Abstract
The mitochondrial respiratory chain (MRC) plays crucial roles in cellular energy production. However, its function in early embryonic development remains largely unknown. To address this issue, GRIM-19, a newly identified MRC complex I subunit, was knocked down in Xenopus laevis embryos. A severe deficiency in heart formation was observed, and the deficiency could be rescued by reintroducing human GRIM-19 mRNA. The mechanism involved was further investigated. We found that the activity of NFAT, a transcription factor family that contributes to early organ development, was downregulated in GRIM-19 knockdown embryos. Furthermore, the expression of a constitutively active form of mouse NFATc4 in these embryos rescued the heart developmental defects. NFAT activity is controlled by a calcium-dependent protein phosphatase, calcineurin, which suggests that calcium signaling may be disrupted by GRIM-19 knockdown. Indeed, both the calcium response and calcium-induced NFAT activity were impaired in the GRIM-19 or NDUFS3 (another complex I subunit) knockdown cell lines. We also showed that NFAT can rescue expression of Nkx2.5, which is one of the key genes for early heart development. Our data demonstrated the essential role of MRC in heart formation and revealed the signal transduction and gene expression cascade involved in this process.
GRIM-19 (gene associated with retinoid-interferon [IFN]-induced mortality 19) was originally identified as a beta IFN (IFN-β)- and retinoic acid (RA)-inducible gene with an apoptotic effect in human cancer cell lines (1). Subsequently, GRIM-19 was found to copurify with mitochondrial NADH:ubiquinone oxidoreductase (complex I) in the bovine heart (13). Through gene targeting in mice, we further demonstrated that GRIM-19 is a new functional subunit of complex I that is essential for complex I assembly and electron transfer activity (20). Based on its primary function in mitochondria, the HUGO Gene Nomenclature Committee has suggested that GRIM-19 be renamed NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 13 (NDUFA13). To explore the functional link of GRIM-19 as a death-related gene and a complex I subunit, we recently reported that not only GRIM-19 but also other subunits of the mitochondrial respiratory chain (MRC) are induced by IFN-β and RA treatment. More importantly, the MRC plays an essential role in the IFN-β/RA-induced cancer cell death (19). These studies support the primary role of GRIM-19 as a complex I protein and also suggest that GRIM-19 is a useful tool for the study of MRC functions.
The MRC plays crucial roles in basic biological processes through the production of cellular energy, generation of reactive oxygen species, and initiation of apoptosis. The mitochondrial defects have been implicated in various degenerative diseases, aging, and cancer (42). However, how the MRC controls early embryonic development in animals is less known. It has been reported that homozygous knockout of mitochondrial transcription factor A (Tfam), a gene necessary for transcription of all 13 mitochondrial DNA-encoded MRC subunits, causes embryonic lethality prior to embryonic day 10.5 (E10.5) by a heart defect in mice (22). Knockout of GRIM-19 in mice also caused embryonic lethality before E9.5 (20). Although these data provide genetic evidence for the essential role of the MRC in embryonic development, the mechanisms involved are largely unknown.
