Abstract
The Notch signaling pathway appears to perform an important function in a wide variety of organisms and cell types. In our present study, we provide evidence that UV irradiation-induced Tip60 proteins reduced Notch1 activity to a marked degree. Accumulated UV irradiation-induced Tip60 suppresses Notch1 transcriptional activity via the dissociation of the Notch1-IC-CSL complex. The binding between endogenous Tip60 and Notch1-IC in UV radiation-exposed cells was verified in this study by coimmunoprecipitation. Interestingly, the physical interaction of Tip60 with Notch1-IC occurs to a more profound degree in the presence of CSL but does not exist in a trimeric complex. Using Notch1-IC and Tip60 deletion mutants, we also determined that the N terminus, which harbors the RAM domain and seven ankyrin repeats of Notch1-IC, interacts with the zinc finger and acetyl coenzyme A domains of Tip60. Furthermore, here we report that Notch1-IC is a direct target of the acetyltransferase activity of Tip60. Collectively, our data suggest that Tip60 is an inhibitor of the Notch1 signaling pathway and that Tip60-dependent acetylation of Notch1-IC may be relevant to the mechanism by which Tip60 suppresses Notch1 signaling.
Notch is a vitally important signaling receptor which modulates cell fate determination and pattern formation in a number of ways during the development of both invertebrate and vertebrate species (2, 3, 19, 46, 54, 64, 70, 74, 75). In vertebrates, Notch proteins comprise a family of four transmembrane receptors (Notch1 to Notch4) which harbor multiple epidermal growth factor-like repeats, followed by conserved cysteine-rich Notch/Lin12 repeats in their extracellular domain, and seven cdc10/ankyrin repeats and a PEST domain within their intracellular domains (41, 57, 73, 74). Interactions between Notch and its proposed ligand Delta or Serrate/Jagged induce a cleavage step near the transmembrane region of the C-terminal protein fragment, which results in the release of the intracellular domain (Notch-IC), followed by its nuclear translocation. An important nuclear target of Notch-IC is the DNA binding protein CSL/CBF-1, the mammalian homologue of Drosophila melanogaster Suppressor of Hairless [Su(H)] (52, 72). Notch1-IC interacts with CSL/Su(H) primarily through the RAM domain, a sequence located N-terminal to the ankyrin repeats, resulting in the activation of target gene transcription (67). Several downstream targets of Notch signaling have also been identified, including Enhancer of split [E(spl)] complex members and the mammalian homologues of Hairy and E(spl), Hes1 and Hes5 (1, 16, 34, 35, 50, 60). These basic helix-loop-helix (bHLH) proteins antagonize other bHLH factors, such as MyoD, which induces muscle differentiation (31, 42, 43, 76). The nuclear activity of the Notch intracellular domain is linked with complexes that regulate chromatin organization via the deacetylation and acetylation of histones. Remodeling of the chromatin template via the inhibition of histone deacetylase activities represents a higher level of Notch expression in the presomitic mesoderm major (14, 32, 36, 45, 53, 80).
The histone acetyltransferase (HAT) Tip60 is a member of the MYST family, which acetylates principally histone H4 of the histone acetyltransferases, and was initially identified as a 60-kDa human immunodeficiency virus (HIV) Tat-interacting protein (13, 65, 81). Tip60 is able to interact with and enhance human androgen receptor activity via the acetylation of the human androgen receptor (7, 24, 25). Other studies have shown that Tip60 associates with alternative transcription factors, including Bcl-3, Myc, TEL/ETV, and ING (17, 18, 23, 56). Tip60 roles have been established in the transcription of the amyloid precursor protein, in interleukin-1β signaling, and in subsequent NF-κB-mediated gene regulation (4, 10). However, Tip60 has also been shown to repress transcription by the phosphorylated cyclic AMP response element binding (CREB) protein and the signal transducer and activator of transcription 3 (Stat3) protein (27, 79). In Drosophila melanogaster chromatin remodeling complexes, Tip60 exists as part of a multiple component complex that includes TRRAP, actin, Baf53a, Epc1, p400/domino, Reptin, Pontin, MrgBP, Brd8, Gas41, YL1, Ing3, and DMAP1 (44). Genetic studies using the fly system have shown that TRRAP and domino have functions in Notch signaling. However, findings do not rule out the possibility that TRRAP and domino can also act independently of Tip60 (20, 26). Cellular responses to DNA damage also involve the HAT Tip60, as the overexpression of a dominant negative HAT-defective Tip60 mutant decreases both DNA repair and apoptosis upon the induction of DNA double-stranded breaks (12, 33, 69). Tip60 levels have been shown to increase significantly as a result of the UV irradiation of Jurkat cells, and the HAT activity of Tip60 is regulated via phosphorylation by cyclin-dependent kinases (30, 47). Numerous studies have demonstrated that Tip60 is able to regulate transcription either positively or negatively, depending on the cell type or the promoter.
In this study, we have demonstrated that Tip60 is involved in the negative regulation of the Notch signaling pathway. Accumulated UV irradiation-induced Tip60 suppresses Notch1 transcriptional activity via the dissociation of the Notch1-IC-CSL complex. Interestingly, the physical interaction of Tip60 with Notch1-IC is much more profound in the presence of CSL but does not involve the formation of a trimeric complex. Using Notch1-IC and Tip60 deletion mutants, we were also able to determine that the N terminus containing seven ankyrin repeats of Notch1-IC interacts with the zinc finger and acetyl (acetyl-coenzyme A CoA) domains of Tip60. Furthermore, we identified four lysine residues in Notch1-IC as direct acetylation sites for Tip60. Collectively, our data show that Tip60 is an inhibitor of the Notch1 signaling pathway and that direct interaction with Notch1-IC may be relevant to the mechanism by which Tip60 suppresses Notch1 signaling.
MATERIALS AND METHODS
Cell culture and transfection.
NIH 3T3 and human embryonic kidney (HEK) 293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% bovine calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). For plasmid DNA transfection, cells were plated at a density of 2 × 106 cells/100-mm dish, grown overnight, and transfected with the appropriate expression vectors in the presence of the indicated combinations of plasmid DNAs, via the calcium phosphate and liposome method (11).
Cloning and preparation of recombinant proteins.
A full-length mouse Tip60 gene, a mouse Notch1 gene, and deletion mutants were constructed via PCR and inserted into either the mammalian expression vector p3XFlag-CMV (Sigma) or the bacterial expression vector pGEX4T-3 (Amersham Pharmacia). The Tip60 deletion mutants constructed for the present study were Tip60-CM (chromodomain; amino acid residues 1 to 258), Tip60-ZF (zinc finger motif; amino acid residues 69 to 290), Tip60-ZCoA (zinc finger motif and acetyl-CoA binding domain; amino acid residues 158 to 395), and Tip60-CoA (acetyl-CoA binding domain; amino acid residues 285 to 513). The Notch1-IC deletion mutants constructed for the present study were Notch1-IC-RAM-ANK (amino acid residues 1744 to 2110), Notch1-IC-RAM (amino acid residues 1744 to 1870), Notch1-IC-OPA (amino acid residues 2076 to 2369), and Notch1-IC-PEST (amino acid residues 2369 to 2531). The C-terminal Myc-tagged constitutively active form of Notch1 (ΔEN1-Myc) and Myc-tagged Notch1-IC (Myc-Notch1-IC) were kind gifts from Raphael Kopan (Washington University, St. Louis, MO). Expression of the recombinant glutathione S-transferase (GST) fusion proteins within the transformed bacteria was induced using 1 mM isopropyl-β-d-thiogalactopyranoside (Sigma). GST fusion proteins were purified with glutathione (GSH)-agarose beads (Sigma), in accordance with the manufacturer's instructions.
Site-directed mutagenesis.
Site-directed mutagenesis of Notch1-IC cDNA was performed with a QuikChange kit (Stratagene), and the mutagenic primers were K2019R (5′-GTGGATGACCTAGGCAgGTCGGCTTTGCATTGG-3′), K2039/2044R (5′-GTGCTCCTGAgGAACGGAGCCAACAgGGACATGCAG-3′), and K2068R (5′-GCTATGAGACTGCCAgAGTGTTGCTGGACC-3′) (mismatches with the Notch1-IC cDNA template are indicated by lowercase letters). The mutations were verified by automatic DNA sequencing.
Luciferase reporter assay.
After 48 h of transfection, the cells were lysed in chemiluminescent lysis buffer (18.3% of 1 M K2HPO4, 1.7% of 1 M KH2PO4, 1 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol) and assayed for luciferase activity using a luciferase assay kit (Promega). The activity of the luciferase reporter protein in the transfected cells was normalized with regard to the β-galactosidase activity in the same cells (61).
Antibodies and reagents.
