Abstract
CHD1 encodes an ATP-dependent chromatin remodeler with two chromodomains. Deletion of CHD1 suppresses the temperature-sensitive growth defect caused by mutations in either SPT16 or POB3, which encode subunits of the yFACT chromatin-reorganizing complex. chd1 also suppresses synthetic defects caused by combining an spt16 mutation with other transcription factor mutations, including the synthetic lethality caused by combining an spt16 mutation with TATA binding protein (TBP) or TFIIA defects. Binding of TBP and RNA polymerase II to the GAL1 promoter is reduced in a pob3 mutant, resulting in low levels of GAL1 expression, and all three defects are suppressed by removing Chd1. These results suggest that Chd1 and yFACT have opposing roles in regulating TBP binding at promoters. Additionally, overexpression of Chd1 is tolerated in wild-type cells but is toxic in spt16 mutants. Further, both the ATPase and chromodomain are required for Chd1 activity in opposing yFACT function. Similar to the suppression by chd1, mutations in the SET2 histone methyltransferase also suppress defects caused by yFACT mutations. chd1 and set2 are additive in suppressing pob3, suggesting that Chd1 and Set2 act in distinct pathways. Although human Chd1 has been shown to bind to H3-K4-Me, we discuss evidence arguing that yeast Chd1 binds to neither H3-K4-Me nor H3-K36-Me.
Chromatin structure limits the accessibility of DNA sequences in eukaryotic chromosomes. Accessibility is enhanced through three major processes in vivo. First, posttranslational histone modifications either change the properties of the chromatin structure or create binding sites for other transcription factors (50). These posttranslational modifications include phosphorylation of serine residues and acetylation, methylation, and ubiquitylation of lysine residues (11). Several transcription factors that recognize specific histone modifications for their recruitment have been described. For example, the bromodomain-containing proteins recognize acetylation of histone proteins (61), and chromodomain-containing proteins are involved in recognizing methylation marks on histone proteins (6, 11). Second, ATP-dependent chromatin-remodeling factors promote accessibility by repositioning nucleosomes (9, 21, 55). These factors utilize the energy from ATP hydrolysis to establish or disrupt repressive chromatin structures (21). The third way by which the DNA sequence is made available is through ATP-independent chromatin-reorganizing factors that change the properties of nucleosomes in a localized manner (17). For example, the yFACT complex changes the properties of nucleosomes without requiring ATP hydrolysis (39, 41). The reorganization by yFACT has been shown to alter DNA accessibility both in vivo and in vitro (5, 14, 17, 30, 44, 45).
The FACT complex (facilitates chromatin transcription) was first identified as a factor that promoted RNA polymerase II (Pol II) transcription in vitro using assembled chromatin as a template (33). The mammalian FACT complex is composed of two subunits, p140 and SSRP1. The homologs of p140 and SSRP1 in yeast are Spt16 and Pob3, respectively (34). The Spt16 and Pob3 proteins are always present in a heterodimer to form the SP complex in yeast (57). Although the N-terminal DNA binding domain of SSRP1 is absent in Pob3, Nhp6, a high-mobility group protein, is thought to serve as the DNA binding activity of the SP complex to form the yFACT complex (7, 18). Genetic and biochemical evidence suggests that yFACT is involved in regulating both transcription and DNA replication (1a, 3, 5, 18, 19, 27, 30, 34, 42, 56, 57). While the association of yFACT with elongation factors (27, 45) and with transcribed regions of genes (30, 42) supports an elongation role, studies also suggest that the FACT complex has a role in transcription initiation (5, 44). We have shown earlier that yFACT has a role in regulating TATA binding protein (TBP) binding during the transcriptional initiation step (5). The evidence for this included synthetic lethality between certain mutations of TBP and TFIIA and defective alleles of SPT16, reduced binding of TBP at some promoters in spt16 mutants, and enhanced binding of TBP to a TATA box within nucleosomal DNA in presence of TFIIA and yFACT.
The yeast chromodomain protein (Chd1) is a member of the Snf2-like subfamily of nucleic acid-stimulated ATPases (21) and has ATP-dependent chromatin-remodeling activity in vitro (29, 40). Chd1 and other CHD proteins have two chromodomains near the N terminus, a centrally located Snf2-related helicase/ATPase domain, and a Myb-related DNA binding domain near the C terminus (59). Chd1 is thought to promote formation of inhibitory chromatin, as extracts derived from cells lacking Chd1 are unable to produce the same level of DNase I resistance at specific loci that results from similar preparations derived from normal cells (40). Genetic interactions have been reported between mutations of CHD1 and mutations in transcription elongation factors such as Spt5, Isw1, and Isw2 (45, 54). Chd1 also physically interacts with several transcription elongation factors, such as members of the Paf1 complex, the Spt4-Spt5 complex, and components of yFACT (24, 27, 45, 54). Recently Chd1 has been shown to physically associate with the SAGA/SLIK complex in yeast and to bind histone tail peptides methylated at K4 (37). However, binding of yeast Chd1 to methylated H3-K4 has not been observed by others (32, 46). Relatively little is known about the functional role of Chd1 in vivo in regulating transcription, although it was recently reported that a chd1 mutation affects chromatin structure of the ADH2 gene and the kinetics of ADH2 activation (60). In this report, we show that part of the role of Chd1 is to oppose the function of the positive transcription factor yFACT. We present evidence which suggests that Chd1 negates yFACT's ability to enhance TBP binding at promoters. We show that in a strain with a yFACT defect, deletion of CHD1 results in increased TBP binding and increased Pol II binding at promoters. Finally, we find that deletion of CHD1 suppresses synthetic lethalities between spt16 mutations and TBP mutations as well as between spt16-11 and TFIIA mutations.
MATERIALS AND METHODS
The yeast strains used are isogenic with W303 (52) and are listed in Table S1 in the supplemental material. Standard genetic methods were used for strain construction (43). Cells were grown in YPD medium (43) at 30°C, except as noted, or in synthetic complete medium (43) with 2% glucose and supplemented with adenine, uracil, and amino acids, as appropriate. For the galactose induction experiments, cells were grown at 25°C in YP medium supplemented with 2% raffinose to mid-log phase and shifted to 30°C for 2 h, and then galactose was added to a final concentration of 2%. Plasmids are listed in Table S2 in the supplemental material. RNA levels were determined with S1 nuclease protection assays as described previously (2, 3).
Chromatin immunoprecipitations (ChIPs) were performed as described previously (1), using the 8WG16 monoclonal antibody against the Pol II C-terminal repeat and a polyclonal anti-TBP serum generously provided by Tony Weil (42a). Real-time PCR was performed as described previously (14a), using the open reading frame (ORF)-free chromosome I region (30a) as a nontranscribed region control.
For all ChIP experiment the value for each ChIP output PCR signal was divided by that for the ChIP output PCR signal for the ORF-free control, and to control for primer pair efficiencies, this ratio was further divided by a similar ratio of target to nontranscribed regions but using input DNA PCR signals, resulting in a ChIP ratio. Each PCR was performed in triplicate, and the normalized mean and standard deviation of the ratio were calculated as described previously (14a).
