY008 (WT), and CMY009 (arg1-ΔTATA) were cultured in synthetic complete medium lacking Ile and Val and treated with sulfometuron (final concentration, 0.5 μg/ml) for 30 min to induce Gcn4p synthesis. Cells were cross-linked with formaldehyde and subjected to ChIP analysis with anti-Myc (B and C) or anti-Rpb3p (D) antibodies. DNA was extracted from immunoprecipitates (IP) and input chromatin (Input) samples and subjected to PCR in the presence of [33P]dATP to amplify radiolabeled fragments from the UAS, TATA, 5′-ORF, middle-ORF, or 3′-ORF regions of ARG1 together with a control fragment from a nontranscribed region of HML. PCR products were resolved by 6% Tris-buffered EDTA polyacrylamide gel electrophoresis and visualized by autoradiography (B) or quantified with a phosphorimager, and the ratios of experimental to control signals in the immunoprecipitate samples were normalized for the corresponding ratios for input samples to yield the occupancy values plotted in histograms (C and D). (E) Strains with genes encoding the relevant Myc-tagged proteins HQY691 (SUA7-myc), CMY005 (CBP20-myc), and CMY006 (CBP80-myc) were cultured in synthetic complete medium lacking histidine with 2% raffinose to an A600 of ≈0.6 and treated with galactose (final concentration, 2%) for 30 min to induce GAL1. Cells were cross-linked with formaldehyde and subjected to ChIP analysis with anti-Myc antibodies as described above, except with the use of primers to amplify the TATA, 5′-ORF, and 3′-ORF sequences at GAL1 corresponding to nucleotides −185 to −57, +422 to + 567, and +1233 to +1355, respectively. The average results obtained from two independent cultures and two PCR amplifications for each culture were plotted in the histograms with standard errors shown as error bars.
Deletion of the TATA element had the expected effect of reducing Pol II (Rpb3p) occupancy at all locations tested at the induced ARG1 gene (Fig. 1D). The same finding was made for Cbp20p occupancy (Fig. 1C), suggesting that transcription initiation at the correct site is required for high-level CBC recruitment to the 5′ end of ARG1. We showed previously that shorter heterogeneous ARG1 mRNAs are produced in the arg1-ΔTATA mutant at a level (combined for all transcripts) below that of native ARG1 mRNA, most likely from inefficient initiation at cryptic promoter sequences in the ARG1 ORF (32). Thus, the fact that the ΔTATA mutation does not eradicate Cbp20p occupancy of the middle, 3′-ORF, and downstream sequences could be explained if Cbp20p is recruited to the capped 5′ ends of shorter transcripts initiating in the ARG1 ORF of the arg1-ΔTATA allele.
Together, these results seem inconsistent with the possibility that CBC is recruited directly by Gcn4p to the UAS prior to its association with the mRNA cap. If this were the case, then deletion of the TATA element might cause an accumulation of CBC in the UAS due to the reduced level of capped transcripts produced by the arg1-ΔTATA allele, whereas we observed nearly background levels of Cbp20p at the UAS in the presence or absence of the TATA element. Consistent with our findings on ARG1, we found that induction of the GAL1 gene by galactose leads to much higher CBC occupancies in the coding sequences compared to the promoter, whereas Myc-tagged TFIIB/Sua7p is recruited exclusively to the promoter region of GAL1 (Fig. 1E).
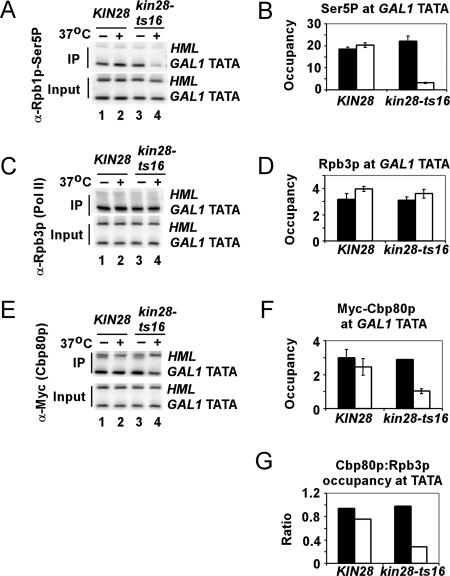
The results presented above suggest that CBC is recruited directly to the capped 5′ end of nascent transcripts. If so, then mutations that reduce 5′ capping of the mRNA should reduce CBC recruitment to the 5′ end of the gene. Moreover, since recruitment of the capping enzymes is stimulated by Ser5 CTD phosphorylation by Kin28p, then reducing Kin28p function by the temperature-sensitive kin28-ts16 mutation should decrease CBC occupancy at the 5′ end of the induced GAL1 gene. ChIP analysis showed that incubation of kin28-ts16 cells at 37°C for 30 min followed by shifting cells from glucose to galactose to induce GAL1 (at 37°C) led to the expected decline in occupancy of Rpb1p-Ser5P at the GAL1 promoter (Fig. 2A and B; open bars are 37°C), whereas total levels of Pol II (Rpb3p) at the promoter did not decrease (Fig. 2C and D). Thus, Pol II hypophosphorylated on Ser5 accumulates at the induced GAL1 promoter in the kin28-ts16 cells at 37°C. Importantly, Cbp80p occupancy in the GAL1 promoter was substantially reduced in the kin28-ts16 mutant at 37°C (Fig. 2E and F), leading to a substantial decline in the ratio of Cbp80p to Pol II (Rpb3p) (Fig. 2G).
FIG. 2.