MRC consists of five multisubunit complexes (complexes I to V) localized in the inner membrane of the mitochondria. The MRC complexes catalyze oxidative phosphorylation (OXPHOS) by transferring two electrons from reducing substrates (NADH-FADH2) to molecular oxygen, thereby generating a proton gradient across the inner mitochondrial membrane. The electrochemical energy of this gradient is then used to drive ATP synthesis by complex V (18, 39). Mitochondria also play an important role in modulating intracellular Ca2+ homeostasis (11, 35). In most cells, Ca2+ functions as an intracellular messenger to regulate various signal pathways and gene expression (2, 5). Binding of hormones and growth factors to their specific receptors on the plasma membrane (PM) leads to the activation of phospholipase C, which catalyzes production of inositol 1,4,5-triphosphate (InsP3). InsP3 binds to its receptors (InsP3R) on the endoplasmic reticulum (ER) and sarcoplasmic reticulum (SR) membrane and opens the Ca2+ channel to allow the ER/SR-stored Ca2+ to be released to the cytoplasm. This initial Ca2+ release triggers a Ca2+ influx through specialized Ca2+ release-activated Ca2+ channels (CRAC) on the PM (34). The resulting elevation of the cytosol Ca2+ concentration is required to activate Ca2+-dependent activities. Mitochondrion function in Ca2+ regulation has been reported as a buffering system that can rapidly take up and slowly release large amounts of Ca2+ to protect cells against cytosolic Ca2+ overloading under pathophysiological conditions (11). Furthermore, refilling Ca2+ from the cytosol into the ER/SR or extruding Ca2+ to the extracellular fluid requires ATP (2). Conversely, the accumulation of mitochondrial Ca2+ increases the OXPHOS rate and ATP synthesis through the activation of several mitochondrial enzymes (16, 28). Defective ATP production has been reported to impair calcium homeostasis and oscillation in mouse eggs (12, 25), but there is no genetic evidence to demonstrate a role of the MRC in Ca2+ regulation.
A Ca2+/calmodulin-dependent serine/threonine protein phosphatase, calcineurin, is one of the major targets controlled by Ca2+ and plays pivotal roles in signaling pathways involved in antigen-dependent T-cell activation and development of the mammalian heart (8). In both cases, calcineurin causes dephosphorylation of the cytoplasmic nuclear factor of activated T cells (NFATc), which drives the nuclear translocation of NFATc. The mouse NFAT family contains four cytosolic members (NFATc1 to NFATc4) that contribute to the development of several organ systems, including the heart (8). Mutation of nfatc1 results in cardiac valve and septal defects (9), whereas double knockout of nfatc3 and nfatc4 in mice leads to a reduced myocardium and vascular defect (4, 15).
In this study, we investigated the role of MRC in early embryonic development by using Xenopus laevis as a model. We found that knockdown (KD) of GRIM-19 in Xenopus embryos caused severe heart deficiencies. These defects, however, can be adequately rescued by the expression of a constitutively activated mouse NFATc4. We provide evidence demonstrating that normal heart formation requires an intact functional MRC that regulates NFAT activation by modulating Ca2+ signaling.
MATERIALS AND METHODS
Cell culture, chemicals, and reagents.
HeLa and MCF-7 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and supplemented with penicillin/streptomycin and l-glutamine. Jurkat cells (ATCC) were cultured in RPMI 1640 medium containing 10% fetal bovine serum, penicillin/streptomycin, 1 mM sodium pyruvate, and 2 mM l-glutamine. Histamine (His) was obtained from Fluka. Fura2-AM, F-127, ionomycin, and antibody against NDUFS3 were purchased from Molecular Probes. Anti-NFATc4 and voltage-dependent anion channel (VDAC) antibodies were purchased from Calbiochem. Anti-CD3 and CD28 antibodies were from eBioscience. Phorbol-12-myristate-13-acetate (PMA) and antibodies against actin and Hsp60 were from Sigma. Antibody against mouse GRIM-19 was generated as described previously (26). It cross-reacts with human and Xenopus GRIM-19.
Plasmids and DNA transfection.
Human GRIM-19 (hG19) cDNA was amplified by reverse transcription (RT)-PCR from HeLa cells and cloned into BamHI and XbaI sites of the pCS2 vector. Mouse NFATc4 full-length cDNA was purchased from ATCC. The constitutively activated NFATc4 (CA-NFATc4) was constructed by deleting the first 317 amino acids from the wild-type (WT) NFATc4 according to a previous report (30). NFATc4 and CA-NFATc4 were subcloned into EcoRI and XhoI sites of either pcDNA3 or pCS2 vector.
Isolation of cDNA clones of X. laevis GRIM-19.
