Abstract
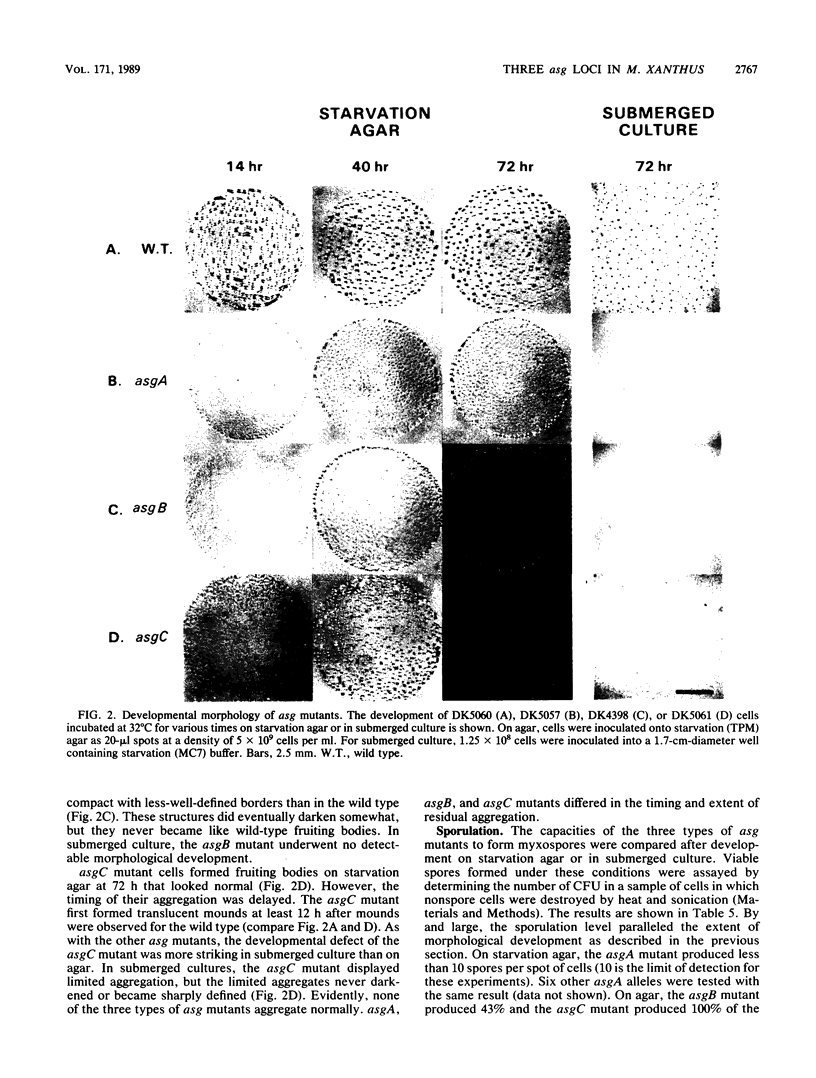
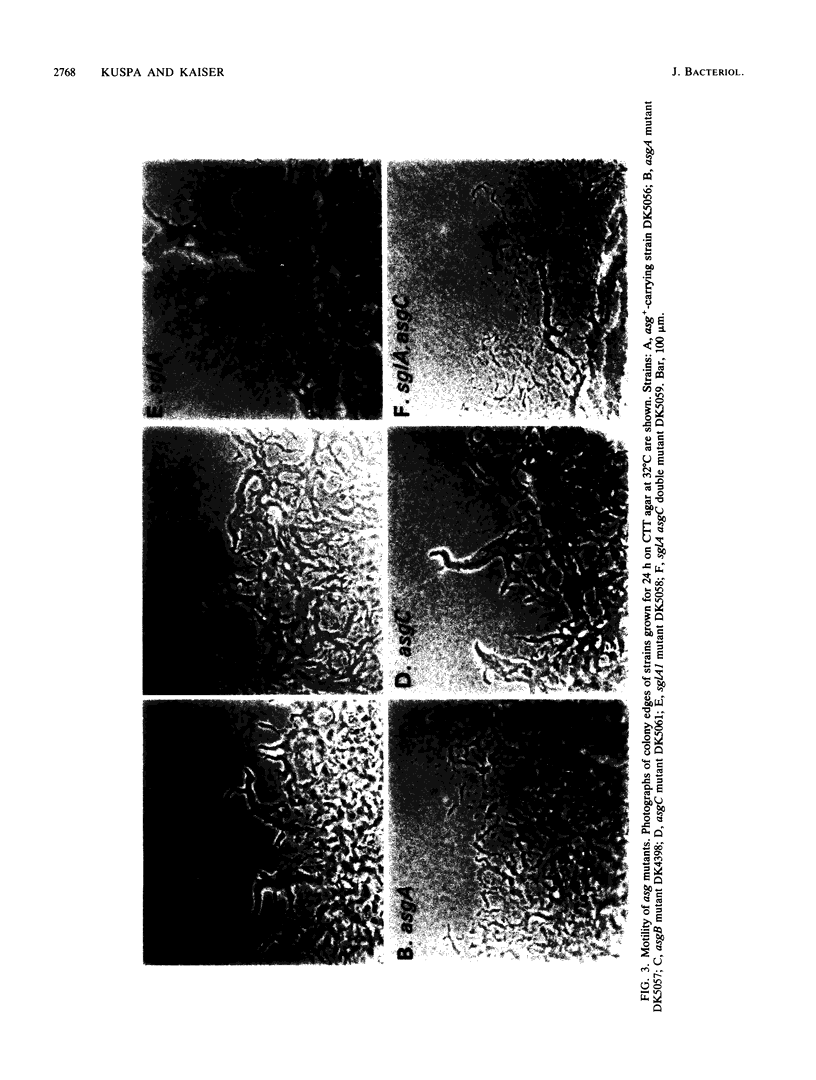
asg-carrying strains of Myxococcus xanthus arose in a selection for mutants defective in cell-cell signalling during fruiting body development. All 15 asg mutations examined were found to lie in one of three genetic loci, asgA, asgB, or asgC. The loci were defined by linkage to different insertions of transposon Tn5 and molecular cloning of asgA. asg mutants of all three types were deficient in the aggregation of cells into mounds of the sort that normally give rise to fruiting bodies. asg mutants were also deficient in spore formation; sporulation is normally one of the last steps in fruiting body development. Consistent with a requirement for cell-to-cell signalling, at 1 to 2 h asg+-carrying cells release a material called A-factor that can rescue development of asg mutants. asgA, asgB, and asgC mutants released 5% or less of the asg+ level of A-factor, as measured by bioassay. The experimental results are consistent with the hypothesis that a deficiency in A-factor production or release is the primary developmental defect in asg mutants and that aggregation and sporulation depend on A-factor. asg mutations at all three loci also changed the color and morphology of growing colonies, and failure to release A-factor may itself arise from a defect in growing cells.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold J. W., Shimkets L. J. Cell surface properties correlated with cohesion in Myxococcus xanthus. J Bacteriol. 1988 Dec;170(12):5771–5777. doi: 10.1128/jb.170.12.5771-5777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J. W., Shimkets L. J. Inhibition of cell-cell interactions in Myxococcus xanthus by congo red. J Bacteriol. 1988 Dec;170(12):5765–5770. doi: 10.1128/jb.170.12.5765-5770.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., Kaiser D. In situ transposon replacement and isolation of a spontaneous tandem genetic duplication. Mol Gen Genet. 1983;191(1):99–109. doi: 10.1007/BF00330896. [DOI] [PubMed] [Google Scholar]