Optimal Cbp80p recruitment to GAL1 requires Kin28p-dependent Ser5 phosphorylation of the Pol II CTD. Isogenic CBP80-myc strains CMY013 (KIN28) and CMY014 (kin28-ts16) were cultured in synthetic complete medium lacking histidine with 2% raffinose at 25°C to an A600 of ≈0.6 and transferred to 37°C for 30 min, and galactose (2%) was added for another 30 min at 37°C (open bars). The same strains were cultured identically except at 25°C rather than 37°C (filled bars). (A to F) ChIP analysis of factor occupancies at GAL1 was conducted as described for Fig. 1E except with the use of H14 antibodies specific for the Rpb1p CTD phosphorylated on Ser5 (Ser5P) (A and B), Rpb3p antibodies (C and D), or Myc antibodies (to detect Myc-Cbp80p) (E and F). (G) Occupancies of Myc-Cbp80p in panel F were normalized to those for Rpb3p in panel D. The average results obtained from two independent cultures and two PCR amplifications for each culture were plotted in the histograms with standard errors shown as error bars.
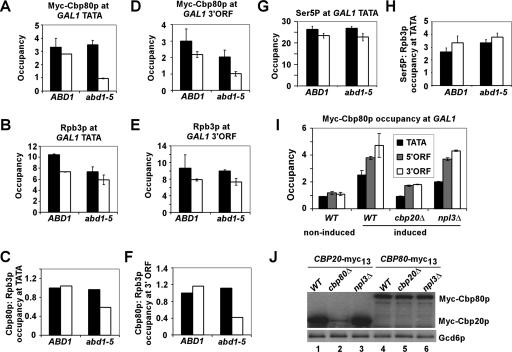
We presumed that the requirement for Rpb1p-Ser5P in efficient CBC recruitment is indirect and reflects the stimulatory effect of Ser5 phosphorylation on recruitment of the capping enzymes. If so, then mutation of a capping enzyme should decrease CBC occupancy without any decrease in Rpb1p-Ser5P levels in the promoter. The temperature-sensitive abd1-5 mutation is perfectly suited to test this prediction because it impairs production of the m7G caps on mRNAs without decreasing Pol II occupancy at the 5′ ends of yeast genes in vivo (40). Consistent with this, incubation of abd1-5 cells at 37°C for 30 min followed by a shift to galactose (at 37°C) produced little or no reduction in Pol II occupancy at the promoter or 3′-ORF sequences at GAL1 compared to the values observed in isogenic WT cells incubated at 37°C (cf. open bars in Fig. 3B and E). By contrast, there were significant reductions in Cbp80p occupancy at both the promoter and the 3′ end of the ORF in the abd1-5 mutant (Fig. 3A and D, open bars), leading to reduced Cbp80p-to-Pol II (Rpb3p) ratios in both GAL1 locations (Fig. 3C and F). The GAL1 mRNA level is not reduced in the abd1-5 mutant grown under nonpermissive conditions (data not shown). Importantly, the abd1-5 mutation also does not lower Rpb1p-Ser5P at GAL1 (Fig. 3G and H), indicating that Ser5 CTD phosphorylation is insufficient for high-level CBC recruitment in the absence of the m7G cap. These results provide the first in vivo evidence that the guanine 7-methylated cap is a prerequisite for efficient cotranscriptional recruitment of the CBC.
FIG. 3.
Optimal Cbp80p recruitment to GAL1 requires the cap guanine 7-methyltransferase Abd1p. (A to H) Isogenic CBP80-myc strains CMY015 (ABD1) and CMY016 (abd1-5) were cultured under the same conditions described for Fig. 2 for galactose induction and heat treatment, and ChIP analysis was conducted as described for Fig. 1 using antibodies to Myc (to detect Myc-Cbp80p) (A and D) or Rpb3p (B and E). Occupancies of Myc-Cbp80p were normalized to those for Rpb3p for the GAL1 TATA (C) and GAL1 3′ ORF (F). ChIP analysis of the same samples was conducted using H14 antibodies specific for the Ser5-phosphorylated CTD (Ser5P) (G). Occupancies of Ser5P were normalized to those for Rpb3p for the GAL1 TATA in panel B (H). Optimal Cbp80p recruitment to GAL1 requires Cbp20p but not Npl3p. (I) Isogenic CBP80-myc strains CMY006 (WT), CMY011 (cbp20Δ), and CMY012 (npl3Δ) were cultured under the conditions described for Fig. 2 for galactose induction except that cells were grown at 30°C, and ChIP analysis was conducted as described for Fig. 1 with the use of antibodies to Myc (to detect Myc-Cbp80p). The average results obtained from two independent cultures and two PCR amplifications for each culture were plotted in the histograms with standard errors shown as error bars. (J) Western blot analysis of Myc-Cbp20p and Myc-Cbp80p expression. Whole-cell extracts of isogenic CBP20-myc strains CMY005 (WT), CMY009 (cbp80Δ), and CMY010 (npl3Δ) and CBP80-myc strains CMY006 (WT), CMY011 (cbp20Δ), and CMY012 (npl3Δ) were subjected to Western blot analysis using antibodies to Myc (to detect Myc-Cbp20p or Myc-Cbp80p) or Gcd6p (loading control).
The results presented above suggest that CBC is recruited directly by the m7G cap on the nascent transcript and that the transcriptional activator, intact promoter, and Ser5 CTD phosphorylation all contribute indirectly to CBC recruitment by promoting formation of the capped nascent transcript. Another result consistent with this model is that recruitment of Cbp80p is strongly dependent on Cbp20p, the cap-binding subunit of the CBC (27). This is shown by the fact that deletion of CBP20 strongly impairs Cbp80p recruitment to GAL1 (Fig. 3I), without reducing the steady-state level of Cbp80p (Fig. 3J, lanes 4 and 5). Hence, binding of Cbp20p to the m7G cap is likely to be required for Cbp80p recruitment to the nascent transcript. As deletion of CBP80 greatly reduces the level of Myc-Cbp20p (Fig. 3J, lanes 1 and 2) (41), we could not address whether Cbp80p is also necessary for recruitment of Cbp20p.