RESULTS
Deletion of CHD1 suppresses phenotypes of yFACT mutant strains.
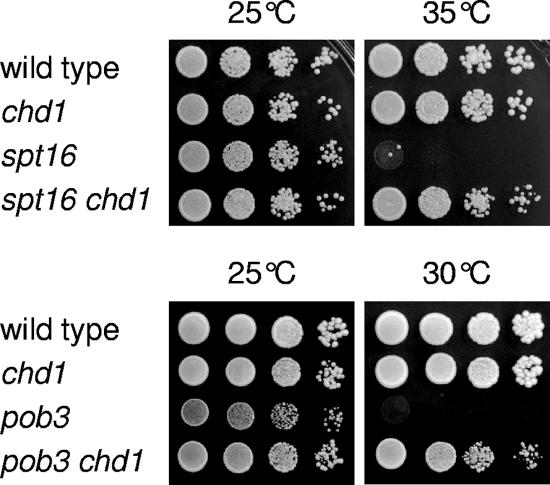
It was previously reported that a chd1 mutation can suppress the growth defects of a pob3-272 mutant (10). We asked whether a chd1 deletion could suppress the temperature-sensitive growth defects of spt16-11 and pob3(L78R) mutations in our strain background. Figure 1 shows that spt16 chd1 and pob3 chd1 strains grow well under conditions where the spt16 and pob3 single mutants are inviable. Thus, the chd1 suppression of yFACT mutant phenotypes suggests that yFACT and Chd1 have opposing roles in regulating transcription.
FIG. 1.
chd1 suppresses spt16 and pob3 phenotypes. (Top panels) Tenfold dilutions of strains DY150 (wild type), DY6957 (chd1), DY8107 (spt16), and DY9151 (spt16 chd1) were plated on complete medium at the indicted temperature for 2 days. (Bottom panels) Tenfold dilutions of strains DY150 (wild type), DY6957 (chd1), DY7379 (pob3), and DY9458 (pob3 chd1) were plated on complete medium at the indicted temperature for 3 days.
The yFACT complex, in addition to Spt16 and Pob3, contains the Nhp6 HMGB (18). Nhp6 is encoded by two redundant genes, NHP6A and NHP6B, and the nhp6ab double mutant strain is temperature sensitive for growth. Based on our observation that a chd1 mutation suppresses the temperature-sensitive growth phenotype of the spt16 and pob3 strains, we asked whether chd1 could also suppress temperature sensitivity of the nhp6ab strain. We constructed a nhp6ab chd1 triple mutant strain, but this strain failed to grow at the restrictive temperature (data not shown). Nhp6 has a role in transcription by Pol III (23) and interacts with other chromatin proteins besides yFACT, including Swi/Snf, RSC, and Ssn6/Tup1 (4, 20, 51). The ability of a chd1 mutation to suppress spt16 and pob3 but not nhp6ab may reflect these additional roles of Nhp6.
Mutations in the ATPase domain and in the chromodomain of Chd1 suppress yFACT mutations.
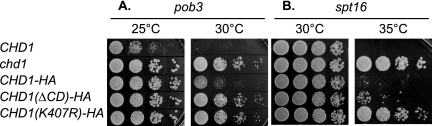
The Chd1 protein has ATPase activity (53), and it also contains two chromodomain sequence motifs (59). We investigated the roles of the ATPase and chromodomains of Chd1 in the genetic suppression of spt16 and pob3 mutations. Strains were constructed with an integrated CHD1(ΔCD)-HA allele, lacking both Chd1 chromodomains, and an integrated CHD1(K407R)-HA allele, with a mutation within the consensus ATP binding motif (45). The control strains had wild-type CHD1-HA integrated and also hemagglutinin (HA) tagged at the 3′ end. In this assay, the strain will not grow if a mutant complements the gene deletion, indicating restoration of the normal ability to oppose yFACT function. Similarly, growth indicates a failure to provide Chd1 function. The pob3 chd1 strain grows at 30°C, while the pob3 CHD1 strain is inviable. The ΔCD and K407R mutant versions of Chd1 allow growth of the pob3 mutant, and thus both the chromodomain and the ATPase are required for the Chd1 activity that, here, is toxic in pob3 mutants (Fig. 2A). With spt16, again both the chromodomain and ATPase mutations in Chd1 allow growth (Fig. 2B), although growth in the spt16 CHD1(ΔCD)-HA strain is less robust, suggesting that the chromodomain plays a less prominent role than the ATPase in generating toxicity in the spt16 strain. Finally, comparing growth of the pob3 CHD1 and the pob3 CHD1-HA strains suggests that the HA-tagged allele is not fully functional. In summary, these experiments suggest that both the ATPase activity and the chromodomain of Chd1 are required for Chd1 to be toxic in yFACT mutants.
FIG. 2.
Both the chromodomain and the ATPase contribute to Chd1 toxicity in yFACT mutants. (A) Tenfold dilutions of strains DY7379 (pob3), DY9458 (pob3 chd1), DY11724 (pob3 chd1 CHD1-HA), DY11736 [pob3 chd1 CHD1(ΔCD)-HA], and DY11770 [pob3 chd1 CHD1(K407R)-HA] were plated on complete medium at 25°C or 30°C for 2 days. (B) Tenfold dilutions of strains DY8107 (spt16), DY9152 (spt16 chd1), DY11614 (spt16 chd1 CHD1-HA), DY11624 (spt16 chd1 CHD1(ΔCD)-HA], and DY11643 [spt16 chd1 CHD1(K407R)-HA] were plated on complete medium at 30°C or 35°C for 2 days.
Deletion of CHD1 suppresses synthetic lethality between spt16 and other transcription factors.
The Isw1 and Isw2 chromatin complexes have been implicated in both transcriptional elongation and repressing transcriptional initiation (31). Additionally, the isw1 isw2 chd1 triple mutant shows additive growth defects at elevated temperatures (54). Based on these results, we looked for genetic interactions between spt16, chd1, isw1, and isw2. The spt16 isw1 double mutant shows a significant growth defect at 33°C (Fig. 3A), and the spt16 isw1 isw2 triple mutant is completely dead at 33°C (Fig. 3B). These synthetic growth defects suggest that the yFACT chromatin-reorganizing complex and the Isw remodeling complexes may perform similar functions in vivo. Importantly, a chd1 mutation suppresses both the spt16 isw1 and the spt16 isw1 isw2 growth defects, supporting the idea that Chd1 acts in opposition to yFACT for the function that overlaps Isw-mediated remodeling.