We purchased antibodies to cleaved Notch1(Val1744) (catalog number 2421; Cell Signaling), Tip60 (catalog number 07-038; Upstate Biotechnology), acetylated-lysine (catalog number 9441; Cell Signaling), Flag M2 (catalog number F-3165; Sigma), Myc (9E10) (catalog number 1667149; Roche Molecular Biochemicals), hemagglutinin (HA) (12CA5) (catalog number 11583816001; Roche Molecular Biochemicals), and β-actin (catalog number SC-47778; Santa Cruz Biotechnology). The secondary antibodies used in our experiment were goat anti-mouse immunoglobulin G (IgG) and goat anti-rabbit IgG and were purchased from Cell Signal Biotechnology. All other reagents were obtained from Sigma.
Knockdown of Tip60 in cells.
The short interfering RNA targeting Tip60 (siTip60) was used as previously described (38). Sham control or Tip60 siRNA was transfected into HEK293 or NIH 3T3 cells by using Fugene6 reagent (Roche), in accordance with the manufacturer's instructions.
RNA isolation and reverse transcriptase PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen). All samples were treated with RNase-free DNase I (Takara) at 37°C for 30 min under the following conditions: 20 μg of RNA, 40 mM Tris-HCl, 8 mM MgCl2, 5 mM dithiothreitol, 0.4 unit/μl RNase inhibitor (Promega), and 10 units of DNase I in a volume of 50 μl. Then, a phenol-chloroform extraction was performed, and the RNA was precipitated. Total RNA (2 μg) was used to synthesize cDNA with Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer's instructions; contaminating DNA was removed by treating the samples with RNase-free DNase. The primers used in PCR analysis were Hes1 forward (5′-CAGCCAGTGTCAACACGACAC-3′), Hes1 reverse (5′-TCGTTCATGCACTCGCTGAA-3′), Hes5 forward (5′-CGC ATC AAC AGC AGC ATA GAG-3′), Hes5 reverse (5′-TGG AAG TGG TAA AGC AGC TTC-3′), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward (5′-GTC ATC ATC TCC GCC CCT TCT GC-3′), GAPDH reverse (5′-GAT GCC TGC TTC ACC ACC TTC TTG-3′), β-actin forward (5′-GTG GGG CGC CCC AGG CAC CA-3′), and β-actin reverse (5′-CTC CTT AAT GTC ACG CAC GAT-3′). GAPDH and β-actin were used as controls for PCR analysis.
Coimmunoprecipitation assays.
The cells were lysed in 1 ml of radioimmunoprecipitation assay buffer for 30 min at 4°C. After 20 min of centrifugation at 12,000 × g, the supernatants were subjected to immunoprecipitation with the appropriate antibodies coupled to protein A-agarose beads. The resulting immunoprecipitates were then washed three times in phosphate-buffered saline (pH 7.4). Laemmli sample buffer was then added to the immunoprecipitated pellets, and the pellets were heated for 5 min at 95°C and analyzed via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Western blotting was conducted using the indicated antibodies.
In vitro binding assay.
The recombinant GST-Notch1-IC protein was expressed in Escherichia coli strain BL21, using the pGEX system as indicated. The GST-Notch1-IC protein was then purified using glutathione-agarose beads (Sigma), in accordance with the manufacturer's instructions. An equal amount of GST or GST-Notch1-IC fusion protein was incubated with the lysates of HEK293 cells, which had been transfected for 3 h with combinations of expression vectors at 4°C, with rotation. After incubation, the beads were washed three times with ice-cold phosphate-buffered saline and boiled with 20 μl of Laemmli sample buffer. The precipitates were separated via SDS-PAGE, and the pull-down proteins were detected via immunoblotting with specific antibodies.
In vitro Notch1-IC acetylation assay.
Notch1-IC acetylation assays were carried out as follows: 20 ml of reaction mixture contained 50 mM Tris-HCl (pH 8.0), 10% glycerol (vol/vol), 0.1 mM EDTA, 1 mM dithiothreitol, 10 mM sodium butyrate, 5 mM acetyl-CoA (Sigma), 100 ng of GST-Notch-IC proteins, and 200 ng of Tip60. The mixture was incubated at 30°C for 60 min and analyzed on SDS-PAGE. Acetylated Notch1-IC was detected by Western blotting using the anti-acetyl-lysine antibody. GST-Notch1-IC proteins were visualized with Ponceau staining using the same membrane as that for Western blotting.
RESULTS
Tip60 inhibits Notch transcriptional activity.
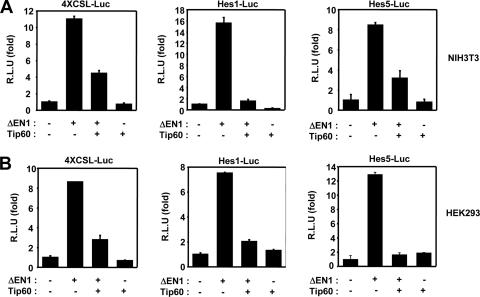
In order to evaluate the possible function of Tip60 in Notch signaling, a reporter assay was conducted with NIH 3T3 and HEK293 cells, using luciferase reporter genes. The NIH 3T3 or HEK293 cells were transfected with 4XCSL-Luc and either the active Notch1 mutant ΔEN1 or an empty vector. As expected, ΔEN1-mediated transcription activity increased in these samples in both cell lines (Fig. 1A and B). We determined that Tip60 attenuated the ability of ΔEN1 to stimulate transcription (Fig. 1A and B). The bHLH proteins Hes1 and Hes5, both of which harbor multiple CSL binding DNA sequences on their promoters, were identified as essential targets of Notch in the epithelial cells (60). Therefore, we have confirmed the effects of Tip60 on the Notch1 signaling pathway, using the Hes1 and Hes5 reporter systems, respectively. Expression of the active form of Notch1 was determined to significantly induce activation of the Hes1 and Hes5 reporter systems (Fig. 1A and B). Tip60 overexpression inhibits constitutively active Notch1-induced natural Hes1 and Hes5 promoter transcriptional activities (Fig. 1A and B).
FIG. 1.
Tip60 inhibits Notch transcriptional activity. (A) NIH 3T3 cells were transfected with expression vectors for 100 ng of 4XCSL-Luc, 100 ng of Hes-1-Luc, 100 ng of Hes-5-Luc, and 100 ng of β-galactosidase, along with 100 ng of ΔEN1 and 100 ng of Tip60, as indicated. (B) HEK293 cells were transfected with expression vector for 100 ng of 4XCSL-Luc, 100 ng of Hes-1-Luc, 100 ng of Hes-5-Luc, and 100 ng of β-galactosidase, along with 100 ng of ΔEN1 and 100 ng of Tip60, as indicated. After 48 h of transfection, the cells were lysed, and the luciferase activity was determined. Data were normalized with β-galactosidase. Results represent the means ± average deviations of three independent experiments. R.L.U, relative luciferase units.
UV suppresses Notch1 transcriptional activity via the induction of Tip60.
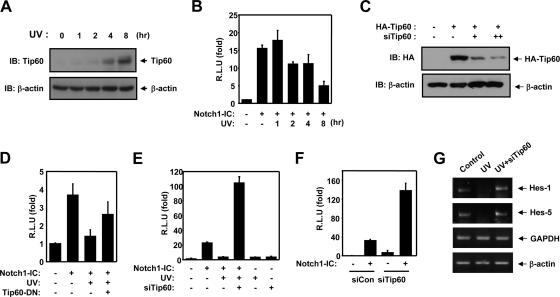
As the Tip60 protein is known to be induced by UV irradiation, we attempted to determine whether UV radiation could modulate Notch1 signaling (47). In our experiment, we observed that Tip60 protein is accumulated in a time-dependent manner by UV radiation-exposed HEK293 cells (Fig. 2A). However, during this time, cell death is not observed (data not shown). Luciferase has a half-life in mammalian cells of approximately 3 h. Due to this short half-life, changes in the promoter activity are rapidly reflected in the luciferase activity since it is not accumulated. As is shown in Fig. 2B, UV radiation profoundly suppresses Notch1 transcriptional activity in a time-dependent manner. Because UV irradiation can induce the expression of a variety of proteins, including Tip60, we attempted to determine whether Tip60 induction was a major requirement for the UV radiation-induced suppression of Notch1 signaling. In order to evaluate the involvement of Tip60 in the UV radiation-induced suppression of Notch signaling, we employed a Tip60 dominant negative mutant and Tip60 siRNA to reduce enzyme activity and down-regulate the expression of Tip60, respectively. Unlike the levels observed for the control short interference plasmid-transfected cells, the cells transfected with short interference Tip60 harbored low levels of Tip60 (Fig. 2C). Whereas Notch1-IC-mediated transcriptional activity was repressed in the UV radiation-exposed HEK293 cells, the UV radiation-induced suppression of Notch1 transcriptional activity was restored via coexpression of the Tip60 dominant negative mutant (Fig. 2D). In order to delineate the possible role for Tip60 in the regulation of Notch1 signaling in intact cells, we assessed the effects of the Tip60 knockdown on Notch signaling. We determined that Tip60 siRNA was able to block the suppressive effects of UV radiation on Notch1-IC-induced transcriptional activity (Fig. 2E). Furthermore, we determined that the transfection of Tip60 siRNA into HEK293 cells resulted in a robust increase of Notch1 transcriptional activity in intact cells but that the control siRNA does not affect Notch activity (Fig. 2F). From these results, it was assumed that UV radiation can decrease the expression of the Notch1 target gene through upregulation of Tip60. In order to demonstrate this assumption, reverse transcriptase PCR (RT-PCR) was performed for Hes1 and Hes5. HEK293 cells were transfected with empty vector or with siTip60 prior to their exposure to UV irradiation. The expression levels of Hes1 and Hes5 were decreased by UV irradiation, as expected, and the UV irradiation-induced suppression of Hes1 and Hes5 was restored via coexpression of the siTip60 (Fig. 2G). Thus, taken together, our results appear to suggest that Tip60 induction is a prerequisite for the suppression of the Notch signaling pathway.
