Abstract
The decoding of specific UGA codons as selenocysteine is specified by the Sec insertion sequence (SECIS) element. Additionally, Sec-tRNA[Ser]Sec and the dedicated Sec-specific elongation factor eEFSec are required but not sufficient for nonsense suppression. SECIS binding protein 2 (SBP2) is also essential for Sec incorporation, but its precise role is unknown. In addition to binding the SECIS element, SBP2 binds stably and quantitatively to ribosomes. To determine the function of the SBP2-ribosome interaction, conserved amino acids throughout the SBP2 L7Ae RNA binding motif were mutated to alanine in clusters of five. Mutant proteins were analyzed for ribosome binding, SECIS element binding, and Sec incorporation activity, allowing us to identify two distinct but interdependent sites within the L7Ae motif: (i) a core L7Ae motif required for SECIS binding and ribosome binding and (ii) an auxiliary motif involved in physical and functional interactions with the ribosome. Structural modeling of SBP2 based on the 15.5-kDa protein-U4 snRNA complex strongly supports a two-site model for L7Ae domain function within SBP2. These results provide evidence that the SBP2-ribosome interaction is essential for Sec incorporation.
The incorporation of the trace element selenium, in the form of the 21st amino acid, selenocysteine (Sec), is required for the function of a variety of proteins. Sec is encoded by what normally serves as a signal to halt protein synthesis, a UGA codon. Several essential factors that are required for recoding UGA from a terminator to a Sec codon have been identified previously. Among these is Sec-tRNA[Ser]Sec (18), which forms a ternary complex with GTP and the Sec-specific elongation factor eEFSec (9, 30). Surprisingly, the eEFSec-GTP-Sec-tRNA[Ser]Sec complex is not sufficient for nonsense suppression, indicating that these factors do not have direct access to the ribosomal A site. To distinguish Sec codons from UGA stop codons at the ends of coding sequences, selenoprotein mRNAs contain Sec insertion sequence (SECIS) elements in their 3′ untranslated regions (3′ UTRs). SECIS elements direct selenocysteine incorporation at in-frame UGA codons that are at least 52 nucleotides upstream of the SECIS (20). SECIS binding protein 2 (SBP2) is also required for Sec incorporation in vitro and in vivo, but its precise role has not been determined (7, 23). Preliminary structure-function analyses indicate that SBP2 has at least three distinct domains: a dispensable N-terminal domain (amino acids [aa] 1 to 398), a central functional domain (aa 399 to 517), and a C-terminal RNA binding domain (aa 517 to 846). Thus, the C-terminal half of SBP2 (CTSBP2; aa 399 to 846) is the minimal fully functional protein in vitro. This region is sufficient for the three known functions of SBP2: (i) SECIS element binding, (ii) ribosome binding, and (iii) Sec incorporation. Truncations in the RNA binding domain perturb ribosome binding and SECIS binding, resulting in a loss of Sec incorporation activity (8).
Within the RNA binding domain is a canonical L7Ae RNA binding motif found in several proteins known to interact specifically with RNA helical structures called kink-turns (K-turns). The mutagenesis of the universally conserved glycine residue in the L7Ae motif (G669 in rat SBP2) was shown to abolish SECIS binding but had no impact on ribosome binding, suggesting that the ribosome binding and SECIS binding domains overlap but are not identical (8). Other factors have also been proposed to be involved in Sec incorporation, notably, the L7Ae motif-containing ribosomal protein L30 (rpL30) (4), but whether these factors are required for Sec incorporation remains unknown.
K-turns are prevalent in the ribosome and are believed to confer structural flexibility during protein synthesis to allow for the transmission of signals over a large distance (16, 24, 25). Sequence comparison of archaeal and mammalian 28S rRNAs suggests that most, if not all, K-turns are conserved. The L7Ae RNA binding motif was originally identified in a computational study, in which it was predicted to form a conserved structural unit despite the low overall level of conservation of residues in this region (17). The presence of this motif in ribosomal proteins, ribosome-associated factors, and the ribosome-modifying enzyme RimK led the authors of this study to predict that this motif may be utilized to deliver additional activities to the ribosome. Interestingly, rpL30 is the only L7Ae motif-containing ribosomal protein that is known to interact with a K-turn (11), suggesting that many K-turns may be available for interactions with nonribosomal proteins.
We previously hypothesized that SBP2 may function by stably binding ribosomes, making them competent for Sec incorporation (8). Further studies showed that excess SECIS elements compete with ribosomes for SBP2 binding (15). Thus, an intriguing possibility is that SBP2 functions by interacting with K-turns on the ribosome to modify the A site and, in a SECIS-dependent fashion, create a high-affinity binding site for the eEFSec-GTP-Sec-tRNA[Ser]Sec ternary complex. Interestingly, the ability of SBP2 to bind ribosomes has previously been reported to be regulated by oxidative stress (23), suggesting that the ribosome binding activity of SBP2 is an important point of regulation during selenoprotein synthesis.
In this study, we analyzed the role of the SBP2 L7Ae RNA binding motif by clustered alanine-scanning mutagenesis and screening for mutant proteins with ribosome binding defects. This screen allowed us to identify a distinct region within the L7Ae motif in which mutations partially disrupt ribosome binding and have a minimal impact on SECIS binding but produce proteins unable to support Sec incorporation, indicating an essential role for ribosome binding. A structural analysis of SBP2 modeled after the 15.5-kDa protein-U4 snRNA cocrystal structure (32) predicts that the identified residues are found in an alpha-helical region that is not in direct contact with RNA and may form a unique surface involved in physical and functional interactions with the ribosome. Finally, competition experiments show that ribosome binding is regulated predominantly by the SECIS element via direct interaction with the core L7Ae motif.
MATERIALS AND METHODS
Mutagenesis.
This study made use of rat CTSBP2 consisting of aa 399 to 846, the coding region for which was TA cloned into pCR3.1 (8). Alanine substitutions were generated by QuikChange site-directed mutagenesis per the protocol of the QuikChange kit manufacturer (Stratagene). Mutant constructs were sequenced completely by automated DNA sequencing.
In vitro translation.
Plasmid DNA coding for wild-type or mutant CTSBP2 was linearized with XbaI or XhoI and used as a template for in vitro transcription with T7 RNA polymerase (mMessage kit; Applied Biosystems). In vitro translation reactions were set up with rabbit reticulocyte lysate in the presence of [35S]Met as described by the lysate manufacturer (Promega). The duration of the translation reaction was optimized to 15 min for CTSBP2 and the mutant forms generated. Translation products were resolved by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and quantitated by phosphorimager analysis. The amount of each protein was determined by the quantitation of known amounts of [35S]Met spotted onto filter paper and calculated based on an endogenous concentration of 5 μM cold Met in the lysate as specified by the manufacturer.