FIG. 3.
chd1 suppresses spt16 synthetic growth defects. (A) Tenfold dilutions of strains DY150 (wild type), DY8107 (spt16), DY9816 (isw1), DY9809 (chd1), DY9827 (chd1 isw1), DY9055 (spt16 isw1), and DY9834 (spt16 isw1 chd1) were plated on complete medium at 33°C for 2 days. (B) Tenfold dilutions of strains DY150 (wild type), DY8107 (spt16), DY9809 (chd1), DY9152 (spt16 chd1), DY7656 (isw1 isw2), DY9823 (chd1 isw1 isw2), DY9820 (spt16 isw1 isw2), and DY9831 (spt16 isw1 isw2 chd1) were plated on complete medium at 33°C for 2 days. (C) Tenfold dilutions of strains DY150 (wild type), DY6612 (nhp6ab), DY8107 (spt16), DY8808 (spt16 nhp6ab), and DY9978 (spt16 nhp6ab chd1) were plated on complete medium at 33°C for 2 days. (D) Tenfold dilutions of strains DY150 (wild type), DY8156 (elp3), DY8107 (spt16), DY8185 (spt16 elp3), and DY9965 (spt16 elp3 chd1) were plated on complete medium at 33°C for 2 days. (E) Tenfold dilutions of strains DY150 (wild type), DY7836 (htz1), DY8107 (spt16), DY9808 (spt16 htz1), and DY9811 (spt16 htz1 chd1) were plated on complete medium at 33°C for 2 days.
As a chd1 mutation suppresses a number of spt16 phenotypes, we asked whether chd1 can also suppress other synthetic lethal phenotypes seen with spt16. spt16 shows marked growth defects when combined with mutations in both nhp6a and nhp6b (18), and an spt16 nhp6ab double mutation is lethal at 33°C. A chd1 mutation suppresses this synthetic lethality, as seen by growth of the spt16 nhp6ab chd1 strain (Fig. 3C). ELP3 encodes a histone acetyltransferase subunit of the elongator complex (58), and elp3 is synthetic lethal with spt16 (19). The spt16 elp3 synthetic lethality is suppressed by a chd1 mutation (Fig. 3D). HTZ1 encodes the yeast H2A.Z histone variant of H2A (12), and we recently showed that htz1 and spt16 are synthetic lethal (3). A chd1 mutation also suppresses the spt16 htz1 synthetic lethality (Fig. 3E). Htz1 is believed to function at promoter regions, as it localizes preferentially at promoter regions of genes (28, 38, 62), and this suppression suggests that Chd1 might influence promoter function.
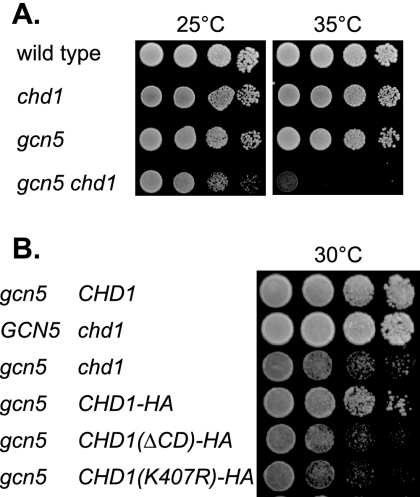
We recently showed that a set2 mutation can suppress many spt16 phenotypes (3). Like chd1, set2 suppresses the spt16 np6ab, spt16 elp3, and spt16 htz1 synthetic lethalities. There are also differences in the suppression profiles, however. While chd1 suppresses the spt16 isw1 isw2 lethality, a set2 mutation does not (data not shown). Conversely, set2 suppresses synthetic lethality of the spt16 gcn5 double mutant (3), but a chd1 mutation does not (data not shown). In fact, a gcn5 chd1 double mutant shows a growth defect at 25°C and is synthetic lethal at 35°C (Fig. 4A). The CHD1(ΔCD)-HA and CHD1(K407R)-HA alleles both also show strong growth defects when combined with the gcn5 disruption (Fig. 4B). This suggests that both the chromodomain and the ATPase activity are required for the Chd1 activity that is needed when Gcn5 is not active. It was reported that Chd1 is present in the SAGA/SLIK coactivator complexes (37), and the synthetic effects of combining gcn5 and chd1 mutations could reflect distinct functions of these two proteins in the same protein complex.
FIG. 4.
chd1 and gcn5 show a synthetic growth defect. (A) Tenfold dilutions of strains DY150 (wild type), DY9809 (chd1), DY5926 (gcn5), and DY9873 (gcn5 chd1) were plated on complete medium at 25°C for 3 days or at 35°C for 2 days. (B) Tenfold dilutions of strains DY5925 (gcn5), DY6957 (chd1), DY11500 (gcn5 chd1), DY11716 (gcn5 chd1 CHD1-HA), DY11694 [gcn5 chd1 CHD1(ΔCD)-HA], and DY11703 [gcn5 chd1 CHD1(K407R)-HA] were plated on complete medium at 30°C for 2 days.
A chd1 mutation suppresses the synthetic lethality of spt16 with set1 or histone H3(K4R) mutations.
Human Chd1 binds to methylated K4 of histone H3 (46). If Chd1 function in yeast requires interaction with methylated H3-K4-Me, then either replacement of this residue or a set1 gene disruption that eliminates the methyltransferase that modifies K4 of histone H3 (8) should lead to suppression of yFACT defects. Instead, combining spt16 and set1 mutations leads to a synthetic defect, lethality at 33°C (3). Thus, the simple idea of the absence of Chd1 binding via methylated K4 of histone H3 is not sufficient to explain the suppression caused by loss of Chd1 (see Discussion).
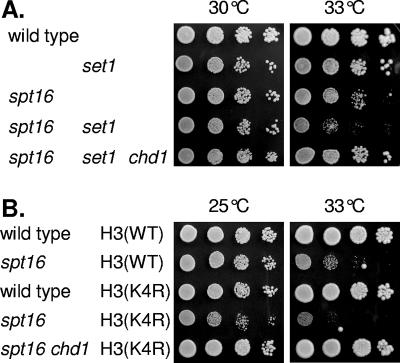
As a chd1 mutation suppresses many spt16 phenotypes, including some synthetic lethal interactions, we constructed an spt16 set1 chd1 triple mutant strain. As shown in Fig. 5A, chd1 suppresses the spt16 set1 growth defect at 33°C, similar to the suppression of spt16 set1 by set2 (3). Similar to set1, a K4R substitution in histone H3 is synthetic lethal with spt16 at 33°C, and this is also suppressed by a chd1 mutation (Fig. 5B). Similar effects can be seen with pob3 mutants, where pob3 set1 and pob3 H3(K4R) mutations are lethal but can be suppressed by chd1 (data not shown). The fact that similar genetic effects are seen with either a set1 or a histone H3(K4R) mutation is consistent with lysine 4 of H3 being the critical target for the Set1 enzyme. These data are also consistent with a recent report showing suppression of the set1 growth defect by chd1 (63). Importantly, we find that the effects of Set1 and H3(K4R) mutations in a yFACT mutant are different from those in a chd1 mutant. This suggests that the mechanism by which Chd1 opposes yFACT does not involve methylation of H3-K4.
FIG. 5.