FIG. 2.
UV suppresses Notch1 transcriptional activity through Tip60 induction. (A) HEK293 cells were exposed to UV light (60 J/m2) and then incubated for an additional 1 to 8 h at 37°C. The cell lysates were then subjected to immunoblotting analysis with anti-Tip60 and anti-β-actin antibody. β-Actin was used as a loading control. (B) HEK293 cells were transfected with expression vector for 100 ng of 4XCSL-Luc and 100 ng of β-galactosidase, along with 100 ng of Notch1-IC, as indicated. After 40 h of transfection, cells were exposed to UV light (60 J/m2) and then incubated for an additional 1 to 8 h at 37°C. (C) HEK293 cells were transfected with the expression vectors for 0.5 μg of HA-Tip60 and 0.5 μg (+) and 1 μg (++) of siTip60, as indicated. Cell lysates were subjected to immunoblotting (IB) analyses with anti-HA or anti-β-actin antibody. (D) NIH 3T3 cells were transfected with expression vector for 100 ng of 4XCSL-Luc, 100 ng of Tip60-DN, and 100 ng of β-galactosidase, along with 100 ng of Notch1-IC, as indicated. After 40 h of transfection, the cells were exposed to UV light (60 J/m2) and then incubated for an additional 8 h at 37°C. (E) HEK293 cells were transfected with expression vector for 100 ng of 4XCSL-Luc, 200 ng of siControl, 200 ng of siTip60, and 100 ng of β-galactosidase, along with 100 ng of Notch1-IC, as indicated. After 40 h of transfection, the cells were exposed to UV light (60 J/m2) and then incubated for an additional 8 h at 37°C. (F) HEK293 cells were transfected with expression vector for 100 ng of 4XCSL-Luc, 200 ng of siControl, 200 ng of siTip60, and 100 ng of β-galactosidase, along with 100 ng of Notch1-IC, as indicated. After 48 h of transfection, the cells were lysed and the luciferase activity was assayed. Data were normalized with β-galactosidase. Results are expressed as the means ± average deviations of three independent experiments. R.L.U, relative luciferase units. (G) HEK293 cells were transfected with 2 μg of siTip60 and exposed to UV light (60 J/m2), and then the total RNA was extracted from the cell lysates. Reverse transcriptase PCR was performed using Hes1, Hes5, GAPDH, and β-actin primers with the cDNA from the total RNA. GAPDH and β-actin were used as a control for RT-PCR.
Fe65 does not affect the Tip60-mediated suppression of Notch1 signaling.
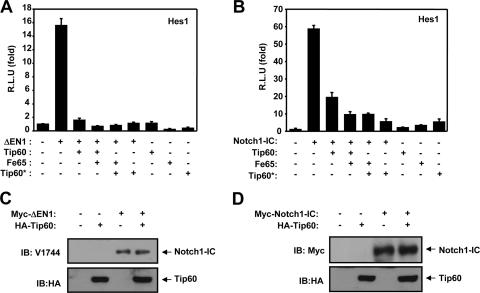
Previous reports have shown that the Tip60 histone acetyltransferase forms a complex with the Fe65 nuclear adaptor protein (10). In order to determine the function of Fe65 in the Tip60-mediated suppression of Notch signaling, we introduced a Tip60 mutant (Tip60*), which is unable to bind to Fe65 (10). The expression of Notch1ΔE (ΔEN1) was found to significantly induce the activation of the Hes1-Luc reporter system (Fig. 3A). When Tip60 was transiently coexpressed in NIH 3T3 cells, constitutively active Notch1ΔE-mediated transcriptional activity was suppressed to a substantial degree (Fig. 3A). However, the forced expression of Fe65 and Tip60* had no effect on the negative role of Tip60 in ΔEN1-mediated transcriptional activity. Moreover, we also verified the effects of Fe65 on the Tip60-induced suppression of Notch signaling, using Notch1-IC. When Tip60 was transiently coexpressed in NIH 3T3 cells, constitutively active Notch1-IC-mediated transcriptional activity was clearly suppressed (Fig. 3B). However, overexpression of Fe65 and Tip60* had no effect on the negative role of Tip60 in Notch1-IC-mediated transcriptional activity (Fig. 3B). Our findings further indicated that the Tip60-induced suppression of Notch activity was not correlated significantly with the inhibition of the γ-secretase-dependent proteolytic cleavage of Notch1ΔE (ΔEN1) (Fig. 3C). Also, the steady-state protein level of Notch1 was not diminished in this case (Fig. 3D), suggesting that at least in part, the Tip60-induced inhibition of Notch can be linked neither to the regulation of cleavage patterns nor to protein stability.
FIG. 3.
Fe65 does not affect Tip60-mediated suppression of Notch signaling. (A) NIH 3T3 cells were transfected with expression vector for 100 ng of Hes-1-Luc and 100 ng of β-galactosidase, along with 100 ng of ΔEN1, 100 ng of Tip60, 100 ng of Fe65, and 100 ng of the Tip60* mutant, as indicated. (B) NIH 3T3 cells were transfected with expression vector for 100 ng of Hes-1-Luc and 100 ng of β-galactosidase, along with 100 ng of Notch1-IC, 100 ng of Tip60, 100 ng of Fe65, and 100 ng of Tip60*, as indicated. After 48 h of transfection, the cells were lysed and the luciferase activity was assayed. Data were normalized with β-galactosidase. Results are expressed as the means ± average deviations of three independent experiments. R.L.U, relative luciferase units. (C and D) HEK293 cells were transfected with expression vector for 1 μg of the ΔEN1 mutant, 1 μg of Notch1-IC, and 1 μg of Tip60, as indicated. Cell lysates were also subjected to immunoblotting (IB) analysis with the indicated antibodies.
HAT activity of Tip60 is required for the transcriptional repression of Notch1-IC.
Two major phosphorylation sites, Ser-86 and Ser-90, were identified, and the phosphorylation of Tip60 was determined to modulate its histone acetyltransferase activity (30). As Tip60 acetyltransferase activity is controlled by phosphorylation, we then attempted to determine whether the phosphorylation of Tip60 might also be linked to the negative regulation of Notch signaling. In the luciferase reporter gene assay conducted with the HEK293 cells, Tip60(S86A), Tip60(S90A), and Tip60(S86/90A) were transfected, and the effects of these transfections on the transcriptional activation of Notch1-IC target genes were then evaluated using 4XCSL-Luc. Notch1-IC transcriptional activity was inhibited by Tip60(S90A) but was not inhibited by Tip60(S86A) and Tip60(S86/90A) (Fig. 4A). We also attempted to determine the effects of the dominant negative form of Tip60 on Tip60-induced suppression of Notch1-IC transcriptional activity. Notch1-IC transcriptional activation was restored by the cotransfection of dominant-negative Tip60 (Tip60-DN) in 4XCSL-Luc reporter genes (Fig. 4B). Results indicated that Tip60 HAT activity was essential to the negative transcriptional regulation of Notch1 target genes.
FIG. 4.
HAT activity of Tip60 is required for the transcriptional repression of Notch1-IC. (A) HEK293 cells were transfected with expression vector for 100 ng of 4XCSL-Luc and 100 ng of β-galactosidase, along with 100 ng of Notch1-IC, 100 ng of Tip60, 100 ng of Tip60(S86A), 100 ng of Tip60(S90A), and 100 ng of Tip60(S86/90A), as indicated. (B) HEK293 cells were transfected with expression vector for 100 ng of 4XCSL-Luc and 100 ng of β-galactosidase, along with the 100 ng of Notch1-IC, 100 ng of Tip60, and 100 ng of Tip60 dominant negative mutants, as indicated. After 48 h of transfection, the cells were lysed and the luciferase activity was assayed. Data were normalized with β-galactosidase. Results are expressed as the means ± average deviations of three independent experiments. R.L.U, relative luciferase units.
Next, we conducted immunofluorescence staining to determine whether Tip60 was colocalized with Notch1-IC and CSL. The nucleus may constitute the primary site of interaction between Notch1-IC and CSL. Tip60 is also located predominantly within the nucleus (10). As expected, the Tip60 stain was localized to the nucleus, as were the stains for CSL and Notch1-IC (data not shown). The image overlay revealed the nuclear colocalization of Notch1-IC, CSL, and Tip60 (data not shown). Furthermore, when the cells were transfected with Tip60 or the mutated Tip60*, Notch1-IC and CSL subcellular localization did not change to any substantial extent (data not shown). This suggests that Tip60 does not significantly regulate the Notch signaling pathway via the disruption of the cellular distribution of Notch1-IC and CSL.