Purification of ribosomes.
Fresh, pretrimmed rat testes purchased from Pel-Freez were minced with a razor, and cold ribosome buffer (100 mM potassium acetate, 20 mM HEPES-KOH [pH 7.6], 2.5 mM magnesium acetate, 2 mM dithiothreitol [DTT], 1 mg of heparin/ml, and 1× Complete protease inhibitors [Roche]) was added at 2 ml/g (wet weight) of tissue. The tissue was briefly homogenized in a blender, followed by homogenization on ice with a high-clearance pestle. Extracts were spun at 17,000 × g for 15 min (all spins were performed at 4°C in a Sorvall S80-AT3 rotor). To pellet the ribosomes, supernatants were spun at 300,000 × g for 45 min. After washing in 0.5 ml of ribosome buffer, pellets were resuspended in 2.5 ml of high-salt buffer (ribosome buffer with 500 mM KCl) and stirred for 30 to 90 min. To clear insoluble material, samples were spun at 14,000 × g for 10 min and the supernatant was transferred into a new tube. This step was performed until no visible pellet could be seen (two to three spins). Cleared ribosomes were layered over a 1.0 M sucrose cushion in high-salt buffer and spun at 300,000 × g for 30 min. Ribosome pellets were resuspended in PBSD (1× phosphate-buffered saline-2 mM DTT) and stored in aliquots at −80°C.
Sucrose cushion assay.
To assay ribosome binding, 10 μl of in vitro-translated CTSBP2 was added to a reaction mixture containing 4 pmol of salt-washed ribosomes, 0.5 mM MgCl2, and 40 mM NaCl and brought to a final volume of 25 μl with PBSD. The reaction mixture was incubated for 5 min at 30°C and then placed on ice for 5 min. Twenty microliters of each reaction mixture was loaded over a 200-μl 20% sucrose cushion made in the same buffer and spun at 300,000 × g for 45 min at 4°C in a Sorvall S80-AT2 rotor. Supernatants were removed, and SDS sample buffer was added to a concentration of 1×. Pellets were resuspended in 100 μl of 1× SDS sample buffer. Supernatants and pellets (5% each) were resolved by 12% SDS-PAGE on a 12% gel. Radiolabeled CTSBP2 was detected by phosphorimaging, and the percentage of ribosome binding was calculated using ImageQuant software (GE Healthcare). To correct for nonspecific binding, the percentage of pelleting obtained with hnRNP F (the average of results from three independent trials) was subtracted from the percentages of pelleting observed with wild-type and mutant CTSBP2. Subsequently, the percentages of pelleting observed with the mutant proteins were normalized relative to the percentage of pelleting observed with wild-type CTSBP2. For the SECIS competition experiments, increasing amounts of the phospholipid hydroperoxide glutathione peroxidase (PHGPx) RNA SECIS element (0 to 2 pmol) were added to the 25-μl reaction mixture and the volume of PBSD was adjusted accordingly. After correcting for nonspecific binding, the percentages of pelleting in the SECIS element-containing reaction mixtures were normalized relative to the percentage of pelleting in reaction mixtures without SECIS RNA.
SECIS probes and EMSA.
32P-labeled wild-type and mutant PHGPx RNA SECIS elements were transcribed from HindIII-linearized plasmids bearing a 217-nucleotide insert derived from the PHGPx RNA 3′ UTR as described elsewhere (6). Electrophoretic mobility shift assays (EMSA) were performed as described previously (8). Briefly, 4 μl of in vitro-translated wild-type or mutant CTSBP2 and 20 fmol of the wild-type or mutant PHGPx RNA 3′ UTRs were mixed with 1× phosphate-buffered saline supplemented with 250 μg of Escherichia coli tRNA per ml, 10 mM DTT, and 5 μg of soybean trypsin inhibitor (Sigma) per ml. Complexes were formed at 37°C for 30 min, resolved on a 4% nondenaturing polyacrylamide gel, and analyzed by phosphorimaging. The percentage of SECIS binding was determined using ImageQuant software (GE Healthcare).
Sec incorporation assay.
Sec incorporation activity was determined using a luciferase construct containing a Sec (UGA) codon at position 258 and a PHGPx RNA SECIS element in the 3′ UTR as described previously (22). The Sec incorporation assay was performed with rabbit reticulocyte lysate (Promega). The total reaction mixture volume was 12 μl, which included 2 μl of the CTSBP2 in vitro-translation reaction mixture and 50 ng of luciferase reporter construct mRNA. Reaction mixtures were incubated for 1 h at 30°C and then diluted in 50 μl of 1× phosphate-buffered saline. Luminescence was detected with a Dynex MLX plate luminometer by using 50 μl of luciferase assay substrate (Promega).
UV cross-linking.
Cross-linking reactions were carried out using 200 ng of Xpress epitope- and His-tagged CTSBP2 (CTXH) as described above for the EMSA with either a wild-type or mutant SECIS element. CTXH was purified from E. coli as described elsewhere (15). Each reaction mixture was treated with UV irradiation by using a Bio-Rad GS Genelinker at 254 nm for 10 min in a 96-well tissue culture plate (Corning). The entire cross-linking reaction mixture was included in a 50-μl ribosome binding assay mixture with 10 pmol of salt-washed purified ribosomes in PBSD. Reaction mixtures were spun as described for the ribosome binding assay. Afterwards, supernatants were removed and pellets were resuspended in 100 μl of 1× PBSD. Supernatants and pellets were then treated with RNase A (10 μg) for 15 min at 37°C. A 4× concentration of SDS sample buffer was added to a final concentration of 1×, and equal amounts (9%) of supernatants and pellets were analyzed by SDS-PAGE on a 12% gel and then subjected to phosphorimaging to determine the location of the SECIS-bound SBP2. A parallel gel was also run, followed by Western blot analysis to determine the location of non-cross-linked CTXH. Western blotting was performed as described elsewhere (15).
RESULTS
The SBP2 L7Ae motif is required for ribosome binding.
The L7Ae motif within SBP2 is known to be required for SECIS binding, but its role in ribosome binding is less clear. To clarify the role of the L7Ae motif in the function of SBP2 and to identify critical residues within this motif that are involved in ribosome binding, SBP2 sequences from multiple species were aligned and the conserved amino acids were mutated to alanine in clusters of five (each cluster of five alanine mutations is hereafter referred to as a penta-alanine mutation) (Fig. 1). A total of 15 penta-alanine mutations were examined in this study.
FIG. 1.
Multiple sequence alignment of the SBP2 L7Ae RNA binding domain. SBP2 amino acid sequences from rat, chicken, puffer fish, and fly were aligned. Residues shaded in black are identical, and residues shaded in gray are conservative substitutions. Residues mutated to penta-alanine are indicated above the corresponding positions in SBP2. Sequences were generated using MultAlin, and conserved residues were shaded using Boxshade.