A chd1 mutation suppresses the synthetic growth defect of spt16 with set1 or histone H3(K4R) mutations. (A) A chd1 mutation suppresses the spt16 set1 synthetic growth defect. Tenfold dilutions of strains DY150 (wild type), DY8875 (set1), DY8107 (spt16), DY9206 (spt16 set1), and DY9271 (spt16 set1 chd1) were plated on complete medium at 30°C or 33°C for 2 days. (B) A chd1 mutation suppresses the spt16 histone H3(K4R) synthetic growth defect. Tenfold dilutions of strains DY8862 [hht1-hhf1 hht2-hhf2 + YCp-TRP1:H3(wild type)-H4(wild type)], DY8865 (spt16 hht1-hhf1 hht2-hhf2 + YCp-TRP1:H3(wild type)-H4(wild type)], DY8863 [hht1-hhf1 hht2-hhf2 + YCp-TRP1:H3(K4R)-H4(wild type)], DY8866 [spt16 hht1-hhf1 hht2-hhf2 + YCp-TRP1:H3(K4R)-H4(wild type)], and DY10472 [spt16 chd1 hht1-hhf1 hht2-hhf2 + YCp-TRP1:H3(K4R)-H4(wild type)] were plated on complete medium at the indicated temperature for 2 days.
CHD1 overexpression is toxic in yFACT mutant strains.
Our experiments suggest that yFACT and Chd1 act in opposition during regulation of transcription. Thus, the activity of Chd1 is toxic in cells that have a partially defective yFACT chromatin-reorganizing factor, and a chd1 mutation relieves this toxicity. This model predicts that Chd1 overexpression could be toxic in strains with yFACT mutations. We transformed wild-type and spt16 mutant strains with a multicopy CHD1 plasmid and assessed growth on selective medium (Fig. 6). CHD1 overexpression has no effect in the wild-type stain but is very toxic in the spt16 strain. Interestingly, a set2 mutation partially reverses the toxicity of CHD1 overexpression in the spt16 mutant. There is no phenotypic consequence of CHD1 overexpression in set1 or set2 single mutant strains (data not shown), and thus the effect appears to be specific to yFACT mutant strains. We conclude that the amount of Chd1 is of critical importance in strains with a defect in the yFACT complex.
FIG. 6.
CHD1 overexpression is toxic in an spt16 mutant. Strains DY150 (wild type), DY8117 (spt16), and DY8799 (spt16 set2) were transformed with either YEp-CHD1 or the empty YEp-URA3 vector and plated on complete medium at 25°C for 3 days or on medium lacking uracil at 30°C for 2 days.
A chd1 mutation suppresses a galactose induction defect in a pob3 strain.
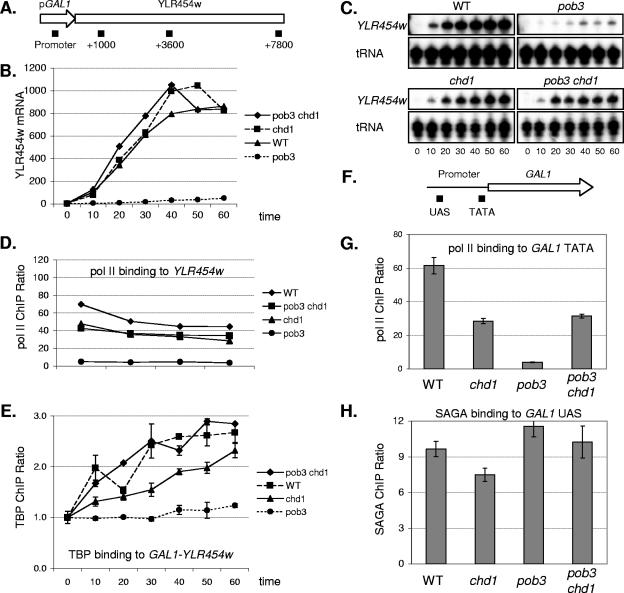
Although genetic and biochemical experiments suggest a role for Chd1 in regulating transcription in eukaryotes, the exact mechanism of Chd1 function is unclear. ChIP experiments showed that Chd1 was bound to the coding regions of the TEF2 and GAL10 genes, suggesting an elongation function (45). However, the ChIP studies also showed that Chd1 was recruited to the GAL10 promoter, consistent with a role in initiation of transcription. We recently showed that a pob3 mutation reduces expression of a GAL1-YLR454w gene fusion and that the pob3 mutation reduces binding of both Pol II and TBP to the GAL1 promoter (3). We performed similar experiments examining the effect of pob3 and chd1 mutations on expression and factor binding at GAL1-YLR454w (Fig. 7). Four isogenic strains were grown first in raffinose medium at 25°C and then shifted for 2 h to 30°C, galactose was added to the medium to induce GAL1-YLR454w expression, and samples were taken at timed intervals for mRNA and ChIP analyses. There is a rapid rise in GAL1-YLR454w mRNA levels following galactose induction in wild-type and chd1 cells (Fig. 7B and C). There is a marked defect in GAL1-YLR454w induction in the pob3 mutant, but this defect is completely suppressed in the pob3 chd1 double mutant. We conclude that Chd1 has a negative role at the GAL1 promoter, opposing the yFACT-dependent transcriptional activation at this promoter.
FIG. 7.
A chd1 mutation suppresses defects in GAL1 induction and Pol II and TBP binding caused by a pob3 mutation. Strains DY9591 (GAL1-YLR454w), DY9959 (chd1 GAL1-YLR454w), DY9972 (pob3 GAL1-YLR454w), and DY10020 (pob3 chd1 GAL1-YLR454w) were grown on YP medium with 2% raffinose. Galactose was added to 2%, and samples were taken at 10-min intervals and processed for ChIP analysis to measure Pol II and TBP binding. (A) Map of the GAL1-YLR454w allele showing the positions of regions amplified by at the promoter and within the gene. (B) YLR454w mRNA levels measured from the GAL1-YLR454w allele, quantified after phosphorimaging of the gels in panel C. WT, wild type. (C) S1 protection assays to measure YLR454w mRNA from the GAL1-YLR454w allele, using probes specific for YLR454w and a tRNA internal control. (D) Distribution of Pol II at 60 min following galactose induction at different GAL1-YLR454w regions in four different strains. Error bars show variance (standard deviations) among replicate PCRs. (E) TBP binding to the GAL1-YLR454w promoter following galactose induction in four different strains. ChIP values were normalized to binding at time zero. Error bars show variance among replicate PCRs. (F) Map of the native GAL1 gene showing the positions of regions amplified by at the upstream activation sequence (UAS) and TATA within the promoter. (G) Pol II binding to the TATA region (positions −190 to + 54) of the native GAL1 promoter at 30 min following galactose induction in four different strains. Error bars show variance among replicate PCRs. (H) SAGA binding to the native UAS region (positions −496 to −316) of the native GAL1 promoter at 30 min following galactose induction in four different strains. Error bars show variance among replicate PCRs.