Tip60 disrupts the binding of Notch1-IC to CSL.
The transcriptional activation of downstream target genes by Notch1-IC depends on the association of Notch1-IC with CSL within the nucleus. Thus, we attempted to demonstrate the effects of Tip60 on Notch1-IC and CSL binding, both in vitro and in intact cells. In the in vitro binding studies, purified GST or GST-Notch1-IC proteins were immobilized onto GSH-agarose. Flag-tagged CSL (Flag-CSL) and HA-tagged Tip60 (HA-Tip60) cells expressing the cell lysates were incubated with either GST or with GST-Notch1-IC, both of which were immobilized onto GSH-agarose. The interaction between GST-Notch1-IC and CSL was previously verified on bead complexes in the absence of Tip60 (Fig. 5A). Formation of the GST-CSL and Notch1-IC complexes was substantially suppressed as the result of Tip60 expression in vitro.
FIG. 5.
Tip60 disrupts the binding of Notch1-IC to CSL. (A) Recombinant GST and GST-Notch1-IC proteins were immobilized onto GSH-agarose, as described in Materials and Methods. HEK293 cells were transfected for 48 h with the expression vectors for 1 μg of Flag-CSL and 1 μg of HA-Tip60, as indicated. Cell lysates were then subjected to GST-pulldown experiments with immobilized GST or GST-Notch1-IC. Proteins bound to GST or GST-Notch1-IC were analyzed via immunoblotting (IB) with anti-Flag antibody. GST or GST-Notch1-IC proteins were visualized via staining with Coomassie brilliant blue. (B) HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors for 1 μg of Myc-Notch1-IC, 1 μg of Flag-CSL, and 1 μg of HA-Tip60. Cell lysates were then subjected to immunoprecipitation (IP) with anti-Myc or anti-Flag antibody, and the resulting precipitates were subjected to immunoblotting analysis with anti-Flag or anti-Myc antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. (C) HEK293 cells were transfected for 40 h with the indicated combinations of expression vectors for 1 μg of Myc-Notch1-IC, 1 μg of Flag-CSL, 2 μg of siControl, and 2 μg of siTip60. Cells were exposed to UV light (60 J/m2) and then incubated for an additional 8 h at 37°C. Cell lysates were then subjected to immunoprecipitation with anti-Flag antibody, and the resulting precipitates were subjected to immunoblotting analysis with anti-Myc antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. (D) HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors for 1 μg of Myc-Notch1-IC, 1 μg of Flag-CSL, 1 μg of HA-Tip60, 1 μg of the HA-Tip60(S86A), 1 μg of HA-Tip60(S90A) and 1 μg of HA-Tip60(S86/90A). Cell lysates were then subjected to immunoprecipitation with anti-Flag antibody, and the resulting precipitates were subjected to immunoblotting analysis with anti-Myc antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. (E) HEK293 cells were transfected for 40 h with the vectors for 2 μg of siControl or 2 μg of siTip60, after which the cells were exposed to UV light (60 J/m2) and incubated for an additional 8 h at 37°C. Cell lysates were then subjected to immunoprecipitation with anti-Notch1-IC or IgG antibody, and the resulting precipitates were subjected to immunoblotting analysis with anti-CSL, anti-β-actin, or anti-Tip60 antibody.
In order to determine the effects of Tip60 on the molecular interactions occurring between Notch1-IC and CSL, coimmunoprecipitation was conducted with HEK293 cells via the cotransfection of Myc-Notch1-IC, Flag-CSL, and HA-Tip60. The transfected cell lysates were immunoprecipitated using anti-Flag M2 antibody and immunoblotted with anti-Myc antibody (9E10) (Fig. 5B). Conversely, under conditions identical to those described above, we analyzed immunocomplexes which had been precipitated against anti-Myc antibody and immunoblotted with anti-Flag antibody. The results indicated that Tip60 interrupts the physical association between Notch1-IC and CSL (Fig. 5B). Furthermore, we also attempted to confirm the effects of Tip60 on the physical association occurring between Notch1-IC and CSL, using UV radiation induction and Tip60 siRNA. Whereas UV irradiation stimulates the dissociation of Notch1-IC and CSL, Tip60 siRNA was found to robustly restore the interaction between Notch1-IC and CSL (Fig. 5C). Furthermore, we also found that loss-of-function Tip60 mutants also increased the binding ability between Notch1-IC and CSL (Fig. 5D). These results have patterns similar to those shown in Fig. 2D and 4A. We can assume that the hyperstimulation of Notch1 transcription activity shown in Fig. 2D and 4A is due to stabilization of the Notch1-IC/CSL complex.
Next, we attempted to ascertain whether Tip60 was able to interrupt the association between Notch1-IC and CSL in intact cells. HEK293 cells were transfected with Tip60 siRNA and then exposed to UV irradiation. The cells were subjected to immunoprecipitation with anti-Notch1-IC, and the resultant immunopellets were examined via immunoblotting analysis with anti-CSL antibody (Fig. 5E). The immunoblot data showed that the accumulated Tip60 protein, which had been induced by UV irradiation, suppressed endogenous interaction between Notch1-IC and CSL in the intact cells (Fig. 5E). However, the physical association between Notch1-IC and CSL in the presence of UV radiation was not substantially diminished by coexpression with Tip60 siRNA (Fig. 5E). These results imply that Tip60 negatively regulated the transactivation of Notch1-IC target genes via the suppression of the interaction between Notch1-IC and CSL in intact cells.
Tip60 interacts directly with Notch1-IC in intact cells.
During our previous investigations, we have noticed that Tip60 may interact with Notch1-IC. In order to delineate more precisely the manner in which Tip60 prevents Notch1-IC- and CSL-mediated transcription, we conducted a series of in vitro binding and coimmunoprecipitation experiments. In the in vitro binding studies, purified GST or GST-Tip60 proteins were immobilized onto GSH-agarose. Flag-CSL and Myc-Notch1-IC expressing the cell lysates were incubated with either GST or GST-Tip60, both of which had been immobilized onto GSH-agarose. The interaction between GST-Tip60 and Notch1-IC has been detected on bead complexes in the absence of CSL (Fig. 6A). However, despite the performance of repeated experiments, no interaction was determined to have occurred between GST-Tip60 and CSL (Fig. 6A). Surprisingly, the strength of the interaction between GST-Tip60 and Notch1-IC was stronger in the presence of CSL than with Notch1-IC alone (Fig. 6A).
FIG. 6.
Notch1-IC interacts directly with Tip60 in intact cells. (A) Recombinant GST and GST-Tip60 proteins were immobilized onto GSH-agarose, as described in Materials and Methods. HEK293 cells were transfected for 48 h with the expression vectors for 1 μg of Myc-Notch1-IC and 1 μg of Flag-CSL, as indicated. Cell lysates were then subjected to GST-pulldown experiments with immobilized GST or GST-Notch1-IC. Proteins bound to GST or GST-Notch1-IC were analyzed via immunoblotting (IB) with anti-Myc or anti-Flag antibody. GST or GST-Tip60 proteins were visualized via staining with Coomassie brilliant blue. (B) HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors for 1 μg of Myc-Notch1-IC, 1 μg of Flag-CSL, and 1 μg of HA-Tip60. Cell lysates were then subjected to immunoprecipitation (IP) with anti-HA antibody, and the resulting precipitates were subjected to immunoblotting analysis with anti-Myc or anti-Flag antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. (C) HEK293 cells were transfected for 48 h with expression vectors for 1 μg of Myc-Notch1-IC and 1 μg of Flag-CSL. Cell lysates were then subjected to immunoprecipitation with anti-Flag antibody, and the resulting precipitates were incubated with either GST or with GST-Tip60 for 1 h on ice. The immunopellets were also subjected to immunoblotting analysis with anti-Myc antibody. GST or GST-Tip60 proteins were visualized via staining with Coomassie brilliant blue. (D) HEK293 cells were transfected for 40 h with the vector for 2 μg of siControl or 2 μg of siTip60, after which the cells were exposed to UV light (60 J/m2) and incubated for an additional 8 h at 37°C. Cell lysates were then subjected to immunoprecipitation with anti-Notch1-IC or IgG antibody, and the resulting precipitates were subjected to immunoblotting analysis with anti-Tip60 or β-actin antibody.