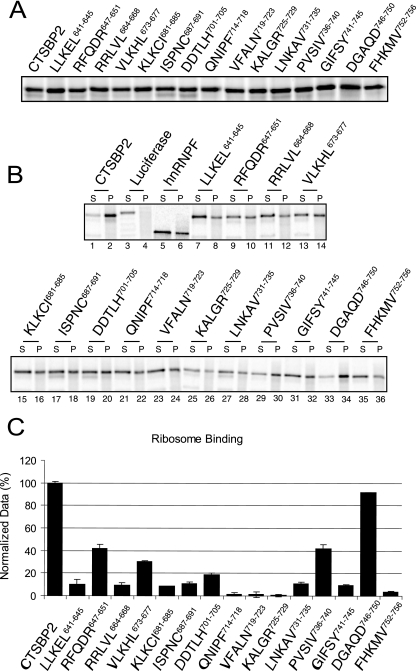
To test CTSBP2 mutant forms for defects in ribosome binding activity, in vitro-translated CTSBP2 was radiolabeled with [35S]Met. Maximal CTSBP2 translation occurs after 15 min of incubation, typically yielding 5 to 10 fmol per μl. Longer translation times yield degraded protein (data not shown). To minimize the negative effects of protein degradation, equal volumes of in vitro-translated protein were immediately analyzed for all functions. All mutant proteins were translated with an efficiency similar to that of the translation of the wild type, with levels of efficiency varying less than twofold (Fig. 2A). In vitro-translated wild-type and mutant proteins were incubated with an excess amount (4 pmol) of salt-washed ribosomes purified from rat testicular extracts. Complexes were centrifuged through a sucrose cushion, and equal portions of the supernatant and pellet were resolved by SDS-PAGE. Approximately 80% of the CTSBP2 was found consistently in the pellet fraction (Fig. 2B, lanes 1 and 2). As a negative control, the luciferase construct was translated and tested to account for nonspecific binding to the ribosome. As expected, 87% of the protein remained in the supernatant fraction (Fig. 2B, lanes 3 and 4). To control for the amount of ribosome binding derived from nonspecific RNA-protein interactions, hnRNP F, which is not known to bind ribosomes but has multiple cellular RNA targets (5, 10, 21), was in vitro translated and analyzed for ribosome binding activity. In this case, 67% of the protein remained in the supernatant fraction (Fig. 2B, lanes 5 and 6). To focus on the dynamic range of the ribosome binding assay, the level of binding obtained with hnRNP F was considered to be background. After subtracting the background from the amounts of binding obtained with the penta-alanine mutant proteins, the binding levels were normalized relative to that for wild-type CTSBP2 (Fig. 2C).
FIG. 2.
Ribosome binding analysis of CTSBP2 penta-alanine mutant proteins. (A) Wild-type and mutant forms of CTSBP2 were translated and [35S]Met labeled in rabbit reticulocyte lysate and resolved by 12% SDS-PAGE. (B) The proteins described in the legend for panel A were incubated with purified rat ribosomes and centrifuged through a 20% sucrose cushion. Equal portions (5%) of the supernatant (S) and pellet (P) were resolved by SDS-PAGE and analyzed by phosphorimaging. The luciferase construct was translated and tested as a negative control. (C) The results presented in panel B were quantitated as the percentage of protein in the pellet and normalized relative to the level of pelleting observed for wild-type CTSBP2. The amount of ribosome binding obtained with in vitro-translated hnRNP F (33% pelleting) was considered background and subtracted. The data shown are the averages ± standard errors of results from at least three independent experiments.
Penta-alanine mutations of 14 of the 15 clusters in the CTSBP2 L7Ae motif resulted in various degrees of reduction in ribosome binding activity. The mutation of the DGAQD746-750 region, which is at the far C-terminal end of the L7Ae motif, was the sole change that had no effect on ribosome binding. Two mutations in the N-terminal portion, those of RFQDR647-651 and VLKHL673-677, as well as the mutation of PVSIV736-739 in the far C-terminal end produced an intermediate defect, with ∼30 to 40% of the ribosome binding activity remaining. All other mutations led to more severe ribosome binding defects, resulting in the retention of ∼20% or less of the ribosome binding activity of wild-type CTSBP2.
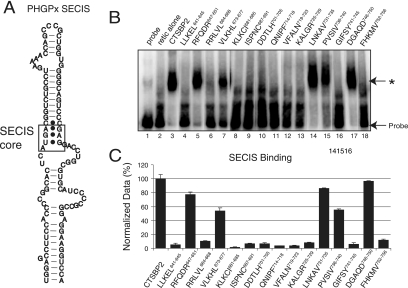
To identify regions in the L7Ae motif that contribute specifically to ribosome binding, the penta-alanine mutant proteins were screened for the ability to bind the SECIS element. To assay SECIS binding, equal volumes of in vitro-translated CTSBP2 were incubated with a [32P]UTP-labeled PHGPx RNA SECIS element (Fig. 3A) and analyzed by EMSA (Fig. 3B). Notably, 5 of the 15 penta-alanine mutant proteins retained a significant level of SECIS binding activity (Fig. 3B, lanes 5, 7, 14, 15, and 17). The largest defects in SECIS binding were apparent in proteins with mutations in the central portion of the L7Ae motif ranging from KLKCI681-685 to KALGR725-729. In parallel with the ribosome binding data, the DGAQD746-750 mutant protein retained 85% of wild-type SECIS binding activity (Fig. 3C). Surprisingly, the RFQDR647-651 and LNKAV731-735 mutant proteins also retained SECIS binding activity (∼80%) despite clear defects in ribosome binding (Fig. 4). The VLKHL673-677 and PVSIV736-739 mutant proteins retained about 50% of the wild-type SECIS binding activity, which parallels their decrease in ribosome binding activity to ∼30 to 40% of the wild-type level. All of the CTSBP2 mutant proteins that bound the SECIS element still showed specificity for the SECIS core (Fig. 3A) as indicated by their inability to shift a mutant SECIS element lacking the core (AUGA) motif (data not shown).
FIG. 3.
SECIS binding analysis of CTSBP2 penta-alanine mutant proteins. (A) Diagram of the PHGPx SECIS element. The 203-nucleotide PHGPx SECIS element was utilized to assay SECIS binding. The SECIS core motif is boxed to indicate the SBP2 binding site. (B) [35S]Met-labeled wild-type and mutant forms (indicated by the regions corresponding to the mutations) of CTSBP2 were incubated with 20 fmol of the wild-type 32P-labeled PHGPx SECIS element, and the complexes were resolved on a 4% nondenaturing gel. The asterisk indicates the position of a SECIS-specific complex. Unsupplemented reticulocyte lysate (retic alone) was tested as a negative control to account for nonspecific binding. (C) The results presented in panel B were quantitated as the percentage of shifted probe, corrected for nonspecific binding, and normalized relative to the level of shift observed for wild-type CTSBP2. The data shown are the averages ± standard errors of results from at least three independent experiments.