Finally, we addressed whether Npl3p promotes CBC recruitment, a possibility prompted by physical and genetic interactions of Npl3p with the CBC (19, 41). Deletion of NPL3 had no significant impact on recruitment of Cbp80p or Cbp20p to GAL1 (Fig. 3I and data not shown). Furthermore, it was reported previously that cbp80Δ does not affect Npl3p recruitment in vivo (24). The independent recruitment of these factors might be explained by the fact that recruitment of Npl3p is mediated by its interactions with Pol II and the nascent mRNA (41) while CBC is recruited directly by the m7G cap.
CBC does not play a critical role in promoting the rate or processivity of elongation by Pol II.
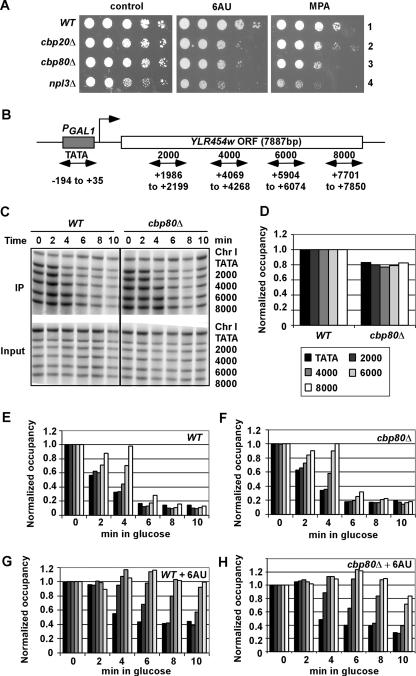
The findings that cbp80Δ confers sensitivity to 6AU and MPA and interacts genetically with deletion of HPR1, a subunit of the THO/TREX complex, suggested a possible defect in transcription elongation. Consistent with this, we found that cbp20Δ also confers sensitivity to 6AU and MPA (Fig. 4A). Hence, we measured the effect of cbp80Δ on the elongation rate and processivity of Pol II in vivo by kinetic ChIP analysis during induction and repression of the ∼8-kb YLR454w ORF placed under the control of the GAL1 promoter (PGAL1-YLR454w) (Fig. 4B). On addition of glucose to cells growing in galactose, recruitment of new Pol II molecules to the GAL1 promoter is blocked and preexisting elongating Pol II molecules finish transcribing the coding sequences. The kinetics of Pol II runoff during this last wave of elongation provides a measure of the elongation rate (26). As expected, following addition of glucose to WT cells, Pol II was lost from the promoter and the sequences at the 5′ end of the PGAL1-YLR454w ORF sooner than it disappeared from the 3′ end of the ORF (Fig. 4C). Analysis of the cbp80Δ mutant revealed no significant difference from WT in the rate of Pol II runoff during glucose repression (Fig. 4C, E, and F). Very similar results were obtained in several replicate experiments. As shown previously (26), addition of 6AU decreased the elongation rate of Pol II in WT cells (Fig. 4G), and we observed a similar decrease in the cbp80Δ mutant (Fig. 4H).
FIG. 4.
cbp80Δ does not detectably affect Pol II elongation rate and processivity in vivo. (A) cbp20Δ and npl3Δ mutants, like cbp80Δ cells, are sensitive to 6AU and MPA. Tenfold serial dilutions of strains BY4741 (WT), 2074 (cbp20Δ), 6566 (cbp80Δ), and 4268 (npl3Δ), transformed with YCplac33, were spotted on synthetic complete medium lacking uracil plates containing 2% glucose supplemented with 6AU (75 μg/ml) or MPA (15 μg/ml). (B) Diagram of the PGAL1-YLR454w gene with PCR primer pairs used to amplify the promoter and indicated coding sequences (numbered relative to ATG). (C) ChIP analysis of Rpb3p occupancies across the PGAL1-YLR454w gene induced with galactose and at different times of glucose repression. CMY017 (WT) or CMY018 (cbp80Δ) strains containing RPB3-myc and PGAL1-YLR454w were induced with galactose for 2 h (time = 0) and treated with 4% glucose for the indicated times. ChIP analysis was conducted using Rpb3p antibodies and primers (TATA to 8000, from top to bottom) shown in panel B. Amplification of a noncoding region on chromosome I (Chr I) was used as a control for nonspecific immunoprecipitation. (D) Quantification of the Rpb3p occupancies measured in panel C at the five indicated positions in WT and cbp80Δ cells grown in galactose medium (time = 0), normalized to the values in the WT strain at each position. (E and F) Quantification of the Rpb3p occupancies measured in panel C at the five positions indicated by the key in panel D in WT (E) or cbp80Δ (F) cells grown in glucose for the indicated times, normalized to the values in galactose (time = 0). (G and H) The assays were performed exactly as described for panels E and F except that cells were grown with 75 μg/ml 6AU. The average results obtained from two independent cultures and two PCR amplifications for each culture were plotted in the histograms.
The processivity of Pol II during elongation can be examined by comparing the Pol II occupancies at the 5′ and 3′ ends of the ∼8-kb YLR454w ORF in cells growing in galactose. Previous results showed that treatment with 6AU or mutations in certain elongation factors (SPT4 and THO components) lead to a progressive decline in Pol II occupancy across the long PGAL1-YLR454w ORF (26). In agreement with that study, we observed similar Pol II occupancies at the 3′ end compared to the 5′ end of the PGAL1-YLR454w ORF in WT cells (Fig. 4D) and a reduction in Pol II occupancy across the ORF in WT cells treated with 6AU (data not shown). However, there was no difference in Pol II occupancy across the ORF in the cbp80Δ mutant (Fig. 4D), indicating no obvious reduction in Pol II processivity in the absence of Cbp80p. We did observe a small reduction in Pol II occupancy at all locations of the PGAL1-YLR454w gene in cbp80Δ cells, which might indicate a defect in PIC assembly that reduces the number of Pol II molecules entering the elongation phase of transcription.