In order to characterize the physical interaction occurring between Tip60 and Notch1-IC or CSL, coimmunoprecipitation was conducted with HEK293 cells via the cotransfection of Myc-Notch1-IC, Flag-CSL, and HA-Tip60. The transfected cell lysates were immunoprecipitated with anti-HA antibody and immunoblotted with anti-Myc (9E10) or anti-Flag antibody (Fig. 6B). The results indicated that Tip60 interacts with Notch1-IC and that this association is significantly stronger in the presence of CSL, although no trimeric complex was detected in this case (Fig. 6B). To address the role of CSL in enhancing the recognition of Notch1-IC by Tip60, we examined whether Tip60 destabilizes preformed CSL/Notch1-IC complexes. Flag-CSL and Myc-Notch1-IC expressing the cell lysates were immunoprecipitated and then incubated with either GST or with GST-Tip60. The formation of the CSL and Notch1-IC complex was substantially suppressed by Tip60 in vitro (Fig. 6C).
Next, we attempted to ascertain whether Tip60 was able to physically associate with Notch1-IC in intact cells. HEK293 cells were transfected with Tip60 siRNA and then exposed to UV irradiation. The cells were subjected to immunoprecipitation with anti-Notch1-IC, and the resultant immunopellets were examined via immunoblotting analysis with anti-Tip60 antibody (Fig. 6D). The immunoblot data showed that the accumulated Tip60 protein, which had been induced by UV irradiation, interacted directly with endogenous Notch1-IC in the intact cells (Fig. 6D). In our experiments, no endogenous Tip60 could be detected, as has also been reported in similar previous studies (30, 48). Furthermore, the physical association occurring between Tip60 and Notch1-IC in the presence of UV radiation was diminished by the coexpression with Tip60 siRNA (Fig. 6D). These findings strongly suggest that a physical interaction between the two endogenous proteins Tip60 and Notch1 does occur in intact cells in response to UV irradiation.
The zinc finger and acetyl-CoA domains of Tip60 are critical for the suppression of Notch1 signaling.
Notch1-IC harbors a CDC domain which includes a RAM domain, seven ankyrin repeats, an OPA domain, and a PEST domain within its structure. We attempted to determine which, if any, of these domains might be involved in the interaction between Notch1-IC and Tip60. We utilized a variety of Flag-Notch1 deletion mutants, Notch1-IC-RAM-ANK, Notch1-IC-RAM, Notch1-IC-OPA, and Notch1-IC-PEST. We conducted an in vitro binding assay in which the deletion mutants expressing the cell lysates were applied to GST-Tip60 immobilized onto GSH-agarose beads (Fig. 7A). Our results indicated that Tip60 bound to Notch1-IC-RAM-ANK and to Notch1-IC-RAM but not to Notch1-IC-OPA or Notch1-IC-PEST. In order to verify these findings, we conducted coimmunoprecipitation assays using the same four Notch1-IC deletion mutants and HA-Tip60. As shown in Fig. 7B, the RAM and RAM-ANK domains of Notch1-IC interact with Tip60. Next, we attempted to ascertain whether Tip60 was able to interrupt the association between Notch1-IC-RAM and CSL. HA-CSL and Flag-Notch1-IC-RAM expressing the cell lysates were immunoprecipitated and then incubated with either GST or GST-Tip60. The formation of the CSL and Notch1-IC-RAM complex was substantially suppressed by Tip60 in vitro (Fig. 7C).
FIG. 7.
The zinc finger and acetyl-CoA domains of Tip60 are critical for the suppression of Notch1 signaling. (A) Recombinant GST-Tip60 proteins were immobilized onto GSH-agarose, as described previously. HEK293 cells were transfected for 48 h with the expression vectors for 1 μg of Flag-RAM-ANK, 1 μg of Flag-RAM, 1 μg of Flag-OPA, and 1 μg of Flag-PEST. Cell lysates were then subjected to GST-pulldown experiments with immobilized GST-Tip60. Proteins bound to GST-Tip60 were analyzed via immunoblotting with anti-Flag antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. (B) HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors for 1 μg of Flag-RAM-ANK, 1 μg of Flag-RAM, 1 μg of Flag-OPA, and 1 μg of Flag-PEST, along with 1 μg of HA-Tip60. Cell lysates were then subjected to immunoprecipitation (IP) with anti-HA antibody, and the resultant precipitates were subjected to immunoblotting (IB) analysis with anti-Flag antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. (C) HEK293 cells were transfected for 48 h with expression vectors for 1 μg of Flag-Notch1-IC-RAM and 1 μg of HA-CSL. Cell lysates were then subjected to immunoprecipitation with anti-HA antibody, and the resulting precipitates were incubated with either GST or with GST-Tip60 for 1 h on ice. The immunopellets were also subjected to immunoblotting analysis with anti-Flag antibody. GST or GST-Tip60 proteins were visualized via staining with Coomassie brilliant blue. (D) Recombinant GST-Notch1-IC proteins were immobilized onto GSH-agarose, as described previously. HEK293 cells were transfected for 48 h with the expression vectors for 1 μg of Flag-CM, 1 μg of Flag-ZF, 1 μg of Flag-ZCoA, and 1 μg of Flag-CoA. Cell lysates were then subjected to GST-pulldown experiments with immobilized GST-Notch1-IC. Proteins bound to GST-Notch1-IC were analyzed via immunoblotting with anti-Flag antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. (E) HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors for 1 μg of Flag-CM, 1 μg of Flag-ZF, 1 μg of Flag-ZCoA, and 1 μg of Flag-CoA, along with 1 μg of Myc-Notch1-IC. Cell lysates were then subjected to immunoprecipitation with anti-Myc antibody, and the resulting precipitates were subjected to immunoblotting analysis with anti-Flag antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. (F) HEK293 cells were transfected with expression vectors for 100 ng of 4XCSL-Luc and 100 ng of β-galactosidase, along with 100 ng of Notch1-IC and 100 ng (+), 200 ng (++), and 300 ng (+++) of Tip60-ZCoA, as indicated. After 48 h of transfection, the cells were lysed, and the luciferase activity was assayed. Data were normalized with β-galactosidase. Results represent the means ± average deviations of three independent experiments. R.L.U, relative luciferase units.
We then attempted to determine which of the domains of Tip60 was required for its association with Notch1-IC. Tip60 harbors a chromodomain, a zinc finger motif, and an acetyl-CoA domain within its structure. We used a variety of Flag-tagged Tip60 deletion mutants, Tip60-CM (chromodomain), Tip60-ZF (zinc finger motif), Tip60-ZCoA (zinc finger motif and acetyl-CoA domains), and Tip60-CoA (acetyl-CoA domain). We conducted an in vitro binding assay in which deletion mutants expressing cell lysates were applied to GST-Notch1-IC immobilized onto GSH-agarose beads (Fig. 7D). Our results indicate that Notch1-IC bound to Tip60-ZCoA but not to Tip60-CM, Tip60-ZF, or Tip60-CoA (Fig. 7D). In order to confirm these findings, we conducted coimmunoprecipitation assays using the same four Tip60 deletion mutants and Myc-Notch1-IC. As shown in Fig. 7E, the zinc finger motif and the acetyl-CoA domain of Tip60 were found to interact with Notch1-IC. This indicates that both the zinc finger motif and the acetyl-CoA domain of Tip60 are relevant to the interaction of this protein with the RAM domain of Notch1-IC.
We then assessed the inhibition of Notch1 activity by the Tip60-ZCoA proteins (Fig. 7F). Notch1-IC transcriptional activity was suppressed almost completely upon treatment with Tip60-ZCoA. These data, therefore, suggest that both the zinc finger motif and the acetyl-CoA domains of Tip60 are critical to the interaction of Tip60 with Notch1-IC, as well as its Notch1 signaling-inhibitory effects.
Tip60-mediated acetylation of lysine residues in Notch1.
Based on our observation that the N-terminal region of Notch1-IC interacts with Tip60 proteins, we asked whether Tip60 could also acetylate Notch1-IC. In order to characterize the acetylation, coimmunoprecipitation was conducted with the HEK293 cells via the transfection of HATip60 or siTip60 and then exposed to UV radiation. The transfected cell lysates were immunoprecipitated with anti-acetyl-lysine antibody and immunoblotted with anti-Notch1-IC antibody (Fig. 8A). This experiment revealed that Tip60 acetylates Notch1-IC protein.
FIG. 8.
Tip60 control Notch transcription activity through acetylation. (A) HEK293 cells were transfected with expression vector for 1 μg of Tip60, 2 μg of siControl (Con) and 2 μg of siTip60, as indicated. After 40 h of transfection, the cells were exposed to UV light (60 J/m2) and then incubated for an additional 8 h at 37°C. Cell lysates were then subjected to immunoprecipitation (IP) with anti-acetyl-lysine antibody, and the resulting precipitates were subjected to immunoblotting analysis with anti-Notch1-IC antibody. Cell lysates were also subjected to immunoblotting (IB) analysis with the indicated antibodies. (B) HEK293 cells were transfected for 48 h with the indicated expression vector for 1 μg of Myc-Notch1-IC (K2019/2068R), 1 μg of Myc-Notch1-IC (K2039/2044R), or 1 μg of Myc-Notch1-IC(K2019/2039/2044/2068R), along with 1 μg of HA-Tip60. Cells were lysed and immunoprecipitated against anti-acetyl-Lys antibody. The immunocomplexes were analyzed via SDS-PAGE and immunoblotting against anti-Myc antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. (C) HEK293 cells were transfected with expression vector for 100 ng of 4XCSL-Luc, 100 ng of β-galactosidase, 100 ng of Notch1-IC(K2019/2068R), 100 ng of Notch1-IC(K2039/2044R), or 100 ng of Notch1-IC(K2019/2039/2044/2068R), along with 100 ng of Tip60, as indicated. Cells were then lysed and assayed for luciferase activity. Results represent the means ± average deviation of three independent experiments. R.L.U, relative luciferase units; WT, wild type.