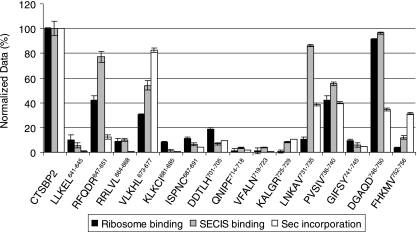
FIG. 4.
Sec incorporation activity of CTSBP2 penta-alanine mutant proteins. The mutant proteins (indicated by the regions corresponding to the mutations) represented in Fig. 1 were added to a reticulocyte lysate assay mixture containing luciferase mRNA with a Sec codon at position 258 and a wild-type PHGPx SECIS element inserted in the 3′ UTR. After normalization for protein expression, the luciferase activity of each mutant protein was normalized relative to the luciferase activity of wild-type CTSBP2, set at 100%. For each SBP2 mutant protein, the average luciferase activity is shown, together with the SECIS and ribosome binding data from Fig. 2 and 3.
The findings with regard to SECIS and ribosome binding indicate that the SBP2 L7Ae motif consists of three subdomains: an N-terminal region including RFQDR647-651 and VLKHL673-677 in which mutations lead to reduced ribosome binding but permit the retention of significant levels of SECIS binding, a central region from KLKCI681-685 to KALGR725-729 in which mutations generate severe SECIS and ribosome binding defects, and finally, a C-terminal region including LNKAV731-735 and PVSIV736-740 in which mutations also permit the retention of substantial levels of SECIS binding but produce severe and intermediate ribosome binding defects, respectively (Fig. 4). Several exceptions to these general observations are noteworthy. The mutant proteins with substitutions in the N-terminal sequences LLKEL641-645 and RRLVL664-668 displayed severe defects in both SECIS binding and ribosome binding. Likewise, mutant proteins harboring substitutions in the C-terminal sequences GIFSY741-745 and FHKMV752-756 also showed severe defects in SECIS binding and ribosome binding. Overall, these results demonstrate that the CTSBP2 L7Ae motif is required for the ribosome binding activity of CTSBP2. These findings also reveal the residues within the CTSBP2 L7Ae motif that contribute to the overlap of SECIS and ribosome binding domains proposed previously (8). More importantly, however, these results have allowed us to identify residues that function primarily in interactions with the ribosome, thus allowing us to determine the functional significance of the CTSBP2-ribosome interaction in Sec incorporation.
SBP2-ribosome interactions are essential for Sec incorporation.
To investigate the impact on Sec incorporation activity, the CTSBP2 mutants described above were tested in an in vitro luciferase assay. A firefly luciferase reporter construct containing a wild-type PHGPx SECIS element and an in-frame UGA codon was translated in rabbit reticulocyte lysate in the presence or absence of CTSBP2. CTSBP2 has been shown to stimulate luciferase activity in a SECIS- and UGA codon-dependent manner by several hundredfold in this assay (22). In addition, the SBP2-dependent increase in luciferase activity is strictly dependent on the presence of a wild-type SECIS element and a UGA codon, indicating that the stop codon read-through represents Sec incorporation. For the analysis of the penta-alanine mutant proteins, the same [35S]Met-labeled CTSBP2 samples that were used for ribosome and SECIS element binding assays were added to luciferase Sec incorporation assay mixtures. Luciferase activity was expressed as a percentage of that obtained with CTSBP2 after normalizing for mutant protein expression. The amounts of SBP2 present in the luciferase reaction mixtures were within the linear range of the assay (data not shown). For the purposes of comparison, the Sec incorporation activity data are shown together with the SECIS and ribosome binding data (Fig. 4).
As expected, most mutant proteins with substitutions that eliminated SECIS binding activity also had little or no detectable Sec incorporation activity and were therefore not considered further in this study. In the case of the FHKMV752-756 mutant protein, 30% of the Sec incorporation activity persisted despite the retention of only 10% of the wild-type level of SECIS binding, indicating that even a low level of SECIS binding is able to support Sec incorporation, as previously shown for a G669A mutation which severely reduced SECIS binding but allowed the retention of Sec incorporation activity (8). The notable exceptions were the RFQDR647-651, LNKAV731-735, and DGAQD746-750 mutant proteins, all of which retained ∼80% of the wild-type level of SECIS binding but 12, 39, and 35% of the wild-type Sec incorporation activity, respectively (Fig. 4). Of these, the RFQDR647-651 and LNKAV731-735 mutant proteins also had a defect in ribosome binding, thus providing evidence that a ribosome binding defect correlates with a reduction in Sec incorporation activity. Interestingly, a similar ribosome binding defect in the VLKHL673-677 mutant protein was observed, but this protein retained ∼80% of the Sec incorporation activity. Overall, these results show that distinct portions of the L7Ae motif contribute differentially to the three activities of SBP2.
SBP2-ribosome contacts involve physical and functional interactions.
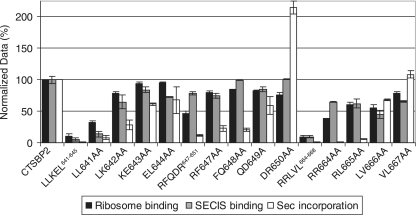
Since the RFQDR647-651 mutation was able to separate SECIS binding activity from ribosome binding activity and had the most severe effect on Sec incorporation, we chose to analyze the RFQDR647-651 region further. To precisely identify which amino acids contributed to the ribosome binding defects described above, we analyzed portions of the RFQDR647-651 segment in four mutant proteins with the following pairwise substitutions: RF647AA, FQ648AA, QD649AA, and DR650AA. The same type of mutagenesis of the flanking segments subjected to penta-alanine mutation, LLKEL641-645 and RRLVL664-668, was performed. Each of these 12 mutant proteins was then analyzed for SECIS binding, ribosome binding, and Sec incorporation activity as described above. While the RFQDR647-651 penta-alanine mutation resulted in a ∼50% reduction in ribosome binding, none of the mutant proteins carrying pairwise substitutions displayed a reduction of more than 20% (Fig. 5). These results illustrate that the major contribution of RFQDR647-651 to ribosome binding can be revealed only when all five residues are simultaneously mutated. Despite the recovery of near-wild-type levels of ribosome binding, however, the restoration of the wild-type Sec incorporation activity was observed only for the DR650AA mutant protein, suggesting that the primary role of these residues may lie in a functional interaction with the ribosome that contributes only partly to the ribosome binding activity. The observed hyperactivity of the DR650AA mutant protein was confirmed when equimolar amounts of wild-type or DR650AA mutant protein were tested. The results showed that the DR650AA protein reached maximal Sec incorporation activity at a lower concentration than wild-type CTSBP2 (data not shown). The LL641AA protein closely resembled the LLKEL641-645 mutant form, while the remaining spectrum of mutant proteins with substitutions in this region, proceeding from the N- to the C-terminal end, gradually regained a portion of all three activities. These results separate the negative effects of the LLKEL641-645 penta-alanine mutation and those of the RFQDR647-651 mutation because all three activities were significantly restored when only the amino acids immediately N-terminal to RFQDR647-651 were mutated (KE643AA and EL644AA). In contrast, the RRLVL664-668 pairwise-mutated RR664AA and RL665AA proteins showed defects generally similar to those seen in the RFQDR647-651 mutant analysis, suggesting that residues within aa 647 to 665 are involved in a ribosome-based function. The RR664AA mutation, therefore, presents the best evidence of an essential ribosome binding requirement since the corresponding mutant protein displays a greater than 90% loss of Sec incorporation activity despite retaining 60% of wild-type SECIS binding activity. Together, these results identify a region within CTSBP2 (aa 647 to 664) in which mutations correspond to a loss in Sec incorporation that directly correlates with a ribosome binding defect.