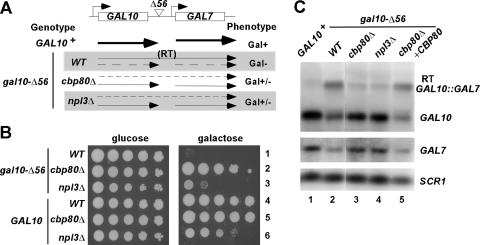
CBC reduces termination/poly(A) addition at the defective gal10-Δ56 terminator in vivo.
Npl3p antagonizes 3′-end formation by impeding the recruitment of polyadenylation/termination factors (7). In view of genetic and physical interactions between CBC and Npl3p and our finding that npl3Δ also confers sensitivity to 6AU and MPA (Fig. 4A), we investigated whether cbp80Δ resembles npl3Δ in elevating 3′-end formation at the gal10-Δ56 allele in vivo. The Δ56 mutation eliminates 55 bp at the 3′ end of GAL10 and reduces the efficiency of 3′-end formation at the correct site, allowing read-through into the GAL7 gene downstream. Gal7p is not expressed from the read-through bicistronic transcript, and the production of native GAL7 mRNA is impaired by transcription elongation across the GAL7 promoter—both factors contributing to the Gal− phenotype of gal10-Δ56 cells (Fig. 5A) (15, 21). It was shown previously that the npl3-120 mutation partially suppresses the Gal− phenotype of gal10-Δ56 (7). We observed the same result for the npl3Δ null allele (Fig. 5B, row 3), which decreases production of the GAL10-GAL7 read-through transcript and increases expression of native GAL7 mRNA (Fig. 5C, lanes 2 and 4). Importantly, cbp80Δ behaved similarly in reducing the read-through transcript and restoring WT levels of GAL7 mRNA, and both effects were complemented by plasmid-borne CBP80 (Fig. 5C, lanes 2, 3, and 5). Also similarly to npl3Δ, the cbp80Δ mutation suppresses the Gal− phenotype of gal10-Δ56 cells (Fig. 5B, rows 1 to 3). These findings are consistent with the idea that Cbp80p antagonizes 3′-end formation at the defective transcription terminator of gal10-Δ56.
FIG. 5.
Cbp80p impedes utilization of the weak gal10-Δ56 terminator. (A) Schematic showing the GAL10-GAL1 gene pair with the Δ56 mutation in the GAL10 terminator. Arrows depict locations and relative abundances (proportional to line thickness) of transcripts produced in strains of the indicated genotypes and Gal phenotypes. RT, read-through. (B) cbp80Δ and npl3Δ partially suppress the Gal− phenotype of gal10-Δ56 cells. Tenfold serial dilutions of cbp80Δ and npl3Δ cells were spotted on yeast extract-peptone plates containing either 2% glucose (left) or 2% galactose (right) as carbon sources for 4 days. Rows 1 to 3, CMY127 (WT), CMY128 (cbp80Δ), and CMY129 (npl3Δ) strains with gal10-Δ56; rows 4 to 6, isogenic GAL10 strains, BY4741 (WT), 6566 (cbp80Δ), and 4268 (npl3Δ). (C) Northern analysis of GAL10, GAL7, and GAL10::GAL7 read-through mRNAs. Total RNAs (20 μg) from wild-type (GAL10+) strain BY4741 and gal10-Δ56 strains CMY127 (WT), CMY128 (cbp80Δ), and CMY129 (npl3Δ) induced with 2% galactose for 3 h were resolved and probed for GAL10 or GAL7 mRNAs and the Pol III transcript SCR1 (as a loading control). The read-through transcript that hybridizes with both GAL10 and GAL7 probes is marked (RT).
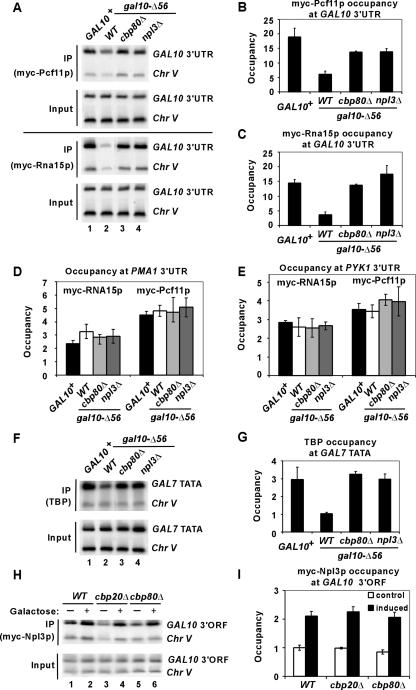
To confirm this interpretation, we conducted ChIP analysis to determine whether cbp80Δ increases recruitment of polyadenylation/termination factors Rna15p and Pcf11p, belonging to the CF IA complex (17), at the defective transcription termination site of gal10-Δ56. The results show that cbp80Δ and npl3Δ lead to comparable increases in Rna15p and Pcf11p occupancies in the 3′ untranslated region (UTR) of gal10-Δ56 (Fig. 6A to C) without affecting Pol II occupancy at this location (data not shown). Moreover, cbp80Δ and npl3Δ produce similar increases in TATA binding protein recruitment to the GAL7 promoter in the gal10-Δ56 background (Fig. 6F and G), providing evidence that they reduce the deleterious effect of read-through transcription on PIC assembly downstream at the GAL7 promoter. The fact that cbp80Δ restores GAL7 mRNA production by eliminating Pol II molecules reading through the defective gal10-Δ56 terminator into the GAL7 promoter shows clearly that cbp80Δ increases recognition of the weak terminator independently of any effect that it might have in stabilizing aberrantly terminated GAL10 transcripts in the nucleus (11).