To identify which lysine residue(s) is acetylated in a Tip60-dependent manner, two purified Notch1-IC deletion mutants, Notch1-IC-RAM-ANK and Notch1-IC-RAM, were incubated with purified Tip60 and then an in vitro acetylation assay was performed using an anti-acetyl-lysine antibody. Acetylation was detected only in Notch1-IC-RAM-ANK, suggesting that the ankyrin repeats of Notch1-IC are possible targets for acetylation (data not shown). In silico studies have shown that Notch1 ankyrin repeats contain five possible conserved lysine residues in vertebrates; four lysine residues (K2019/2039/2044/2068) are accessible and are located between ankyrin repeats; one residue (K1935) is buried inside of an ankyrin repeat. Thus, we estimated that those four lysine residues could be the target of acetylation modification. To identify the precise targets of acetylation, Notch1-IC mutants with lysine-to-arginine substitutions were tested for Tip60-dependent acetylation with HEK293 cells. Immunoprecipitation and Western blotting analysis showed that proteins of the mutant carrying all four lysine substitution residues (K2019/2039/2044/2068R) exhibited a marked decrease in acetylation (Fig. 8B). This result implies that the residues K2019, K2039, K2044, and K2068 of Notch1-IC are the major targets of the acetyltransferase activity of Tip60.
Next, we examined the effect of Tip60 on the transcriptional activity of Notch1-IC mutants in HEK293 cells. Whereas Tip60 inhibits Notch1-IC(K2019/2068R)- and Notch1-IC(K2039/2044R)-induced transcriptional activity, Tip60 did not prevent Notch1-IC(K2019/2039/2044/2068R)-induced transcriptional activity (Fig. 8C). These data suggest that Tip60-mediated acetylation is the important mechanism by which Tip60 suppresses Notch1-IC activation in intact cells.
DISCUSSION
In the present study, we have demonstrated that UV radiation-induced Tip60 physically interacts with Notch1-IC in intact cells, thereby inhibiting Notch1 transcriptional activity. Accumulated Tip60 induced by UV irradiation suppresses Notch1 transcriptional activity via dissociation of the Notch1-IC-CSL complex. Furthermore, the HAT activity of Tip60 is critical to the binding and inhibition of the Notch1 signaling pathway. To our knowledge, this is the first demonstration of the HAT Tip60-mediated negative regulation of Notch signaling.
To allow the Notch signal to be deployed in numerous contexts, a host of distinct mechanisms have evolved in order to regulate the level, duration, and spatial distribution of Notch activity. Regulation occurs at multiple levels, including patterns of ligand and receptor expression, Notch-ligand interactions, receptor and ligand trafficking, and a variety of covalent modifications, including glycosylation, phosphorylation, ubiquitination, and protein-protein interaction (5, 6, 21, 22, 28, 29, 37, 40, 51, 55, 58, 62, 63, 66, 68, 71, 78). Our results showed that Notch1 transcriptional activity was inhibited by Tip60 in intact cells, which suggests that Tip60 may also be involved in the suppression of Notch1 transcriptional activity.
In Drosophila melanogaster chromatin remodeling complexes, Tip60 exists as part of a multiple component complex that includes TRRAP, actin, Baf53a, Epc1, p400/domino, Reptin, Pontin, MrgBP, Brd8, Gas41, YL1, Ing3, and DMAP1 (44). Recent reports have shown that using genetic studies in flies, the Nipped-A/Tra1/TRRAP proteins and the SRCAP/DOM proteins are required for the activity of Notch through its coactivator protein, mastermind, during wing development and are key component of the Tip60 histone acetylase complex (20, 26, 44). The Notch intracellular domain was not identified among the proteins associated with Nipped-A/Tra1/TRRAP and SRCAP/DOM in the Tip60 complexes (9). However, we found that Tip60 binds to the Notch intracellular domain as a single factor (39). It seems likely then that Tip60 participates in one or more protein complexes, distinct from the currently characterized complexes. Tip60 has recently been determined to be involved in DNA repair and apoptosis in response to DNA damage (12, 33, 69). The HAT activity of Tip60 also appears to be important, as the overexpression of a catalytically dead Tip60 mutant has been shown to impair cellular apoptotic responses to genotoxic stress (33). UV irradiation facilitates the degradation of Mdm2 and thereby prevents the degradation of Tip60 (47). Therefore, the cellular expression of Tip60 is increased in response to UV irradiation, resulting in an accumulation of Tip60 in Jurkat cells (47). Consistent with the results of a previous report, we determined that the exposure of NIH 3T3 cells to UV irradiation induced an accumulation of Tip60 in living cells, thereby suppressing Notch1 transcriptional activity. In our experiments, no endogenous Tip60 was detected in the cell lines, as was the case in other studies (30, 48). The downregulation of endogenous Tip60 expression, using Tip60 siRNA, results in an increase in Notch1 transcriptional activity. Thus, the functional role of endogenous Tip60 levels in Notch1 signaling can be estimated from our result.
In the present study, we observed both in vivo and in vitro binding between the Tip60 and Notch1-IC proteins. Surprisingly, the interaction between the Tip60 and Notch1-IC proteins was more profound in the presence of CSL than in the presence of Notch1-IC alone. However, we did not detect any formation of the Tip60-Notch1-IC-CSL trimeric complex in this case. The manner in which CSL is directly involved in the association between Notch1-IC and Tip60 remains unclear. Furthermore, Tip60 was found to inhibit the interaction between Notch1-IC and CSL in intact cells. On the basis of these results, we propose that endogenous Tip60, which is induced by UV irradiation, may prevent Notch1 transcriptional activity via binding to Notch1-IC, as the result of the dissociation of Notch1-IC and CSL.
The activity of HATs is known to be critical for proper gene expression control (8, 77). Many mechanisms relevant to the regulation of enzyme activity have thus far been described (15, 49, 59). This can occur via posttranslational modifications, as has been shown for Tip60, which is activated by phosphorylation (30). We determined that the phosphorylation-deficient forms of Tip60 mutants profoundly induced Notch1 transcriptional activity. Our data indicate that the HAT activity of Tip60, which is regulated by phosphorylation, plays a critical role in the suppression of Notch1 signaling. Tip60 harbors a chromodomain, a zinc finger motif, and an acetyl-CoA binding domain. The zinc finger motif and acetyl-CoA binding domain, in particular, play pivotal roles in the HAT activity of Tip60. Interestingly, our results appear to show that the zinc finger motif and acetyl-CoA domain of Tip60 are capable of inhibiting Notch1 transcriptional activity via direct binding, whereas the chromodomain, zinc finger, and acetyl-CoA binding domains alone neither bind to Notch1 nor inhibit Notch1 transcriptional activity. Thus, both the zinc finger motif and the acetyl-CoA binding domain are essential for the binding of Tip60 to Notch1 and the Tip60-induced inhibition of Notch1 signaling, which suggests that the suppression of Notch1 signaling by Tip60 is dependent on its well-characterized HAT activity. In the present study, we observed both in vivo and in vitro acetylation of Notch1-IC by Tip60. However, despite the important role of Tip60 in acetylation and the inhibition of Notch1 signaling, the precise relevant mechanisms in this phenomenon remain obscure. From our results, however, we can postulate that Notch1-IC is a direct substrate of Tip60 and acetylation is one of the key factors in the regulation of the Notch1 signaling pathway (Fig. 9).
FIG. 9.
Model for the role of Tip60 in the regulation of Notch transcription activity. At the active gene, Notch1-IC interacts with CSL and with coactivator complexes to promote target gene expression. The HAT Tip60 proteins are accumulated when cells were exposed to genotoxic stress such as UV irradiation. Notch1-IC is a substrate for the HAT activity of Tip60. Acetylation of Notch1-IC by Tip60 may induce and then dislodge it from Notch1-IC-CSL complex.
In summary, we have described the suppression of Notch signaling by UV radiation-induced Tip60, occurring as the consequence of direct physical interaction and acetylation. The dysregulation of Notch1 activity appears to be relevant to the development of tumors, and a host of studies have been targeted toward the development of effective therapeutics that might inhibit the relevant signaling pathways. Tip60 has been linked closely to the regulation of the metastasis suppressor gene in prostate cancer (38). The finding that the HAT activity of Tip60 is crucial to the suppression of Notch1 signaling indicates that loss-of-function Tip60 mutants may contribute to the development of disease states. Further studies, however, will be required in order to better understand the biological implications of Tip60 as a target for the treatment of various diseases.