FIG. 5.
Analysis of CTSBP2 forms with pairwise subdivisions of penta-alanine mutations. The RFQDR647-651 segment and the flanking segments subjected to penta-alanine mutation (LLKEL641-645 and RRLVL664-668) were each analyzed in four mutant proteins (indicated by the corresponding mutation) with pairwise substitutions of alanine. Each mutant protein harboring a pairwise substitution, along with the corresponding penta-alanine mutant protein (indicated by the region corresponding to the mutation), was assayed for ribosome binding, SECIS binding, and Sec incorporation activity. The data are the means (± standard errors) of results from at least three independent experiments.
Three-dimensional modeling of SBP2 supports the idea of a distinct interaction surface.
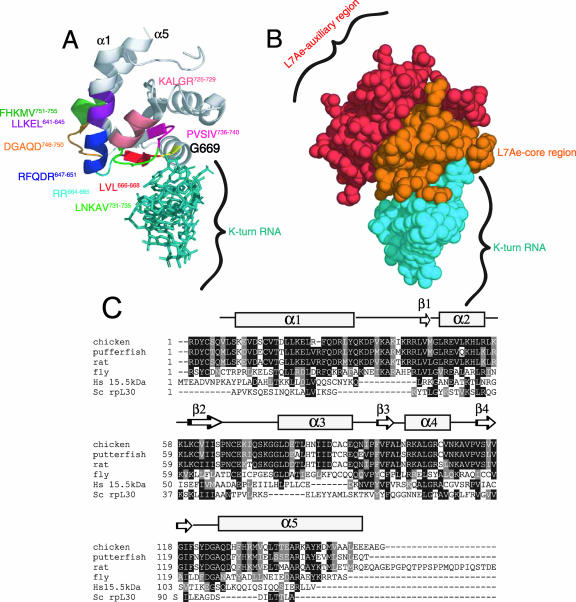
The structures of several proteins in the L7Ae family have been solved previously by X-ray crystallography analyses that have revealed a characteristic α-β-α topology (2, 3, 32). Comparative structural analyses of proteins within this family have revealed a substantial level of structural homology with a root mean square deviation of 1.104 Å for the comparison of the human 15.5-kDa protein with Methanococcus species L7Ae (28). In addition, several conserved residues within the L7Ae motif have previously been shown to affect the SECIS binding activity of SBP2 (1). These results support the idea that SBP2 shares this characteristic α-β-α fold. To analyze SBP2 mutant proteins in a three-dimensional context, we used the cocrystal structure of the human 15.5-kDa protein and U4 snRNA as a model for the SBP2-SECIS interaction (Fig. 6A). To predict the positions of SBP2 residues within this structure, we utilized alignments of SBP2 with several proteins in the L7Ae family, including rpL30 and the 15.5-kDa protein (Fig. 6C). This alignment was consistent with that obtained with a large number of L7Ae family members (28), thus providing a solid foundation for the assignment of SBP2 residues on the 15.5-kDa-protein structure.
FIG. 6.
Structural modeling of SBP2. (A) The cocrystal structure of the 15.5-kDa snRNA binding protein bound to its cognate K-turn U4 RNA (Protein Data Bank coordinates 1E7K) was used to predict the positions of the following regions within CTSBP2: RFQDR647-651, DGAQD746-750, G669 (yellow), LVL666-668, RR664, KALGR725-729, LLKEL641-645, FHKMV752-756, LNKAV731-735, and PVSIV736-740. (B) Space-filled model of the structure presented in panel A, highlighting the auxiliary and core functional regions within the L7Ae domain as noted. The annotation of the structure was performed using MacPyMOL. (C) Alignment of the L7Ae motifs from the following K-turn binding proteins: Saccharomyces cerevisiae (Sc) rpL30, the human (Hs) 15.5-kDa protein, and the SBP2 sequences in Fig. 1. Residues shaded in black are identical, and residues shaded in gray are conservative substitutions. A secondary structure prediction for SBP2 (developed using PredictProtein) (26) is shown above the sequences.
When the SBP2 sequence was modeled on this structure, it was apparent that amino acids RFQDR647-651 lie towards the bottom of an alpha-helical region that does not directly contact K-turn RNA (α1) (Fig. 6A). The corresponding region in any protein of the L7Ae family has not been shown to make contact with RNA (2, 3, 32). The RR664 segment localizes to loop 2 and is critical for the ribosome binding activity of SBP2 but, similar to RFQDR647-651, is likely not directly involved in SECIS contacts. The LVL666-668 region comprises β1 and is likely involved in direct SECIS contacts as predicted by this model and its proximity to the conserved G669. Interestingly, RFQDR647-651 and RR664 are in close physical proximity to DGAQD746-750, the mutation of which also resulted in a loss of Sec incorporation activity despite the retention of SECIS and ribosome binding activity. In addition, the RFQDR647-651 and DGAQD746-750 residues, while within the conserved structural motif, are not highly conserved among K-turn binding proteins (Fig. 6C). Thus, residues in the lower portion of α1 (RFQDR647-651), the loop between α1 and β1 (RR664), and the bottom of α5 (DGAQD746-750) all appear to form a separate interaction surface utilized for ribosome binding and Sec incorporation but not SECIS binding.