To examine the molecular mechanism of suppression by chd1 of the defect in transcriptional induction in the pob3 mutant, we used ChIP experiments to measure Pol II occupancy following galactose induction. Samples were harvested at various times after induction with formaldehyde to cross-link and were processed for ChIP. We used PCR probes specific for four different regions of the 8-kb-long YLR454w gene: the GAL1-YLR454w promoter, 1 kb downstream of start codon, the middle of the YLR454w ORF (position +3600), and the 3′ end of the gene (position +7800) (Fig. 7A). The ChIP results shown in Fig. 7D show Pol II occupancy at 60 min after galactose induction at different regions of the GAL1-YLR454w gene. A mutation with an elongation defect should cause decreased Pol II binding along the gene, but Pol II binding at the promoter should not be affected. In contrast, the pob3 mutation sharply reduces Pol II binding at all regions of the gene, including the promoter, suggesting that the pob3 mutation affects recruitment of Pol II to the promoter. Importantly, Pol II binding is effectively restored in the pob3 chd1 double mutant. Similar results were seen at the native GAL1 promoter, where Pol II binding is reduced in a pob3 mutant but restored in the pob3 chd1 double mutant (Fig. 7G). It is not clear why a chd1 mutation, alone, results in reduced Pol II binding to the promoter, compared to the wild type.
Next, we used ChIP assays to measure binding of TBP to the GAL1-YLR454w promoter following galactose induction (Fig. 7H). TBP binding was severely reduced in the pob3 mutant, and TBP binding approached wild-type levels in the pob3 chd1 double mutant strain. These results are consistent with our earlier data suggesting that yFACT has a role in facilitating formation of the TBP-TFIIA complex on DNA. The observation that deletion of CHD1 overcomes the defect in TBP binding in the pob3 strain suggests that Chd1 has a negative role regulating TBP binding at the GAL1 promoter region.
The SAGA complex is required for activation of GAL1, and mutations in SAGA prevent binding of TBP (13). We used ChIP assays to examine binding of the Ada2-Myc subunit of SAGA. The results show no defect in SAGA binding to GAL1 in a pob3 mutant (Fig. 7F), and thus the defect that we observe in TBP binding at GAL1 is not due to failure of SAGA to be recruited to the promoter.
Deletion of CHD1 suppresses the synthetic lethality between spt16-11 and TBP mutations as well as between spt16-11 and TFIIA mutations.
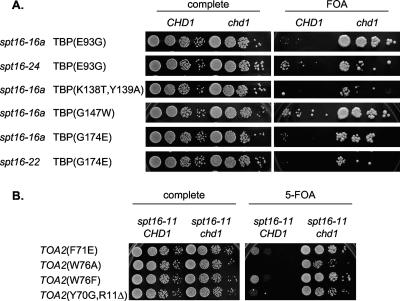
It has been shown earlier that several transcriptional coactivators regulate transcription initiation by regulating formation of the TBP-TFIIA complex. The Swi/Snf chromatin-remodeling complex uses the energy from ATP hydrolysis to regulate TBP binding both in vivo and in vitro (4, 22). Our genetic and biochemical data also showed that yFACT has a role in regulating TBP-TFIIA complex formation. As the defect in TBP binding to the GAL1 promoter caused by a yFACT mutation can be suppressed by deletion of CHD1, we asked whether a chd1 mutation can also suppress the synthetic lethalities between TBP mutations and spt16 mutations that we have described earlier (5). We used a plasmid shuffle assay to address this question. We constructed two isogenic strains containing the wild-type CHD1 gene and deletion of the CHD1 gene. In both of these strains the TBP gene and SPT16 genes were disrupted. (The SPT15 gene encodes TBP, but we will refer to it as the TBP gene to avoid confusion.) Since these genes are essential for cell viability, the strains were kept alive by providing these genes on YCp-URA3 plasmids. We transformed these strains with the TBP plasmid and spt16 plasmid combination that showed synthetic lethality in our earlier genetic assays (5). The transformants were grown on medium containing 5-fluoroorotic acid (5-FOA) so that the strains are required to lose the parental YCp-URA3 plasmid containing both the wild-type TBP gene and SPT16 for their growth. The strain transformed with empty vectors could not grow on a 5-FOA plate (5). However, these strains transformed with wild-type copies of TBP and SPT16 plasmids could grow on medium containing 5-FOA. The introduction of some combinations of TBP mutations and spt16 mutations resulted in either synthetic lethality or a synthetic growth defect in a CHD1 strain background. Interestingly, deletion of CHD1 rescued some of these synthetic lethalities or synthetic growth defects (compare CHD1 with chd1 on 5-FOA plates). This in vivo evidence strongly suggests that Chd1 has a negative role in yFACT-mediated TBP binding. A deletion of this negative factor rescues the synthetic lethal or synthetic growth defect phenotypes associated with spt16 mutations and TBP mutations.
During transcriptional initiation, TBP binding is followed by TFIIA binding to form a stable TBP-TFIIA complex on DNA. Yeast TFIIA is a heterodimer composed of the Toa1 and Toa2 subunits. Some toa2 mutations that abolished the TFIIA interaction with TBP when assayed in an in vitro binding reaction were described previously (35). We have earlier shown that some of these toa2 mutations are synthetic lethal with the spt16-11 mutation (5). Since our data presented here strongly suggest that Chd1 has a negative role in regulating TBP binding in vivo, we asked whether a chd1 mutation would also suppress the synthetic lethal interactions between spt16-11 and toa2 mutations. Two isogenic strains, the spt16-11 toa2 and spt16-11 toa2 chd1 strains, were constructed. Since TOA2 is an essential gene for cell viability, the strains were kept alive by providing the TOA2 gene on a YCp-URA3 plasmid. Both strains were transformed with plasmids containing toa2 mutations that showed a synthetic growth defect or synthetic lethal phenotype with spt16-11. The transformants were grown on a 5-FOA plate so that the strains are required to lose the parental YCp-URA3-TOA2 plasmid and depend on the mutant toa2 plasmid for their growth. Some toa2 mutations show a synthetic lethal phenotype with spt16-11 mutation in the presence of wild-type CHD1. Importantly, deletion of CHD1 rescued these synthetic lethalities between spt16-11 and toa2 mutations (Fig. 8B). We also have observed a synthetic growth defect with some toa2 mutations in combination with the spt16-11 mutation. Deletion of CHD1 also restored this synthetic growth defect between spt16-11 and toa2 mutations (Fig. 8B). Collectively, these data once again strongly suggest a negative role played by Chd1 in yFACT-mediated TBP binding during the transcriptional initiation step.
FIG. 8.