FIG. 6.
Cbp80p and Npl3p impede recruitment of polyadenylation/termination factors Pcf11p and Rna15p to the gal10-Δ56 terminator. (A to C) ChIP analysis of Myc-Pcf11p or Myc-Rna15p occupancies was conducted as described for Fig. 1E except using primers to amplify the GAL10 3′ UTR, employing PCF11-myc strains CMY019 (WT), CMY020 (gal10-Δ56), CMY021 (cbp80Δ gal10-Δ56), and CMY022 (npl3Δ gal10-Δ56) and RNA15-myc strains CMY023 (WT), CMY024 (gal10-Δ56), CMY025 (cbp80Δ gal10-Δ56), and CMY026 (npl3Δ gal10-Δ56). (D and E) Cbp80p and Npl3p do not impede recruitment of polyadenylation/termination factors Pcf11p and Rna15p to the PMA1 and PYK1 terminators. Myc-Pcf11p or Myc-Rna15p occupancies of the PMA1 and PYK1 3′ UTRs were analyzed in the same chromatin samples analyzed for panels A to C. (F and G) TATA binding protein (TBP) occupancies of the GAL7 TATA region were analyzed in the same chromatin samples analyzed for panels A to C. (H and I) Myc-Npl3p occupancy of the GAL10 3′ ORF is unaffected by cbp20Δ and cbp80Δ. ChIP analysis of Myc-Npl3p was done for NPL3-myc strains CMY027 (WT), CMY028 (cbp20Δ), and CMY029 (cbp80Δ) as described for Fig. 1E. Cells were cultured either in 2% raffinose (−) as a control or in 2% galactose (+) for induction of GAL10 expression. The average results obtained from two independent cultures and two PCR amplifications for each culture were plotted in the histograms with standard errors shown as error bars.
It was important to investigate whether the antiterminator functions of CBC and Npl3p apply only to weak termination sites or are fundamental to the mechanism of 3′-end processing. Therefore, ChIP analysis was performed to measure the possible effects of cbp80Δ and npl3Δ mutations on Pcf11p and Rna14p occupancies of the native termination regions at the PMA1 and PYK1 genes (22). The results show that cbp80Δ and npl3Δ do not increase the Pcf11p and Rna14p occupancies in the 3′ UTRs of these genes (Fig. 6D and E), suggesting that CBC and Npl3p effectively impede recruitment of CF IA only at weak terminators, such as that present at gal10-Δ56.
Consistent with previous findings (24), we found that the occupancy of Myc-Npl3p at the 3′ end of GAL10 is not reduced by cbp80Δ or cbp20Δ mutations (Fig. 6H and I), confirming that the effect of the latter mutations on 3′-end formation is not an indirect effect of impaired association of Npl3p with the nascent transcript. Thus, we conclude that CBC and Npl3p collaborate in suppressing utilization of the weak gal10-Δ56 termination site.
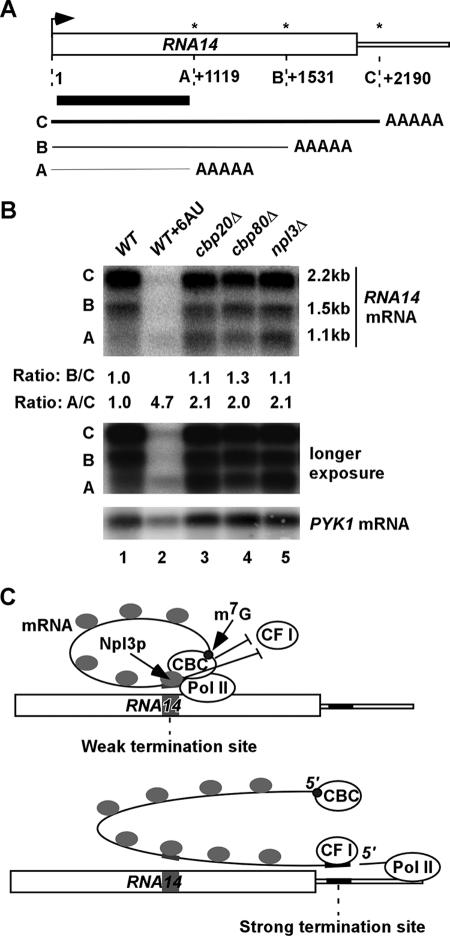
Finally, it was important to determine whether CBC antagonizes termination at a naturally occurring cryptic termination site in a WT gene. Rna14p is a component of CF IA required for cleavage and polyadenylation during mRNA 3′-end processing (30). Interestingly, the RNA14 gene produces three stable mRNAs, the full-length 2.2-kb transcript and truncated 1.5- and 1.1-kb transcripts that terminate in the coding sequences (Fig. 7A). Defects in transcription termination lead to production of only the full-length transcript (2, 5, 43), and growth under respiratory conditions increases the truncated 1.1-kb transcript at the expense of the 1.5- and 2.2-kb transcripts (43). Accordingly, we asked whether cbp20Δ, cbp80Δ, and npl3Δ mutants would increase production of the truncated 1.1-kb RNA14 mRNA by increasing utilization of the weak terminator for this transcript.
FIG. 7.