Acknowledgments
We thank Thomas Südhof (University of Texas Southwestern), S.-H. Baek (Seoul National University), and Saadi Khochbin (INSERM) for providing the Tip60 constructs; Raphael Kopan (Washington University School of Medicine) for the Notch-related constructs; and K.-W. Kim (Seoul National University) for the anti-acetyl-Lys antibody.
This research was supported by a Korea Research Foundation grant (KRF-2004-C00141) funded by the Korean government (MOEHRD, Basic Research Promotion Fund) (to H.-S.P.).
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Abu-Issa, R., and S. Cavicchi. 1996. Genetic interactions among vestigial, hairy, and Notch suggest a role of vestigial in the differentiation of epidermal and neural cells of the wing and halter of Drosophila melanogaster. J. Neurogenet. 10:239-246. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas, S., and P. Simpson. 1991. Choosing a cell fate: a view from the Notch locus. Trends Genet. 7:403-408. [DOI] [PubMed] [Google Scholar]
- 4.Baek, S. H., K. A. Ohgi, D. W. Rose, E. H. Koo, C. K. Glass, and M. G. Rosenfeld. 2002. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell 110:55-67. [DOI] [PubMed] [Google Scholar]
- 5.Baron, M., H. Aslam, M. Flasza, M. Fostier, J. E. Higgs, S. L. Mazaleyrat, and M. B. Wilkin. 2002. Multiple levels of Notch signal regulation (review). Mol. Membr. Biol. 19:27-38. [DOI] [PubMed] [Google Scholar]
- 6.Bash, J., W. X. Zong, S. Banga, A. Rivera, D. W. Ballard, Y. Ron, and C. Gelinas. 1999. Rel/NF-kappaB can trigger the Notch signaling pathway by inducing the expression of Jagged1, a ligand for Notch receptors. EMBO J. 18:2803-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brady, M. E., D. M. Ozanne, L. Gaughan, I. Waite, S. Cook, D. E. Neal, and C. N. Robson. 1999. Tip60 is a nuclear hormone receptor coactivator. J. Biol. Chem. 274:17599-17604. [DOI] [PubMed] [Google Scholar]
- 8.Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 9.Cai, Y., J. Jin, L. Florens, S. K. Swanson, T. Kusch, B. Li, J. L. Workman, M. P. Washburn, R. C. Conaway, and J. W. Conaway. 2005. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 280:13665-13670. [DOI] [PubMed] [Google Scholar]
- 10.Cao, X., and T. C. Sudhof. 2001. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293:115-120. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Col, E., C. Caron, C. Chable-Bessia, G. Legube, S. Gazzeri, Y. Komatsu, M. Yoshida, M. Benkirane, D. Trouche, and S. Khochbin. 2005. HIV-1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. EMBO J. 24:2634-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creaven, M., F. Hans, V. Mutskov, E. Col, C. Caron, S. Dimitrov, and S. Khochbin. 1999. Control of the histone-acetyltransferase activity of Tip60 by the HIV-1 transactivator protein, Tat. Biochemistry 38:8826-8830. [DOI] [PubMed] [Google Scholar]
- 14.Cunliffe, V. T. 2004. Histone deacetylase 1 is required to repress Notch target gene expression during zebrafish neurogenesis and to maintain the production of motoneurones in response to hedgehog signalling. Development 131:2983-2995. [DOI] [PubMed] [Google Scholar]
- 15.Davie, J. R., and V. A. Spencer. 1999. Control of histone modifications. J. Cell Biochem. Suppl. 32-33:141-148. [DOI] [PubMed] [Google Scholar]
- 16.de Celis, J. F., J. de Celis, P. Ligoxygakis, A. Preiss, C. Delidakis, and S. Bray. 1996. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development 122:2719-2728. [DOI] [PubMed] [Google Scholar]
- 17.Dechend, R., F. Hirano, K. Lehmann, V. Heissmeyer, S. Ansieau, F. G. Wulczyn, C. Scheidereit, and A. Leutz. 1999. The Bcl-3 oncoprotein acts as a bridging factor between NF-kappaB/Rel and nuclear co-regulators. Oncogene 18:3316-3323. [DOI] [PubMed] [Google Scholar]
- 18.Doyon, Y., W. Selleck, W. S. Lane, S. Tan, and J. Cote. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24:1884-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duffy, J. B., and N. Perrimon. 1996. Recent advances in understanding signal transduction pathways in worms and flies. Curr. Opin. Cell Biol. 8:231-238. [DOI] [PubMed] [Google Scholar]
- 20.Eissenberg, J. C., M. Wong, and J. C. Chrivia. 2005. Human SRCAP and Drosophila melanogaster DOM are homologs that function in the Notch signaling pathway. Mol. Cell. Biol. 25:6559-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espinosa, L., J. Ingles-Esteve, C. Aguilera, and A. Bigas. 2003. Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J. Biol. Chem. 278:32227-32235. [DOI] [PubMed] [Google Scholar]
- 22.Foltz, D. R., M. C. Santiago, B. E. Berechid, and J. S. Nye. 2002. Glycogen synthase kinase-3beta modulates notch signaling and stability. Curr. Biol. 12:1006-1011. [DOI] [PubMed] [Google Scholar]
- 23.Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaughan, L., M. E. Brady, S. Cook, D. E. Neal, and C. N. Robson. 2001. Tip60 is a co-activator specific for class I nuclear hormone receptors. J. Biol. Chem. 276:46841-46848. [DOI] [PubMed] [Google Scholar]
- 25.Gaughan, L., I. R. Logan, S. Cook, D. E. Neal, and C. N. Robson. 2002. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J. Biol. Chem. 277:25904-25913. [DOI] [PubMed] [Google Scholar]
- 26.Gause, M., J. C. Eissenberg, A. F. Macrae, M. Dorsett, Z. Misulovin, and D. Dorsett. 2006. Nipped-A, the Tra1/TRRAP subunit of the Drosophila SAGA and Tip60 complexes, has multiple roles in Notch signaling during wing development. Mol. Cell. Biol. 26:2347-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gavaravarapu, S., and J. Kamine. 2000. Tip60 inhibits activation of CREB protein by protein kinase A. Biochem. Biophys. Res. Commun. 269:758-766. [DOI] [PubMed] [Google Scholar]
- 28.Goto, S., M. Taniguchi, M. Muraoka, H. Toyoda, Y. Sado, M. Kawakita, and S. Hayashi. 2001. UDP-sugar transporter implicated in glycosylation and processing of Notch. Nat. Cell Biol. 3:816-822. [DOI] [PubMed] [Google Scholar]
- 29.Haines, N., and K. D. Irvine. 2003. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell Biol. 4:786-797. [DOI] [PubMed] [Google Scholar]
- 30.Hass, M. R., and B. A. Yankner. 2005. A {gamma}-secretase-independent mechanism of signal transduction by the amyloid precursor protein. J. Biol. Chem. 280:36895-36904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirsinger, E., P. Malapert, J. Dubrulle, M. C. Delfini, D. Duprez, D. Henrique, D. Ish-Horowicz, and O. Pourquie. 2001. Notch signalling acts in postmitotic avian myogenic cells to control MyoD activation. Development 128:107-116. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 34.Jennings, B., A. Preiss, C. Delidakis, and S. Bray. 1994. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development 120:3537-3548. [DOI] [PubMed] [Google Scholar]
- 35.Jouve, C., I. Palmeirim, D. Henrique, J. Beckers, A. Gossler, D. Ish-Horowicz, and O. Pourquie. 2000. Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development 127:1421-1429. [DOI] [PubMed] [Google Scholar]
- 36.Kao, H. Y., P. Ordentlich, N. Koyano-Nakagawa, Z. Tang, M. Downes, C. R. Kintner, R. M. Evans, and T. Kadesch. 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 12:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlstrom, H., P. Beatus, K. Dannaeus, G. Chapman, U. Lendahl, and J. Lundkvist. 2002. A CADASIL-mutated Notch 3 receptor exhibits impaired intracellular trafficking and maturation but normal ligand-induced signaling. Proc. Natl. Acad. Sci. USA 99:17119-17124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, J. H., B. Kim, L. Cai, H. J. Choi, K. A. Ohgi, C. Tran, C. Chen, C. H. Chung, O. Huber, D. W. Rose, C. L. Sawyers, M. G. Rosenfeld, and S. H. Baek. 2005. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature 434:921-926. [DOI] [PubMed] [Google Scholar]
- 39.Kim, S. Y., M. Y. Kim, J. S. Mo, and H. S. Park. 2007. Notch1 intracellular domain suppresses APP intracellular domain-Tip60-Fe65 complex mediated signaling through physical interaction. Biochim. Biophys. Acta 1773:736-746. [DOI] [PubMed] [Google Scholar]
- 40.Kimble, J., and P. Simpson. 1997. The LIN-12/Notch signaling pathway and its regulation. Annu. Rev. Cell Dev. Biol. 13:333-361. [DOI] [PubMed] [Google Scholar]