A chd1 mutation suppresses the synthetic lethality of an spt16 mutation with either TBP or TFIIA mutations. (A) Strains DY8552 (spt15 spt16 + YCp-URA3-TBP-Spt16) (indicated as “CHD1”) and DY10141 (spt15 spt16 chd1 + YCp-URA3-TBP-Spt16) (indicated as “chd1”) were transformed with two plasmids, a YCp-TRP1 plasmid encoding a TBP mutant and a YCp-LEU2 plasmid with either wild-type SPT16 or spt16 mutations, and 10-fold dilutions were plated at 33°C either on complete medium for 2 days or on FOA medium for 3 days. (B) Strains DY8700 (spt16-11 toa2 + YCp-URA3-TOA2) (indicated as “CHD1”) and DY10214 (spt16-11 chd1 toa2 + YCp-URA3-TOA2) (indicated as “chd1”) were transformed with a YCp-LEU2 plasmid with the indicated toa2 mutant, and 10-fold dilutions were plated for 2 days on complete medium at 25°C and on FOA medium at 30°C.
This work and previous studies suggest a dual role played by yFACT and Chd1 in regulating transcriptional initiation and elongation. Consistent with these observations, other elongation factors such as the Rtf1 component of the PAF complex (49) and TFIIS (36) have also shown genetic interactions with TBP, thereby suggesting a dual role played by these factors in regulating transcription.
Chd1 and Set2 act in different pathways in vivo.
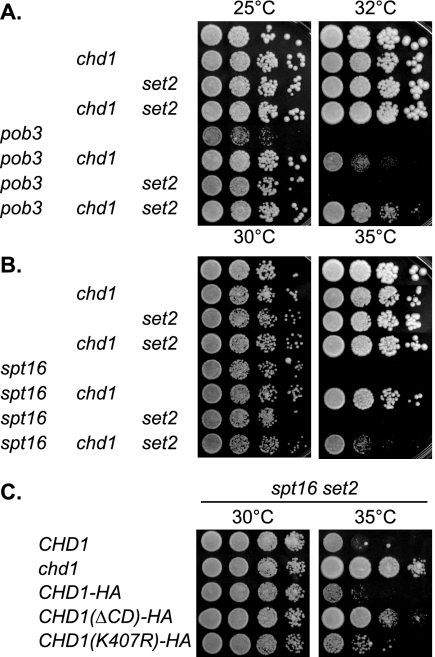
We find that the defects caused by yFACT mutations can be similarly suppressed by chd1 and set2 mutations. These similar suppressive effects could mean that Chd1 and Set2 function in a similar pathway, possibly with the Chd1 chromodomain recognizing the H3-K36 residue methylated by Set2 or other modified histone residues. To explore this possibility, we introduced both chd1 and set2 disruptions into strains with yFACT mutations. At 32°C a pob3 strain does not grow at all, while the pob3 chd1 strain grows weakly (Fig. 9A). The pob3 set2 double mutant does not grow at 32°C (Fig. 9A), but it does grow at 30°C (5), a temperature at which the pob3 single mutant does not grow. The pob3 chd1 set2 triple mutant shows much better growth than either double mutant. This additive defect shows that Chd1 and Set2 act in different pathways. A slightly different result is seen with the spt16 mutant, where chd1 suppresses well the 35°C growth defect but set2 does not suppress at all at this temperature (Fig. 9B). Interestingly, the spt16 chd1 set2 triple mutant shows an intermediate phenotype. The difference in the spt16 response to chd1 and set2 is consistent with these two regulators functioning in distinct pathways, although we do not see an additive effect here.
FIG. 9.
Additive suppression of pob3 by chd1 and set2 mutations. (A) Tenfold dilutions of strains DY150 (wild type), DY9809 (chd1), DY8690 (set2), DY9838 (chd1 set2), DY7379 (pob3), DY9458 (pob3 chd1), DY8878 (pob3 set2), and DY9547 (pob3 chd1 set2) were plated on complete medium at 25°C or 32°C for 3 days. (B) Tenfold dilutions of strains DY150 (wild type), DY9809 (chd1), DY8690 (set2), DY9838 (chd1 set2), DY8107 (spt16), DY9152 (spt16 chd1), DY8777 (spt16 set2), and DY9153 (spt16 chd1 set2) were plated on complete medium at either 30°C or 35°C for 2 days. (C) Tenfold dilutions of strains DY8777 (spt16 set2), DY9153 (spt16 set2 chd1), DY11619 (spt16 set2 chd1 CHD1-HA), DY11626 [spt16 set2 chd1 CHD1(ΔCD)-HA], and DY11645 [spt16 set2 chd1 CHD1(K407R)-HA] were plated on complete medium at 30°C for 2 days or at 35°C for 3 days.
Thus, the pob3(L78R) and spt16-11 alleles differ in terms of additive suppression by chd1 and set2. This is not completely unexpected, as the pob3(L78R) and spt16-11 mutants show opposite effects in response to changes in copy number of histone genes or to an rpd3 mutation (19).
We also examined the CHD1 alleles with mutations in the chromodomain and ATPase domains for effects in the spt16 set2 strain (Fig. 9C). The CHD1 spt16 set2 strain fails to grow at 35°C, while the chd1 spt16 set2 strain is alive. If a CHD1 mutant complements the strain will not grow; failure to complement will result in growth. The ΔCD and K407R mutant versions of Chd1 both allow partial growth of the spt16 set2 mutant at 35°C, and thus both the chromodomain and the ATPase are required for the Chd1 activity that is toxic in the spt16 set2 strain.
In summary, the additive effect of chd1 and set2 disruptions in suppressing the pob3 growth defect demonstrates that Chd1 and Set2 function in different pathways. We also show that both the chromodomain and the ATPase contribute to this function of Chd1.
Consistent with our observations that Chd1 and Set2 both play negative roles in regulating yFACT-mediated transcription, a negative role for these factors in transcriptional elongation was suggested by an earlier study. The Bur1 kinase is thought to promote elongation by phosphorylating Pol II (26), and the severe growth defect caused by a bur1 deletion can be suppressed by disruption of either CHD1 or SET2 (25). Since Bur1 promotes elongation, those authors concluded that Chd1 and Set2 act negatively on elongation.
DISCUSSION
The yeast Chd1 protein has ATPase activity and two chromodomains, and the protein functions as an ATP-dependent chromatin remodeler in vitro (29, 48). We describe yFACT as a chromatin-“reorganizing” complex because its activity is ATP independent, to distinguish it from the chromatin remodelers that require ATP. We find that a chd1 gene disruption suppresses numerous phenotypes caused by mutations in the Spt16 and Pob3 subunits of yFACT, including temperature-sensitive growth, and synthetic growth defects in combination with other transcriptional regulators. This suppression by chd1 mutations suggests that Chd1 and yFACT act in opposition at the steps involved in causing these phenotypes. Previous studies have suggested that Chd1 and yFACT both function in transcriptional elongation (19, 27, 45, 47). However, our results here show that the defect in GAL1 transcription caused by a pob3 mutation can be suppressed by chd1 and that this suppression includes restoration of binding by the TBP basal transcription factor and Pol II. Additionally, spt16 mutations show synthetic lethality in combination with mutations in TBP or TFIIA, and this synthetic lethality can be suppressed by deletion of the CHD1 gene. These results suggest that both Chd1 and yFACT function at promoters, regulating chromatin accessibility for DNA binding by TBP and TFIIA. However, the possibility remains that the effect on TBP recruitment is indirect. Mason and Struhl (30) found that inactivation of a thermosensitive Spt16 results in reduced binding of TBP and TFIIB at promoters, and those authors suggested that this reduced TBP binding is an indirect result of inappropriate TBP binding to cryptic sites elsewhere in the genome.