The CBC inhibits termination at an internal poly(A) addition site in the RNA14 ORF. (A) Schematic of the RNA14 gene and its three transcripts (A, B, and C) generated by the major poly(A) sites labeled with asterisks (43). The position of the DNA fragment used to probe all three transcripts by Northern blot analysis is indicated by the black bar. (B) Strains BY4741 (WT), 2074 (cbp20Δ), 6566 (cbp80Δ), and 4268 (npl3Δ), transformed with YCplac33, were grown in synthetic complete medium lacking uracil with 2% glucose; BY4741 (WT) was treated separately with 6AU (at 50 μg/ml) for 4 h; and poly(A)+ mRNAs were isolated from 150 μg total RNA and subjected to Northern blot analysis, with probing for RNA14 and PYK1 (as loading control). The hybridization signals for the RNA14 transcripts were quantified by phosphorimaging analysis, and the ratios of the B and A transcripts relative to the C transcript were calculated and normalized to the corresponding ratios measured in WT cells. The middle panel shows a longer exposure of the upper panel. (C) A hypothetical model depicting cooperation between the CBC and Npl3p in preventing transcription termination at weak termination sites. (Top) CBC is recruited cotranscriptionally to the capped 5′ end of the nascent transcript and interacts with Pol II during elongation, forming a pseudocircular mRNA. CBC also interacts with Npl3p and stabilizes Npl3p binding to weak termination sites in the nascent mRNA (gray). Npl3p competes with CF I for recruitment to weak terminators, preventing transcript cleavage. The circularization of the mRNA by CBC might also impede cleavage by CF I. (Bottom) At the strong termination site, CF I displaces Npl3p and cleaves the mRNA to prepare the 3′ end for polyadenylation. CBC dissociates from Pol II by an unknown mechanism.
Northern blot analysis of the WT strain showed that the 2.2-kb RNA transcript is the most abundant, while the 1.1-kb RNA transcript occurs at the lowest level, among the three RNA14 transcripts (Fig. 7B, lane 1). Thus, under our experimental conditions, the most distal poly(A) site is used most frequently to produce full-length RNA14 mRNA. Importantly, cbp20Δ, cbp80Δ, and npl3Δ mutants all exhibit higher 1.1-kb to 2.2-kb transcript ratios than do WT cells (Fig. 7B, Ratio: A/C, lanes 3 to 5). These data suggest that CBC and Npl3p cooperate to suppress cleavage and polyadenylation at position +1119 within the native RNA14 coding sequences.
Interestingly, while all three RNA14 transcripts were reduced by 6AU treatment (16), the proportional amount of the 1.1-kb transcript increased in WT cells treated with this inhibitor (Fig. 7B, lane 2). Thus, in addition to decreasing elongation rate and processivity (26), it appears that 6AU also enhances the selection of weak poly(A) sites for transcription termination. Perhaps the increased dwell time of Pol II due to depletion of UTP pools by 6AU increases the probability of transcript cleavage and termination at a weak poly(A) site. The fact that 6AU and the cbp20Δ, cbp80Δ, and npl3Δ mutations all enhance utilization of a weak termination site helps to explain the 6AU sensitivity of the cbp20Δ, cbp80Δ, and npl3Δ mutants (Fig. 4), even though CBC and Npl3p play no significant role in transcription elongation (7).
DISCUSSION
In this study, we have examined the mechanism of CBC recruitment to the cap structure and evaluated the contributions of CBC to transcription elongation and 3′-end formation. We first explored the possibility that the CBC could be recruited directly by transcriptional activators to the UAS, or to the promoter by the Ser5-phosphorylated CTD of Rpb1p, and then transferred to the cap once the latter appeared on the nascent transcript. A prominent role for CBC recruitment by the activator Gcn4p seems unlikely because the UAS occupancy of CBC at ARG1 is only slightly above background. One could propose that CBC is recruited to the UAS but is transferred to the cap so rapidly that its steady-state level at the UAS remains low. If so, then a reduction in the rate of transcript initiation and cap formation should increase CBC occupancy in the UAS. However, we found that deleting the TATA element at ARG1, which reduces transcriptional output and CBC association with the coding sequences, did not increase CBC occupancy in the UAS.
We found that the ts16 mutation in Kin28p, and attendant decline in Rpb1p-Ser5P, lowered CBC recruitment at the 5′ end of the GAL1 ORF without any decrease in Pol II occupancy. This result is consistent with the known requirement for Ser5 CTD phosphorylation in recruitment of the capping enzymes to nascent transcripts in vivo (39). It could also reflect a role for the phosphorylated CTD in direct recruitment of CBC; however, we found that the abd1-5 mutation in the cap guanine 7-methyltransferase lowers recruitment of CBC without decreasing Rpb1p-Ser5P occupancy. In addition, CBC occupancy is higher in the ORF than at the promoter, whereas Rpb1p-Ser5P and the capping enzymes Ceg1p/Cet1p show the opposite pattern. Thus, it seems likely that direct recruitment of CBC to the phosphorylated CTD is inefficient at best, and the major pathway is direct binding of CBC to the cap structure on nascent mRNA. Nevertheless, we cannot eliminate the possibility that CBC interacts transiently with Ser5-phosphorylated Pol II at the promoter (or with the activator at the UAS) in a manner that, while undetectable by ChIP, facilitates its subsequent binding to the m7G cap.
It has been shown that the CBC is required for efficient cotranscriptional assembly of the spliceosome, but its participation in other transcription-coupled functions was not well defined. Although cbp80Δ and cbp20Δ mutants are sensitive to 6AU and MPA, we did not observe any obvious defect in the rate or processivity of Pol II elongation during induced transcription of the ∼8-kb PGAL1-YLR454w coding sequences. This is consistent with the fact that cbp80Δ cells are not defective for induction of IMP dehydrogenase (IMD2) mRNA by 6AU or MPA, a response severely impaired by mutations in known elongation factors, such as Dst1p/TFIIS (36). Of course, it is possible that CBC promotes elongation rate or processivity in a manner that is fully redundant with the functions of other factors. Indeed, no defect in elongation of the PGAL1-YLR454w construct was observed previously in mutants lacking TFIIS or intact Paf1 complex (26).