Most of the mutations in residues that localize near the RNA-protein interface (aa 666 to 668 and 681 to 785) severely diminished ribosome binding and SECIS binding activity. Based on the structural model, these residues either make direct contact with RNA or are in a location that would likely disrupt the proximity of a neighboring region involved in a direct RNA interaction. The KALGR725-729 residues are directly downstream of R724, which has previously been shown to disrupt SECIS binding when mutated in human SBP2 (1). In contrast, the LLKEL641-645 and FHKMV752-756 mutant proteins (Fig. 6A), despite being distant from the RNA-protein interface, also showed severe defects in ribosome binding and SECIS binding activity, suggesting that the mutation of the corresponding residues may result in global misfolding of the protein. On the other hand, the LNKAV731-735 and PVSIV736-740 mutant proteins retained significant SECIS binding activity, despite the close proximity of these amino acid residues to the RNA. Interestingly, these residues are located primarily in loop 9, which has previously been described as a variable region among proteins in the L7Ae family that may contribute to RNA binding specificity (12). Combined with the mutagenesis data, this model indicates that there are likely two distinct interaction surfaces within the L7Ae motif. Figure 6B shows a representation of the L7Ae domain divided into two major sections: the L7Ae core motif that is required for both SECIS and ribosome binding and an L7Ae auxiliary motif that is not required for SECIS binding but primarily for optimal ribosome binding and Sec incorporation activity. The mutagenesis results described above indicate that these two regions are interdependent because mutations in the L7Ae core motif completely eliminate ribosome binding activity, i.e., the L7Ae auxiliary motif is unable to support ribosome binding on its own.
The SECIS binding site is a dominant regulator of ribosome binding.
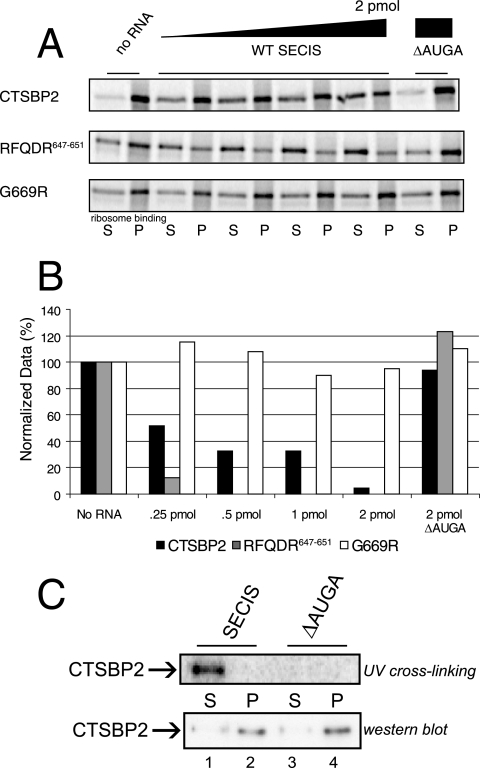
As noted above, mutations in the L7Ae core motif (aa 681 to 729) reduced ribosome binding to background levels, while the mutation of RFQDR647-651 did not. This finding suggests that residues in the L7Ae core region have a dominant effect on ribosome binding which may serve to modulate the function of residues in the L7Ae auxiliary region. To analyze the contribution of the L7Ae auxiliary motif to the total ribosome binding activity of SBP2, the ribosome binding activities of wild-type and RFQDR647-651 and G669R mutant versions of CTSBP2 were challenged with SECIS elements in a competition assay. For this experiment, increasing amounts of wild-type PHGPx SECIS elements were incubated simultaneously with CTSBP2 and purified ribosomes, and the amount of competition was determined by the sucrose cushion assay described above. As shown in Fig. 7A, 2 pmol of the PHGPx SECIS element was able to compete with ribosomes for binding to CTSBP2, yielding background levels of ribosome binding while fourfold less of the SECIS element was required to reduce RFQDR647-651 mutant protein binding to the same extent. This result confirms that the RFQDR647-651 region represents a significant component of the total ribosome binding activity. The G669R mutant form, however, was not forced off the ribosome by SECIS RNA, consistent with the fact that it cannot interact with the SECIS element (8) and confirming that all of the SECIS element competition effects are the result of direct competition at the L7Ae motif and not another as-yet-undefined function of the SECIS element. A graphical representation of these results is shown in Fig. 7B.
FIG. 7.
SECIS elements modulate the SBP2-ribosome interaction. (A) [35S]Met-labeled wild-type CTSBP2 and the RFQDR647-651 and G669R mutant forms were incubated with increasing amounts (0, 0.25, 0.5, 1.0, and 2.0 pmol) of a wild-type (WT) PHGPx SECIS element or a mutant element lacking the core motif (ΔAUGA) and analyzed for ribosome binding as described in the legend to Fig. 2B. S, supernatant; P, pellet. (B) The data obtained as described for panel A were quantitated by phosphorimaging, normalized for nonspecific binding, and graphed as the percentage of ribosome binding relative to that for the no-SECIS reaction mixture. (C) Four picomoles of recombinant CTXH was subjected to UV cross-linking to 20 fmol of the PHGPx SECIS element. Following cross-linking, reaction mixtures were incubated with excess salt-washed purified ribosomes (10 pmol) and spun as described in the legend to Fig. 2. Equal portions of the supernatant and pellet (9%) were resolved on an SDS-PAGE gel and analyzed by phosphorimaging (top panel) or by Western blot analysis (bottom panel). A reaction mixture containing a mutant SECIS element (ΔAUGA) was used as a negative control.
The results shown in Fig. 7A are consistent with the findings of a previous study demonstrating that SECIS elements are efficient competitors with ribosomes for SBP2 binding and that SBP2 is unable to bind stably to the SECIS element and the ribosome simultaneously (15). However, the use of excess SECIS elements in the previous study may have prevented the observation of transient or low-affinity simultaneous interactions. To determine if an SBP2 molecule in its SECIS-bound conformation could interact with the ribosome when the amount of the SECIS element was kept limited, recombinant CTSBP2 was subjected to UV cross-linking to the PHGPx SECIS element, thus creating a small pool of recombinant CTSBP2 covalently linked to the SECIS element. Prior to RNase treatment, the UV cross-linking reaction mixture was combined with an excess of salt-washed ribosomes and complexes were centrifuged through a sucrose cushion as described above. Equal portions of the supernatant and pellet were resolved by SDS-PAGE, followed by phosphorimaging to determine the location of the SECIS-bound CTSBP2 (Fig. 7C, top panel). Another portion was resolved by SDS-PAGE and subjected to Western blot analysis to allow the detection of non-cross-linked CTSBP2, thus confirming that UV cross-linking did not disrupt the ability of CTSBP2 to bind the ribosome (Fig. 7C, bottom panel). All of the SECIS-cross-linked CTSBP2 remained in the supernatant, while the much larger portion of non-cross-linked CTSBP2 was found almost exclusively in the pellet fractions (Fig. 7C, lanes 1 and 2, compare top and bottom panels). These results clearly show that SECIS-bound SBP2 is unable to interact stably with purified ribosomes, indicating that the L7Ae core region is the primary determinant of ribosome binding and that the L7Ae auxiliary motif is involved in a functional interaction with the ribosome. Additionally, the results indicate that the L7Ae auxiliary region is modulated by the SECIS element via this L7Ae core region.