- Bretscher A. P., Kaiser D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol. 1978 Feb;133(2):763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard R. P., Dworkin M. Light-induced lysis and carotenogenesis in Myxococcus xanthus. J Bacteriol. 1966 Feb;91(2):535–545. doi: 10.1128/jb.91.2.535-545.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J. M., Geisselsoder J., Zusman D. R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- Dworkin M., Kaiser D. Cell interactions in myxobacterial growth and development. Science. 1985 Oct 4;230(4721):18–24. doi: 10.1126/science.3929384. [DOI] [PubMed] [Google Scholar]
- Gill R. E., Cull M. G., Fly S. Genetic identification and cloning of a gene required for developmental cell interactions in Myxococcus xanthus. J Bacteriol. 1988 Nov;170(11):5279–5288. doi: 10.1128/jb.170.11.5279-5288.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen G. R., Dworkin M. Cell-cell interactions in developmental lysis of Myxococcus xanthus. Dev Biol. 1985 Nov;112(1):194–202. doi: 10.1016/0012-1606(85)90133-2. [DOI] [PubMed] [Google Scholar]
- Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987 Oct;1(8):840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kuspa A., Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986 Sep;117(1):252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- Kuner J. M., Kaiser D. Fruiting body morphogenesis in submerged cultures of Myxococcus xanthus. J Bacteriol. 1982 Jul;151(1):458–461. doi: 10.1128/jb.151.1.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner J. M., Kaiser D. Introduction of transposon Tn5 into Myxococcus for analysis of developmental and other nonselectable mutants. Proc Natl Acad Sci U S A. 1981 Jan;78(1):425–429. doi: 10.1073/pnas.78.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A., Kroos L., Kaiser D. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev Biol. 1986 Sep;117(1):267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- LaRossa R., Kuner J., Hagen D., Manoil C., Kaiser D. Developmental cell interactions of Myxococcus xanthus: analysis of mutants. J Bacteriol. 1983 Mar;153(3):1394–1404. doi: 10.1128/jb.153.3.1394-1404.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S., Sodergren E., Masuda T., Kaiser D. Systematic isolation of transducing phages for Myxococcus xanthus. Virology. 1978 Jul 1;88(1):44–53. doi: 10.1016/0042-6822(78)90108-3. [DOI] [PubMed] [Google Scholar]
- Nelson D. R., Zusman D. R. Transport and localization of protein S, a spore coat protein, during fruiting body formation by Myxococcus xanthus. J Bacteriol. 1983 May;154(2):547–553. doi: 10.1128/jb.154.2.547-553.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Keller K. H., Dworkin M. Cell density-dependent growth of Myxococcus xanthus on casein. J Bacteriol. 1977 Feb;129(2):770–777. doi: 10.1128/jb.129.2.770-777.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J. Control of morphogenesis in myxobacteria. Crit Rev Microbiol. 1987;14(3):195–227. doi: 10.3109/10408418709104439. [DOI] [PubMed] [Google Scholar]
- Shimkets L. J. Correlation of energy-dependent cell cohesion with social motility in Myxococcus xanthus. J Bacteriol. 1986 Jun;166(3):837–841. doi: 10.1128/jb.166.3.837-841.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J., Gill R. E., Kaiser D. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
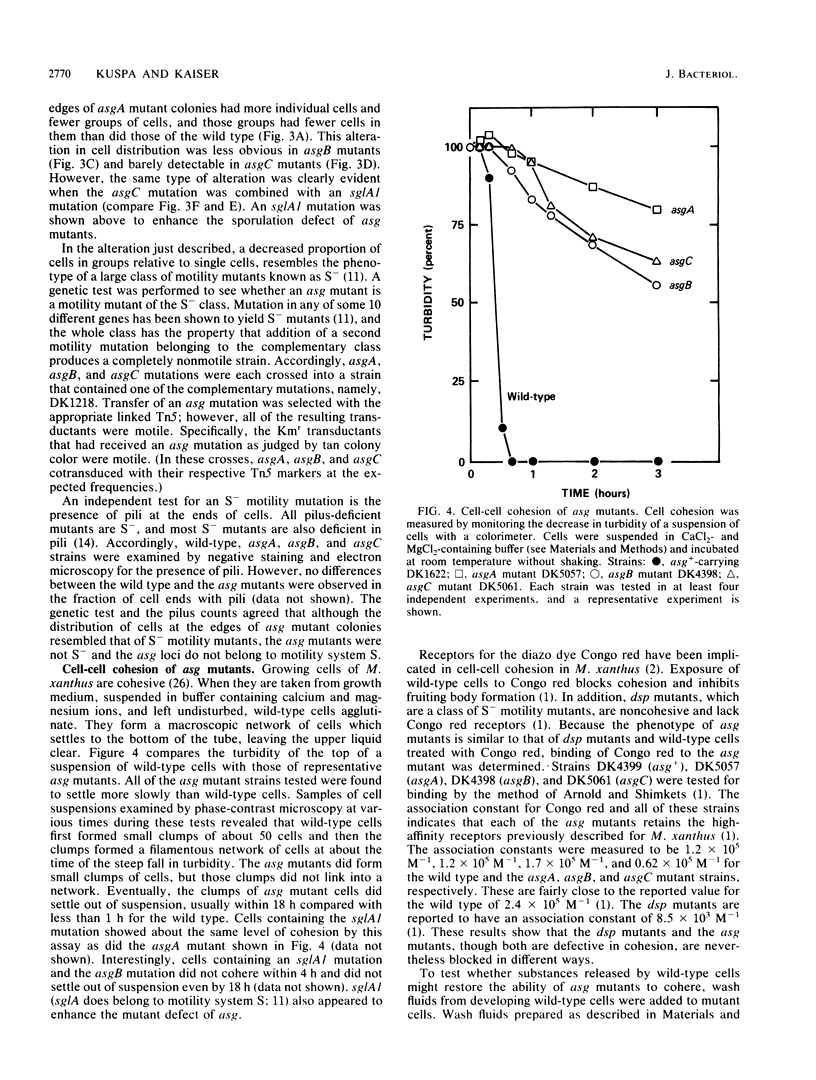
- Shimkets L. J. Role of cell cohesion in Myxococcus xanthus fruiting body formation. J Bacteriol. 1986 Jun;166(3):842–848. doi: 10.1128/jb.166.3.842-848.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodergren E., Kaiser D. Insertions of Tn5 near genes that govern stimulatable cell motility in Myxococcus. J Mol Biol. 1983 Jun 25;167(2):295–310. doi: 10.1016/s0022-2836(83)80337-4. [DOI] [PubMed] [Google Scholar]