On the other hand, we observed a clear defect in 3′-end formation in cbp80Δ cells similar to that described recently (7) for an npl3-120 point mutant (and confirmed here for an npl3Δ null mutant) wherein 3′-end formation occurs more efficiently at the defective termination site of the gal10-Δ56 allele. This can be attributed to the increased recruitment of polyadenylation/termination factors Rna15p and Pcf11p, both CF IA components, to the defective termination site that we observed in cbp80Δ cells. These findings imply that, like Npl3p, CBC is an antagonist of 3′-end formation that functions at least partly by impeding recruitment of CF IA. Thus, termination/poly(A) addition should occur inappropriately at naturally occurring weak termination sites in cbp80Δ cells. Our analysis of termination of RNA14 transcription provides direct confirmation of this prediction, as deletions of CBP20, CBP80, and NPL3 all increase termination at a native weak poly(A) addition site within the RNA14 coding sequences. The fact that 6AU also enhances this weak termination suggests that the 6AU sensitivity of these mutants results from additive increases in utilization of weak termination sites in coding regions.
The idea that CBC suppresses termination/poly(A) addition at weak terminators was suggested previously by the finding that inactivation of CBP80 partially suppressed the deleterious effect of the cyc1-512 mutation on production of stable CYC1 transcripts. Like the gal10-Δ56 mutation, cyc1-512 leads to aberrant, longer transcripts with 3′ ends downstream of the normal poly(A) site. It appeared likely that cbp80Δ increased 3′-end formation at the correct site during transcription of cyc1-512, rather than merely stabilizing aberrantly terminated transcripts (11). Our analysis of the gal10-Δ56 allele provides more direct evidence that cbp80Δ restores termination at the correct site at gal10-Δ56, as cbp80Δ not only increases correctly terminated GAL10 transcripts but eliminates the interference of read-through transcription on PIC assembly downstream at the GAL7 promoter, with an attendant increase in GAL7 mRNA. We also investigated the molecular mechanism of antitermination by CBC and showed directly (by ChIP assays) that cbp80Δ increases the recruitment of subunits of termination/polyadenylation factor CF IA to the 3′ region of gal10-Δ56.
Most pre-mRNAs are processed at their 3′ ends by a two-step process. The first step is cleavage of the nascent transcript ∼20 to 30 bases downstream of a conserved poly(A) addition site. This endonucleolytic cleavage requires the cooperation of several multisubunit cleavage factors, namely, CF IA (Pcf11p, Rna14p, Rna15p, and Clp1p), CF IB (Hrp1), and cleavage/polyadenylation factor (CPF). In the second step, the poly(A) tail is added to the 3′ end of the cleaved transcript by poly(A) polymerase. Surprisingly, the recruitment of some CPFs, e.g., CF IB and Ssu72p, is not restricted to the 3′ ends of genes but also occurs at the promoter and throughout the coding sequences. To prevent premature termination, it is thought that antitermination factors are associated with elongating Pol II that prevent recognition of weak poly(A) addition signals located upstream of the authentic terminator (31).
It was proposed that Npl3p, an RNA binding protein, antagonizes termination at weak poly(A) addition sites in the nascent transcript by competing effectively with the RNA binding termination factors Rna15p and Hrp1p (7). This could be particularly important in budding yeast, where the consensus sequence for polyadenylation/termination is weak However, as Rna15p and Hrp1p show greater specificity than Npl3p does for AU-rich RNA sequences (17, 18), these components of CF IA and CF IB, respectively, could displace Npl3p at the AU-rich consensus elements of strong termination sites and function with CPF to cleave and polyadenylate the transcript.
How might CBC function in blocking weak terminators? As demonstrated here and elsewhere (24), the CBC does not antagonize 3′-end formation indirectly by stimulating recruitment of Npl3p to sites of transcription. However, CBC might stabilize Npl3p binding specifically to weak termination sequences in the nascent transcript. In this view, Npl3p would still be recruited to the nascent mRNA in CBC mutants (as observed in ChIP experiments) but would shift its occupancy toward sequences unrelated to termination signals. Hence, we envision that CBC bound to the cap maintains physical association with elongating Pol II and that simultaneous interaction of CBC with both Npl3p (41) and Pol II, and of Npl3p with Pol II (24), stabilizes Npl3p binding to weak termination signals in the nascent transcript as they emerge from elongating Pol II (Fig. 7C, top). This assembly would be disrupted at strong terminators by the competition of Rna15p and Hrp1p with Npl3p for binding to consensus termination signals, as described above (Fig. 7C, bottom).
Considering that simultaneous deletion of CBP80 and NPL3 is lethal (28, 41), the CBC might also function independently of Npl3p to help suppress cryptic termination sites. CBC could interfere directly with the recruitment or function of a component of the termination/polyadenylation machinery. Alternatively, as circular or pseudocircular RNA precursors are resistant to cleavage by CPF/CF I (45), the hypothetical mRNA loop formed by the interaction of CBC with Pol II in our model (Fig. 7C) could make the nascent transcript topologically resistant to cleavage.
Acknowledgments
We are grateful to S. Shuman, F. Winston, K. Struhl, and I. Mattaj for strains and plasmids and T. Dever for helpful discussions.
C.-M.W. was supported by a fellowship from the Croucher Foundation. This work is supported in part by the Intramural Research Program of the NIH.