DISCUSSION
The presence of a suppressor tRNA in a cell results in nonsense suppression by competing with the termination machinery for access to the ribosomal A site, resulting in stop codon read-through. In addition, the suppression of stop codons can result from the misincorporation of near-cognate tRNAs. The latter mechanism is enhanced by mutations in the ribosome and by drugs that target the ribosome (14, 19). In these cases, the modification of the ribosome promotes nonsense suppression by interfering with translation fidelity and/or by decreasing the efficiency of translation termination (27). Similar defects in translation fidelity when translation elongation or termination factors are mutated have been observed previously (reviewed in reference 31). Sec incorporation represents a modification of this process in that it increases the translational read-through of specific UGA codons in select mRNAs. In order to provide a high level of specificity to this process, the eEFSec-GTP-Sec-tRNA[Ser]Sec ternary complex is not able to suppress UGA codons without the presence of at least two additional factors. This indicates that unlike the canonical elongation factor ternary complex (eEF1A-tRNA-GTP), the eEFSec-tRNA complex likely does not have direct access to the ribosomal A site. The factors providing specificity to the UGA suppression event are SBP2 and its RNA target, the SECIS element. While SBP2 is known to bind specifically to both the ribosome and the SECIS element, its exact role in this process is unknown (7). Here, we have examined the contribution of the conserved L7Ae domain within SBP2 to all three of the protein's known functions: SECIS element binding, ribosome binding, and Sec incorporation.
Evidence for an SBP2-ribosome interaction requirement.
While the initial structure-function study of SBP2 clearly showed that the L7Ae RNA binding domain is required for function, these conclusions were based on either mutations that eliminated SECIS binding (G669R) or truncations that reduced both SECIS and ribosome binding (8). While those results demonstrate a requirement for SECIS binding, they do not address the functional significance of the SBP2-ribosome interaction. Our mutagenic approach led to the identification of a discrete stretch of amino acids (RFQDR647-651) that is required for ribosome binding and Sec incorporation but not for SECIS binding. Surprisingly, pairwise mutations within this sequence of five residues showed that although the ribosome binding defect was eliminated, Sec incorporation activity was restored only for the DR650AA mutant protein. These mutations within RFQDR647-651 were similar to truncations in the SBP2 functional domain that abolish Sec incorporation activity but have no impact on ribosome binding or SECIS element binding (8). These results indicate that SBP2 makes functional contacts with the ribosome that are required for UGA recoding, but whether this phenotype is related to the role of the SBP2 functional domain remains to be determined. Additional evidence for a ribosome binding requirement was derived from the RR664AA mutant protein, which was unable to support Sec incorporation. The RR664AA protein retained 60% of the SECIS element binding activity but only 40% of ribosome binding activity, a defect that is likely to be responsible for the loss of Sec incorporation activity. This finding is in contrast to the loss of SECIS binding as a result of a G669A mutation that produced an 80% loss in SECIS binding activity while permitting the retention of 20% of the Sec incorporation activity (8).
Due to the high level of structural conservation among proteins in the L7Ae family, we sought to determine the positions of the amino acids RFQDR647-651 relative to the universally conserved glycine residue 669. Strikingly, the RFQDR647-651 region does not appear to make any contacts with K-turn RNA, consistent with the ability of SBP2 to bind the SECIS element when these residues are mutated. This observation lends further support to the notion that the SECIS binding and ribosome binding domains overlap but are not identical. Overall, the combination of mutagenesis and structural modeling suggests that the L7Ae domain within SBP2 consists of two interdependent motifs: the L7Ae core motif and the L7Ae auxiliary motif.
Interestingly, however, despite a distinct set of amino acids that are involved predominantly in ribosome binding, SBP2 does not simultaneously interact with the SECIS element and the ribosome (15). This finding was apparent even when SBP2 was cross-linked to the SECIS element and incubated with excess ribosomes (Fig. 7C). Competition studies revealed that a wild-type SECIS element was more effective at competing with the ribosome binding activity of the RFQDR647-651 mutant protein than with that of wild-type SBP2. This confirms that the RFQDR647-651 region contributes to ribosome binding and that this secondary site for ribosome interactions is regulated by SECIS binding. Taken together, these results indicate that SBP2 contacts the ribosome at two distinct but interdependent sites, both of which are regulated by the SECIS element via its interaction with the L7Ae core domain in the L7Ae motif.
Communication with the ribosomal A site.
The aminoglycoside G418 has been shown to promote amino acid misincorporation at UGA Sec codons in vivo in a dose-dependent manner (13). Remarkably, the presence of a functional Sec incorporation complex, as indicated by the presence of a wild-type SECIS element, was able to significantly reduce G418-induced read-through. One possible interpretation of these findings is that the Sec incorporation machinery is able to communicate with the ribosomal A site. We have determined in vitro that a wild-type SECIS element is not sufficient to block G418-induced read-through (our unpublished results), consistent with the inability of the SECIS element to bind directly to the ribosome (15). These results strongly suggest that the SECIS-dependent communication with the A site is mediated by SBP2. Our results clearly show that SECIS-bound SBP2 is unable to interact stably with purified ribosomes, leaving open the question of how SBP2 can exert its seemingly requisite ribosome-proximal effects during a Sec incorporation cycle. One possibility is that SBP2 function is dynamic and that the movement of SBP2 relative to the ribosome, as dictated by the SECIS element, is responsible for the conformational changes on the ribosome that trigger Sec insertion.
The K-turns on the ribosome make an attractive target for SBP2 since they not only resemble the SECIS element in structure but are also predicted to be flexible and thus capable of transmitting signals over a large distance (24, 25). While crystal structure analysis of the archaebacterial ribosome has identified K-turns in both the 28S rRNA and the 18S rRNA (16), the vast majority are in the large subunit rRNA. In addition, preliminary data suggest that SBP2 binds specifically to 28S rRNA (8). Notably, under the more stringent conditions of our present ribosome binding assay, the G669R mutant protein displayed a ribosome binding defect (Fig. 7A) that was not apparent in the original SBP2 structure-function study (8). Because this residue is universally conserved in the L7Ae motif, this defect in ribosome binding further supports the idea that SBP2 interacts with a K-turn on the ribosome.
Conclusions.