- 41.Kopan, R., and R. Cagan. 1997. Notch on the cutting edge. Trends Genet. 13:465-467. [DOI] [PubMed] [Google Scholar]
- 42.Kopan, R., J. S. Nye, and H. Weintraub. 1994. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development 120:2385-2396. [DOI] [PubMed] [Google Scholar]
- 43.Kuroda, K., S. Tani, K. Tamura, S. Minoguchi, H. Kurooka, and T. Honjo. 1999. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J. Biol. Chem. 274:7238-7244. [DOI] [PubMed] [Google Scholar]
- 44.Kusch, T., L. Florens, W. H. Macdonald, S. K. Swanson, R. L. Glaser, J. R. Yates III, S. M. Abmayr, M. P. Washburn, and J. L. Workman. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306:2084-2087. [DOI] [PubMed] [Google Scholar]
- 45.Lai, E. C. 2002. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 3:840-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai, E. C. 2004. Notch signaling: control of cell communication and cell fate. Development 131:965-973. [DOI] [PubMed] [Google Scholar]
- 47.Legube, G., L. K. Linares, C. Lemercier, M. Scheffner, S. Khochbin, and D. Trouche. 2002. Tip60 is targeted to proteasome-mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J. 21:1704-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Legube, G., L. K. Linares, S. Tyteca, C. Caron, M. Scheffner, M. Chevillard-Briet, and D. Trouche. 2004. Role of the histone acetyl transferase Tip60 in the p53 pathway. J. Biol. Chem. 279:44825-44833. [DOI] [PubMed] [Google Scholar]
- 49.Legube, G., and D. Trouche. 2003. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 4:944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ligoxygakis, P., S. Y. Yu, C. Delidakis, and N. E. Baker. 1998. A subset of notch functions during Drosophila eye development require Su(H) and the E(spl) gene complex. Development 125:2893-2900. [DOI] [PubMed] [Google Scholar]
- 51.Mitsiadis, T. A., K. Fried, and C. Goridis. 1999. Reactivation of Delta-Notch signaling after injury: complementary expression patterns of ligand and receptor in dental pulp. Exp. Cell Res. 246:312-318. [DOI] [PubMed] [Google Scholar]
- 52.Mumm, J. S., and R. Kopan. 2000. Notch signaling: from the outside in. Dev. Biol. 228:151-165. [DOI] [PubMed] [Google Scholar]
- 53.Nervi, C., U. Borello, F. Fazi, V. Buffa, P. G. Pelicci, and G. Cossu. 2001. Inhibition of histone deacetylase activity by trichostatin A modulates gene expression during mouse embryogenesis without apparent toxicity. Cancer Res. 61:1247-1249. [PubMed] [Google Scholar]
- 54.Neumann, C. J. 2001. Pattern formation in the zebrafish retina. Semin. Cell Dev. Biol. 12:485-490. [DOI] [PubMed] [Google Scholar]
- 55.Nijjar, S. S., L. Wallace, H. A. Crosby, S. G. Hubscher, and A. J. Strain. 2002. Altered Notch ligand expression in human liver disease: further evidence for a role of the Notch signaling pathway in hepatic neovascularization and biliary ductular defects. Am. J. Pathol. 160:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nordentoft, I., and P. Jorgensen. 2003. The acetyltransferase 60 kDa trans-acting regulatory protein of HIV type 1-interacting protein (Tip60) interacts with the translocation E26 transforming-specific leukaemia gene (TEL) and functions as a transcriptional co-repressor. Biochem. J. 374:165-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nye, J. S., and R. Kopan. 1995. Developmental signaling. Vertebrate ligands for Notch. Curr. Biol. 5:966-969. [DOI] [PubMed] [Google Scholar]
- 58.Oberg, C., J. Li, A. Pauley, E. Wolf, M. Gurney, and U. Lendahl. 2001. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 276:35847-35853. [DOI] [PubMed] [Google Scholar]
- 59.Ogryzko, V. V. 2001. Mammalian histone acetyltransferases and their complexes. Cell. Mol. Life Sci. 58:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohtsuka, T., M. Ishibashi, G. Gradwohl, S. Nakanishi, F. Guillemot, and R. Kageyama. 1999. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 18:2196-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park, H. S., J. S. Lee, S. H. Huh, J. S. Seo, and E. J. Choi. 2001. Hsp72 functions as a natural inhibitory protein of c-Jun N-terminal kinase. EMBO J. 20:446-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfister, S., G. K. Przemeck, J. K. Gerber, J. Beckers, J. Adamski, and M. Hrabe de Angelis. 2003. Interaction of the MAGUK family member Acvrinp1 and the cytoplasmic domain of the Notch ligand Delta1. J. Mol. Biol. 333:229-235. [DOI] [PubMed] [Google Scholar]
- 63.Qiu, L., C. Joazeiro, N. Fang, H. Y. Wang, C. Elly, Y. Altman, D. Fang, T. Hunter, and Y. C. Liu. 2000. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 275:35734-35737. [DOI] [PubMed] [Google Scholar]
- 64.Robey, E. 1999. Regulation of T cell fate by Notch. Annu. Rev. Immunol. 17:283-295. [DOI] [PubMed] [Google Scholar]
- 65.Sapountzi, V., I. R. Logan, and C. N. Robson. 2006. Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 38:1496-1509. [DOI] [PubMed] [Google Scholar]
- 66.Shaye, D. D., and I. Greenwald. 2005. LIN-12/Notch trafficking and regulation of DSL ligand activity during vulval induction in Caenorhabditis elegans. Development 132:5081-5092. [DOI] [PubMed] [Google Scholar]
- 67.Tamura, K., Y. Taniguchi, S. Minoguchi, T. Sakai, T. Tun, T. Furukawa, and T. Honjo. 1995. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr. Biol. 5:1416-1423. [DOI] [PubMed] [Google Scholar]
- 68.Taniguchi, Y., M. Sato, O. Tanaka, M. Sekiguchi, H. Inoko, and M. Kimura. 2001. HOXD3 regulates expression of JAGGED1, a ligand for Notch receptors. Nucleic Acids Res. Suppl. 1:43-44. [DOI] [PubMed] [Google Scholar]
- 69.Tyteca, S., M. Vandromme, G. Legube, M. Chevillard-Briet, and D. Trouche. 2006. Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression. EMBO J. 25:1680-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, M., and P. W. Sternberg. 2001. Pattern formation during C. elegans vulval induction. Curr. Top. Dev. Biol. 51:189-220. [DOI] [PubMed] [Google Scholar]
- 71.Weber, J. M., S. R. Forsythe, C. A. Christianson, B. J. Frisch, B. J. Gigliotti, C. T. Jordan, L. A. Milner, M. L. Guzman, and L. M. Calvi. 2006. Parathyroid hormone stimulates expression of the Notch ligand Jagged1 in osteoblastic cells. Bone 39:485-493. [DOI] [PubMed] [Google Scholar]
- 72.Weinmaster, G. 1997. The ins and outs of notch signaling. Mol. Cell. Neurosci. 9:91-102. [DOI] [PubMed] [Google Scholar]
- 73.Weinmaster, G. 2000. Notch signal transduction: a real rip and more. Curr. Opin. Genet. Dev. 10:363-369. [DOI] [PubMed] [Google Scholar]
- 74.Weinmaster, G. 1998. Notch signaling: direct or what? Curr. Opin. Genet. Dev. 8:436-442. [DOI] [PubMed] [Google Scholar]
- 75.Whitford, K. L., P. Dijkhuizen, F. Polleux, and A. Ghosh. 2002. Molecular control of cortical dendrite development. Annu. Rev. Neurosci. 25:127-149. [DOI] [PubMed] [Google Scholar]
- 76.Wittenberger, T., O. C. Steinbach, A. Authaler, R. Kopan, and R. A. Rupp. 1999. MyoD stimulates delta-1 transcription and triggers notch signaling in the Xenopus gastrula. EMBO J. 18:1915-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu, C. 1997. Chromatin remodeling and the control of gene expression. J. Biol. Chem. 272:28171-28174. [DOI] [PubMed] [Google Scholar]
- 78.Wu, G., S. Lyapina, I. Das, J. Li, M. Gurney, A. Pauley, I. Chui, R. J. Deshaies, and J. Kitajewski. 2001. SEL-10 is an inhibitor of Notch signaling that targets Notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 21:7403-7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiao, H., J. Chung, H. Y. Kao, and Y. C. Yang. 2003. Tip60 is a co-repressor for STAT3. J. Biol. Chem. 278:11197-11204. [DOI] [PubMed] [Google Scholar]
- 80.Yamaguchi, M., N. Tonou-Fujimori, A. Komori, R. Maeda, Y. Nojima, H. Li, H. Okamoto, and I. Masai. 2005. Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development 132:3027-3043. [DOI] [PubMed] [Google Scholar]
- 81.Yamamoto, T., and M. Horikoshi. 1997. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J. Biol. Chem. 272:30595-30598. [DOI] [PubMed] [Google Scholar]