Our genetic experiments suggest that Chd1 and yFACT act in opposition, with Chd1 being toxic in spt16 or pob3 mutants with a partially defective yFACT chromatin-reorganizing factor. In support of this model, we showed that overexpression of Chd1 is toxic in an spt16 mutant; importantly, Chd1 overexpression is not detrimental in an SPT16 strain (Fig. 6).
We have shown that disruption of the SET2 gene, encoding a histone methyltransferase acting on K36 of histone H3, can also suppress growth defects caused by spt16 and pob3 mutations (3). Similar to a chd1 mutation, set2 also suppresses defects in GAL1 transcription, TBP binding, and Pol II binding, as well as the synthetic lethality seen with TBP or TFIIA mutants. Thus, chd1 and set2 have very similar effects in suppressing yFACT mutants. A simple model would have Chd1 and Set2 functioning in a similar pathway, possibly with the Chd1 chromodomain recognizing the H3-K36 residue methylated by Set2 or other modified histone residues. However, we find that chd1 and set2 show additivity in their ability to suppress the pob3 growth defect (Fig. 9A). This genetic experiment clearly shows that Chd1 and Set2 function in distinct pathways.
Structural work has shown that the two chromodomains of human Chd1 form a single structural unit and that this double chromodomain binds to the histone H3 tail with methylated K4 (16). The results with yeast Chd1 are controversial, with one group showing yeast Chd1 binding to H3-K4-Me (37) and two labs failing to detect this interaction (32, 46). Our genetic experiments argue strongly against the idea that Chd1 binds H3-K4-Me. While a chd1 mutation suppresses yFACT defects, either an H3-K4R substitution or disruption of SET1, encoding the H3-K4 methyltransferase, shows strong synthetic defects when combined with either spt16 or pob3 (3). Thus, chd1 has opposite effects from those of either H3-K4R or set1, and thus it seems unlikely that the ability of Chd1 to oppose yFACT requires binding of Chd1 to H3-K4-Me. Like the suppression by chd1, H3-K36R or set2 mutations also suppress yFACT defects (3). However, the additive effect seen by chd1 and set2 in suppressing the pob3 growth defect also makes it unlikely that Chd1 binds to H3-K36-Me. Consistent with these genetic data, a recent structure of yeast Chd1 shows that it lacks aromatic residues involved in binding methyl-lysine and suggests that it will not bind this modified residue (15).
Chd1 has two chromodomains along with its ATPase domain, and we used Chd1 mutants to test whether these protein functions are required for the toxicity of Chd1 in yFACT mutants. We used two mutants, CHD1(ΔCD), where the chromodomain has been deleted, and CHD1(K407R), with a mutation in a lysine residue required for ATPase activity. Our experiments show that the chromodomain and ATPase mutations in Chd1 both partially relieve the growth defect in pob3 and spt16 mutants (Fig. 2). These experiments suggest that the ATPase activity and the chromodomain of Chd1 are both required for Chd1 toxicity in yFACT mutants. We found a strong growth defect in the gcn5 chd1 double mutant, and this growth defect is also seen in gcn5 mutants with either the CHD1(ΔCD) or CHD1(K407R) allele affecting the chromodomain or the ATPase (Fig. 4). Thus, both the ATPase activity and the chromodomain are required for the Chd1 activity that opposes yFACT.
Our genetic experiments show that Chd1 and yFACT act in opposition in regulating transcription, and this may involve regulating TBP binding at promoters. An association between Chd1 and yFACT has been shown by purification of TAP-tagged proteins and by immunoprecipitation (27, 45). Further work is needed to understand how these two chromatin factors function and what is the role of the chromodomain in Chd1.
Supplementary Material
Acknowledgments
We thank Patrick Grant, Grant Hartzog, Sandy Johnson, and Laurie Stargell for providing plasmids; Tony Weil for antibody; and Tim Formosa for many helpful discussions.
This work was supported by grants from the National Institutes of Health.
Footnotes
Published ahead of print on 9 July 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. E. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Wiley and Sons, New York, NY.
- 1a.Belotserkovskaya, R., S. Oh, V. A. Bondarenko, G. Orphanides, V. M. Studitsky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science 301:1090-1093. [DOI] [PubMed] [Google Scholar]
- 2.Bhoite, L. T., and D. J. Stillman. 1998. Residues in the Swi5 zinc finger protein that mediate cooperative DNA-binding with the Pho2 homeodomain protein. Mol. Cell. Biol. 18:6436-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas, D., R. Dutta-Biswas, D. Mitra, Y. Shibata, B. D. Strahl, T. Formosa, and D. J. Stillman. 2006. Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J. 25:4479-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas, D., A. N. Imbalzano, P. Eriksson, Y. Yu, and D. J. Stillman. 2004. Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex. Mol. Cell. Biol. 24:8312-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas, D., Y. Yu, M. Prall, T. Formosa, and D. J. Stillman. 2005. The yeast FACT complex has a role in transcriptional initiation. Mol. Cell. Biol. 25:5812-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm, A., K. R. Tufteland, R. Aasland, and P. B. Becker. 2004. The many colours of chromodomains. Bioessays 26:133-140. [DOI] [PubMed] [Google Scholar]
- 7.Brewster, N. K., G. C. Johnston, and R. A. Singer. 2001. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21:3491-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. Dent, F. Winston, and C. D. Allis. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cairns, B. R. 2005. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15:185-190. [DOI] [PubMed] [Google Scholar]
- 10.Costa, P. J., and K. M. Arndt. 2000. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156:535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Cruz, X., S. Lois, S. Sanchez-Molina, and M. A. Martinez-Balbas. 2005. Do protein motifs read the histone code? Bioessays 27:164-175. [DOI] [PubMed] [Google Scholar]
- 12.Dryhurst, D., A. A. Thambirajah, and J. Ausio. 2004. New twists on H2A.Z: a histone variant with a controversial structural and functional past. Biochem. Cell Biol. 82:490-497. [DOI] [PubMed] [Google Scholar]
- 13.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duroux, M., A. Houben, K. Ruzicka, J. Friml, and K. D. Grasser. 2004. The chromatin remodelling complex FACT associates with actively transcribed regions of the Arabidopsis genome. Plant J. 40:660-671. [DOI] [PubMed] [Google Scholar]
- 14a.Eriksson, P., D. Biswas, Y. Yu, J. M. Stewart, and D. J. Stillman. 2004. TATA-binding protein mutants that are lethal in the absence of the Nhp6 high-mobility-group protein. Mol. Cell. Biol. 24:6419-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flanagan, J. F., B. J. Blus, Y. Kim, K. L. Clines, F. Rastinejad, and S. Khorasanizadeh. 2007. Molecular implications of evolutionary differences in CHD double chromodomains. J. Mol. Biol. 369:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flanagan, J. F., L. Z. Mi, M. Chruszcz, M. Cymborowski, K. L. Clines, Y. Kim, W. Minor, F. Rastinejad, and S. Khorasanizadeh. 2005. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438:1181-1185. [DOI] [PubMed] [Google Scholar]
- 17.Formosa, T. 2003. Changing the DNA landscape: putting a SPN on chromatin. Curr. Top. Microbiol. Immunol. 274:171-201. [DOI] [PubMed] [Google Scholar]