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Aguilera, A. 2005. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr. Opin. Cell Biol. 17:242-250. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 3.Aviv, H., and P. Leder. 1972. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc. Natl. Acad. Sci. USA 69:1408-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brendolise, C., J. M. Rouillard, M. E. Dufour, and F. Lacroute. 2002. Expression analysis of RNA14, a gene involved in mRNA 3′ end maturation in yeast: characterization of the rna14-5 mutant strain. Mol. Genet. Genomics 267:515-525. [DOI] [PubMed] [Google Scholar]
- 6.Bryant, G. O., and M. Ptashne. 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 11:1301-1319. [DOI] [PubMed] [Google Scholar]
- 7.Bucheli, M. E., and S. Buratowski. 2005. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J. 24:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cigan, A. M., M. Foiani, E. M. Hannig, and A. G. Hinnebusch. 1991. Complex formation by positive and negative translational regulators of GCN4. Mol. Cell. Biol. 11:3217-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colot, H. V., F. Stutz, and M. Rosbash. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 10:1699-1708. [DOI] [PubMed] [Google Scholar]
- 10.Das, B., J. S. Butler, and F. Sherman. 2003. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol. Cell. Biol. 23:5502-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, B., Z. Guo, P. Russo, P. Chartrand, and F. Sherman. 2000. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Biol. 20:2827-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 13.Gornemann, J., K. M. Kotovic, K. Hujer, and K. M. Neugebauer. 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell 19:53-63. [DOI] [PubMed] [Google Scholar]
- 14.Govind, C. K., S. Yoon, H. Qiu, S. Govind, and A. G. Hinnebusch. 2005. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol. Cell. Biol. 25:5626-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greger, I. H., and N. J. Proudfoot. 1998. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 17:4771-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigull, J., S. Mnaimneh, J. Pootoolal, M. D. Robinson, and T. R. Hughes. 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 24:5534-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross, S., and C. L. Moore. 2001. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 21:8045-8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guisbert, K. S., H. Li, and C. Guthrie. 2006. Alternative 3′ pre-mRNA processing in Saccharomyces cerevisiae is modulated by Nab4/Hrp1 in vivo. PLoS Biol. 5:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurt, E., M. J. Luo, S. Rother, R. Reed, and K. Strasser. 2004. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl. Acad. Sci. USA 101:1858-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izaurralde, E., and S. Adam. 1998. Transport of macromolecules between the nucleus and the cytoplasm. RNA 4:351-364. [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan, C. D., M. J. Holland, and F. Winston. 2005. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 280:913-922. [DOI] [PubMed] [Google Scholar]
- 22.Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt, and S. Buratowski. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23:354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei, E. P., H. Krebber, and P. A. Silveeer. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15:1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lejeune, F., Y. Ishigaki, X. Li, and L. E. Maquat. 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 21:3536-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mason, P. B., and K. Struhl. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17:831-840. [DOI] [PubMed] [Google Scholar]
- 27.Mazza, C., A. Segref, I. W. Mattaj, and S. Cusack. 2002. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21:5548-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride, A. E., J. T. Cook, E. A. Stemmler, K. L. Rutledge, K. A. McGrath, and J. A. Rubens. 2005. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J. Biol. Chem. 280:30888-30898. [DOI] [PubMed] [Google Scholar]
- 29.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preker, P. J., J. Lingner, L. Minvielle-Sebastia, and W. Keller. 1995. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell 81:379-389. [DOI] [PubMed] [Google Scholar]
- 31.Proudfoot, N. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16:272-278. [DOI] [PubMed] [Google Scholar]
- 32.Qiu, H., C. Hu, C. M. Wong, and A. G. Hinnebusch. 2006. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Mol. Cell. Biol. 26:3135-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu, H., C. Hu, S. Yoon, K. Natarajan, M. J. Swanson, and A. G. Hinnebusch. 2004. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell. Biol. 24:4104-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu, H., C. Hu, F. Zhang, G. J. Hwang, M. J. Swanson, C. Boonchird, and A. G. Hinnebusch. 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 25:3461-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reid, G. A., and G. Schatz. 1982. Import of proteins into mitochondria. J. Biol. Chem. 257:13062-13067. [PubMed] [Google Scholar]
- 36.Riles, L., R. J. Shaw, M. Johnston, and D. Reines. 2004. Large-scale screening of yeast mutants for sensitivity to the IMP dehydrogenase inhibitor 6-azauracil. Yeast 21:241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneiter, R., C. E. Guerra, M. Lampl, G. Gogg, S. D. Kohlwein, and H. L. Klein. 1999. The Saccharomyces cerevisiae hyperrecombination mutant hpr1D is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol. Cell. Biol. 19:3415-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schroder, P. A., and M. J. Moore. 2005. Association of ribosomal proteins with nascent transcripts in S. cerevisiae. RNA 11:1521-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroeder, S. C., D. A. Zorio, B. Schwer, S. Shuman, and D. Bentley. 2004. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell 13:377-387. [DOI] [PubMed] [Google Scholar]
- 41.Shen, E. C., T. Stage-Zimmermann, P. Chui, and P. A. Silver. 2000. The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J. Biol. Chem. 275:23718-23724. [DOI] [PubMed] [Google Scholar]
- 42.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 43.Sparks, K. A., and C. L. Dieckmann. 1998. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 26:4676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304-308. [DOI] [PubMed] [Google Scholar]
- 45.Stumpf, G., A. Goppelt, and H. Domdey. 1996. Pre-mRNA topology is important for 3′-end formation in Saccharomyces cerevisiae and mammals. Mol. Cell. Biol. 16:2204-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swanson, M. J., H. Qiu, L. Sumibcay, A. Krueger, S.-J. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23:2800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uemura, H., S. Pandit, Y. Jigami, and R. Sternglanz. 1996. Mutations in GCR3, a gene involved in the expression of glycolytic genes in Saccharomyces cerevisiae, suppress the temperature-sensitive growth of hpr1 mutants. Genetics 142:1095-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. E. Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. J. Hegemann, T. M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-McDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 49.Zenklusen, D., P. Vinciguerra, J. C. Wyss, and F. Stutz. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22:8241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, D., and M. Rosbash. 1999. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 13:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]