Collectively, our results support the model that the SBP2-ribosome interaction is an essential aspect of the Sec incorporation process. An attractive but speculative model of SBP2 function is that the primary contact of SBP2 with the ribosome is mediated by the L7Ae core motif. This interaction is modulated upon SECIS binding, thus allowing the L7Ae auxiliary motif to link indirectly to the A site conformation. This change in A site conformation should promote eEFSec ternary complex binding and Sec-tRNA[Ser]Sec delivery. These results set the stage for an analysis of the SBP2 binding site on the ribosome and the conformational changes on the ribosome that lead to Sec incorporation. In addition to enhancing our understanding of the Sec incorporation process, these studies will improve our knowledge of ribosome dynamics and K-turn function.
Acknowledgments
We thank Terri Kinzy, Stephane Gross, and Kim Kandl for a critical reading of the manuscript.
This work was supported by Public Health Service grant GM068077 (P.R.C.) and the Montclair State University-Graduate School of Biomedical Sciences Bridge to the Doctoral Degree GM66338 (K.C.) from the National Institutes of Health.
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Allmang, C., P. Carbon, and A. Krol. 2002. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA 8:1308-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 3.Chao, J. A., and J. R. Williamson. 2004. Joint X-ray and NMR refinement of the yeast L30e-mRNA complex. Structure 12:1165-1176. [DOI] [PubMed] [Google Scholar]
- 4.Chavatte, L., B. A. Brown, and D. M. Driscoll. 2005. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 12:408-416. [DOI] [PubMed] [Google Scholar]
- 5.Chou, M. Y., N. Rooke, C. W. Turck, and D. L. Black. 1999. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 19:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copeland, P. R., and D. M. Driscoll. 1999. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J. Biol. Chem. 274:25447-25454. [DOI] [PubMed] [Google Scholar]
- 7.Copeland, P. R., J. E. Fletcher, B. A. Carlson, D. L. Hatfield, and D. M. Driscoll. 2000. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 19:306-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copeland, P. R., V. A. Stepanik, and D. M. Driscoll. 2001. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol. Cell. Biol. 21:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagegaltier, D., N. Hubert, K. Yamada, T. Mizutani, P. Carbon, and A. Krol. 2000. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 19:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamberi, C., E. Izaurralde, C. Beisel, and I. W. Mattaj. 1997. Interaction between the human nuclear cap-binding protein complex and hnRNP F. Mol. Cell. Biol. 17:2587-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halic, M., T. Becker, J. Frank, C. M. Spahn, and R. Beckmann. 2005. Localization and dynamic behavior of ribosomal protein L30e. Nat. Struct. Mol. Biol. 12:467-468. [DOI] [PubMed] [Google Scholar]
- 12.Hamma, T., and A. R. Ferre-D'Amare. 2004. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 A resolution. Structure 12:893-903. [DOI] [PubMed] [Google Scholar]
- 13.Handy, D. E., G. Hang, J. Scolaro, N. Metes, N. Razaq, Y. Yang, and J. Loscalzo. 2006. Aminoglycosides decrease glutathione peroxidase-1 activity by interfering with selenocysteine incorporation. J. Biol. Chem. 281:3382-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jemiolo, D. K., F. T. Pagel, and E. J. Murgola. 1995. UGA suppression by a mutant RNA of the large ribosomal subunit. Proc. Natl. Acad. Sci. USA 92:12309-12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinzy, S. A., K. Caban, and P. R. Copeland. 2005. Characterization of the SECIS binding protein 2 complex required for the co-translational insertion of selenocysteine in mammals. Nucleic Acids Res. 33:5172-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein, D. J., T. M. Schmeing, P. B. Moore, and T. A. Steitz. 2001. The kink-turn: a new RNA secondary structure motif. EMBO J. 20:4214-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koonin, E. V., P. Bork, and C. Sander. 1994. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 22:2166-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, B. J., M. Rajagopalan, Y. S. Kim, K. H. You, K. B. Jacobson, and D. Hatfield. 1990. Selenocysteine tRNA[Ser]Sec gene is ubiquitous within the animal kingdom. Mol. Cell. Biol. 10:1940-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manuvakhova, M., K. Keeling, and D. M. Bedwell. 2000. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA 6:1044-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin, G. W., III, J. W. Harney, and M. J. Berry. 1996. Selenocysteine incorporation in eukaryotes: insights into mechanism and efficiency from sequence, structure, and spacing proximity studies of the type 1 deiodinase SECIS element. RNA 2:171-182. [PMC free article] [PubMed] [Google Scholar]
- 21.Matunis, M. J., J. Xing, and G. Dreyfuss. 1994. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 22:1059-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta, A., C. M. Rebsch, S. A. Kinzy, J. E. Fletcher, and P. R. Copeland. 2004. Efficiency of mammalian selenocysteine incorporation. J. Biol. Chem. 279:37852-37859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papp, L. V., J. Lu, F. Striebel, D. Kennedy, A. Holmgren, and K. K. Khanna. 2006. The redox state of SECIS binding protein 2 controls its localization and selenocysteine incorporation function. Mol. Cell. Biol. 26:4895-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Razga, F., N. Spackova, K. Reblova, J. Koca, N. B. Leontis, and J. Sponer. 2004. Ribosomal RNA kink-turn motif: a flexible molecular hinge. J. Biomol. Struct. Dyn. 22:183-194. [DOI] [PubMed] [Google Scholar]
- 25.Razga, F., M. Zacharias, K. Reblova, J. Koca, and J. Sponer. 2006. RNA kink-turns as molecular elbows: hydration, cation binding, and large-scale dynamics. Structure 14:825-835. [DOI] [PubMed] [Google Scholar]
- 26.Rost, B., G. Yachdav, and J. Liu. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321-W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salas-Marco, J., and D. M. Bedwell. 2005. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J. Mol. Biol. 348:801-815. [DOI] [PubMed] [Google Scholar]
- 28.Suryadi, J., E. J. Tran, E. S. Maxwell, and B. A. Brown II. 2005. The crystal structure of the Methanocaldococcus jannaschii multifunctional L7Ae RNA-binding protein reveals an induced-fit interaction with the box C/D RNAs. Biochemistry 44:9657-9672. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Tujebajeva, R. M., P. R. Copeland, X. M. Xu, B. A. Carlson, J. W. Harney, D. M. Driscoll, D. L. Hatfield, and M. J. Berry. 2000. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 1:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valente, L., and T. G. Kinzy. 2003. Yeast as a sensor of factors affecting the accuracy of protein synthesis. Cell. Mol. Life Sci. 60:2115-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vidovic, I., S. Nottrott, K. Hartmuth, R. Luhrmann, and R. Ficner. 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell 6:1331-1342. [DOI] [PubMed] [Google Scholar]