- 18.Formosa, T., P. Eriksson, J. Wittmeyer, J. Ginn, Y. Yu, and D. J. Stillman. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20:3506-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Formosa, T., S. Ruone, M. D. Adams, A. E. Olsen, P. Eriksson, Y. Yu, A. R. Rhoades, P. D. Kaufman, and D. J. Stillman. 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway. Polymerase passage may degrade chromatin structure. Genetics 162:1557-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fragiadakis, G. S., D. Tzamarias, and D. Alexandraki. 2004. Nhp6 facilitates Aft1 binding and Ssn6 recruitment, both essential for FRE2 transcriptional activation. EMBO J. 23:333-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Havas, K., I. Whitehouse, and T. Owen-Hughes. 2001. ATP-dependent chromatin remodeling activities. Cell Mol. Life Sci. 58:673-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imbalzano, A. N., H. Kwon, M. R. Green, and R. E. Kingston. 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370:481-485. [DOI] [PubMed] [Google Scholar]
- 23.Kassavetis, G. A., and D. F. Steiner. 2006. NHP6 is a transcriptional initiation fidelity factor for RNA polymerase III transcription in vitro and in vivo. J. Biol. Chem. 281:7445-7451. [DOI] [PubMed] [Google Scholar]
- 24.Kelley, D. E., D. G. Stokes, and R. P. Perry. 1999. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma 108:10-25. [DOI] [PubMed] [Google Scholar]
- 25.Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, N. J. Thompson, C. Boone, A. Emili, J. S. Weissman, T. R. Hughes, B. D. Strahl, M. Grunstein, J. F. Greenblatt, S. Buratowski, and N. J. Krogan. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593-605. [DOI] [PubMed] [Google Scholar]
- 26.Keogh, M. C., V. Podolny, and S. Buratowski. 2003. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 23:7005-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen, C. Seidel, J. Gerton, and J. L. Workman. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 102:18385-18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lusser, A., D. L. Urwin, and J. T. Kadonaga. 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 12:160-166. [DOI] [PubMed] [Google Scholar]
- 30.Mason, P. B., and K. Struhl. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23:8323-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Mason, P. B., and K. Struhl. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17:831-840. [DOI] [PubMed] [Google Scholar]
- 31.Mellor, J., and A. Morillon. 2004. ISWI complexes in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1677:100-112. [DOI] [PubMed] [Google Scholar]
- 32.Okuda, M., M. Horikoshi, and Y. Nishimura. 2006. Structural Polymorphism of Chromodomains in Chd1. J. Mol. Biol. 365:1047-1062. [DOI] [PubMed] [Google Scholar]
- 33.Orphanides, G., G. LeRoy, C. H. Chang, D. S. Luse, and D. Reinberg. 1998. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92:105-116. [DOI] [PubMed] [Google Scholar]
- 34.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 35.Ozer, J., L. E. Lezina, J. Ewing, S. Audi, and P. M. Lieberman. 1998. Association of transcription factor IIA with TATA binding protein is required for transcriptional activation of a subset of promoters and cell cycle progression in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:2559-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prather, D. M., E. Larschan, and F. Winston. 2005. Evidence that the elongation factor TFIIS plays a role in transcription initiation at GAL1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates III, and P. A. Grant. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433:434-438. [DOI] [PubMed] [Google Scholar]
- 38.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhoades, A. R., S. Ruone, and T. Formosa. 2004. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol. 24:3907-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson, K. M., and M. C. Schultz. 2003. Replication-independent assembly of nucleosome arrays in a novel yeast chromatin reconstitution system involves antisilencing factor Asf1p and chromodomain protein Chd1p. Mol. Cell. Biol. 23:7937-7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruone, S., A. R. Rhoades, and T. Formosa. 2003. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J. Biol. Chem. 278:45288-45295. [DOI] [PubMed] [Google Scholar]
- 42.Saunders, A., J. Werner, E. D. Andrulis, T. Nakayama, S. Hirose, D. Reinberg, and J. T. Lis. 2003. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301:1094-1096. [DOI] [PubMed] [Google Scholar]
- 42a.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:1-21. [DOI] [PubMed] [Google Scholar]
- 44.Shimojima, T., M. Okada, T. Nakayama, H. Ueda, K. Okawa, A. Iwamatsu, H. Handa, and S. Hirose. 2003. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 17:1605-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa, A. D. Johnson, G. A. Hartzog, and K. M. Arndt. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22:1846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sims, R. J., III, C. F. Chen, H. Santos-Rosa, T. Kouzarides, S. S. Patel, and D. Reinberg. 2005. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 280:41789-41792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stockdale, C., A. Flaus, H. Ferreira, and T. Owen-Hughes. 2006. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J. Biol. Chem. 281:16279-16288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stolinski, L. A., D. M. Eisenmann, and K. M. Arndt. 1997. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4490-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 51.Szerlong, H., A. Saha, and B. R. Cairns. 2003. The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 22:3175-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 53.Tran, H. G., D. J. Steger, V. R. Iyer, and A. D. Johnson. 2000. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. EMBO J. 19:2323-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, W. 2003. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr. Top. Microbiol. Immunol. 274:143-169. [DOI] [PubMed] [Google Scholar]
- 56.Wittmeyer, J., and T. Formosa. 1997. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 17:4178-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wittmeyer, J., L. Joss, and T. Formosa. 1999. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 38:8961-8971. [DOI] [PubMed] [Google Scholar]
- 58.Wittschieben, B. O., G. Otero, T. de Bizemont, J. Fellows, H. Erdjument-Bromage, R. Ohba, Y. Li, C. D. Allis, P. Tempst, and J. Q. Svejstrup. 1999. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4:123-128. [DOI] [PubMed] [Google Scholar]
- 59.Woodage, T., M. A. Basrai, A. D. Baxevanis, P. Hieter, and F. S. Collins. 1997. Characterization of the CHD family of proteins. Proc. Natl. Acad. Sci. USA 94:11472-11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xella, B., C. Goding, E. Agricola, E. Di Mauro, and M. Caserta. 2006. The ISWI and CHD1 chromatin remodelling activities influence ADH2 expression and chromatin organization. Mol. Microbiol. 59:1531-1541. [DOI] [PubMed] [Google Scholar]
- 61.Yang, X. J. 2004. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 26:1076-1087. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, H., D. N. Roberts, and B. R. Cairns. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123:219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, L., S. Schroeder, N. Fong, and D. L. Bentley. 2005. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress.’ EMBO J. 24:2379-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.