An X. laevis oocyte cDNA library (ATCC) was screened using 32P-labeled Xenopus tropicalis GRIM-19 cDNA (ATCC) as a probe. The XGRIM-19 cDNA was cloned into HindIII and EcoRI sites of both pcDNA3 and pDrive vector (QIAGEN) for in vitro transcription/translation assays and probes for in situ hybridization, respectively.
In situ hybridization and histological analysis.
Whole-mount and on-section in situ hybridizations were performed as previously described (17) using BM Purple as a substrate for alkaline phosphatase. Full-length cDNA of Nkx2.5, MLC, and cardiac actin were amplified from stage 28 Xenopus embryo mRNA by RT-PCR and cloned into pDrive vector. Probes were made from the above constructs with a DIG RNA labeling kit (Roche) by following the manufacturer's instructions.
In vitro transcription and translation.
XGRIM-19 protein was synthesized using a TnT quick coupled transcription/translation system. Plasmid pcDNA-XGRIM-19 (0.5 μg) was incubated with 40 μl of TnT T7 Quick master mix and 20 μM methionine in the presence or absence of 4 to 20 ng/μl morpholino oligonucleotides (MOs). Translation was carried out at 30°C for 90 min.
Embryo manipulation.
The Xenopus protocol was approved by the Biological Resource Center Institutional Animal Care and Use Committee, Biopolis, Singapore. Embryos were fertilized in vitro, dejellied in 2% cysteine-HCl (pH 7.8), and maintained in 0.1× MMR (0.1 M NaCl, 2.0 mM KCl, 1 mM MgSO4, 2 mM CaCl2, 5 mM HEPES [pH 7.8], and 0.1 mM EDTA) supplemented with 50 μg/ml gentamicin. To inhibit complex I activity, 0.5 μM rotenone or dimethyl sulfoxide (DMSO; solvent for rotenone) was added to the 0.1× MMR at stage 12. To inhibit NFAT activity, the vitelline membranes of Xenopus embryos were removed and the embryos were treated with solvent (17% Tween 20, 83% ethanol) or 2 to 5 μM of FK506 and cyclosporine (CsA) in 0.1× MMR plus 6% Ficoll continuously from stage 15. Heart development was examined under the microscope at stage 42. Embryos were staged according to the method described by Nieuwkoop and Faber (31). The antisense XGRIM-19 MO was purchased from Gene Tools. The sequences for MOs are as follows: XGRIM-19 MO1, 5′-AGCTTAGTTTCTGTCGAAGACCTCT-3′; XGRIM-19 MO2, 5′-CCTGTTTCACCTTGGACGCCGCCAT-3′; control MO, 5′-AGGTTACTTTGTG TGGAAGACGTCT-3′. Capped mRNA for injection was transcribed from plasmids pCS2-hGRIM-19 and pCS2-CA-NFATc4 by SP6 polymerase (Roche). For morpholino KD experiments, 20 ng of MOs was injected into stage 1 embryos. For the rescue experiments, 500 pg of hG19 or CA-NFATc4 mRNA was injected into two dorsal cells of stage 3 embryos after the MO injection at stage 1. A fluorescent dye FDA was coinjected with mRNA to monitor the correct delivery of mRNA into the dorsal region.
Luciferase reporter assay.
For embryos injected with 20 ng of MOs at stage 1, two cells of each embryo at stage 2 were coinjected with luciferase reporter construct pNFAT-TA-luc (40 pg/blastomere), purchased from BD Bioscience, and Renilla control plasmid (20 pg/blastomere). For each sample, three pools containing four whole embryos each were collected at stages indicated in Fig. 5A and homogenized in passive lysis buffer (Promega), and dual luciferase reporter assays were performed as described previously (26). To check NFAT activity in the cardiac region, tissues from the cardiac region of stage 28 embryos were removed by microdissection using an eyebrow knife. For each sample, three pools containing six cardiac tissues each were collected for luciferase assays. The cultured cells were transfected using Lipofectamime 2000. For Jurkat cells, after transfection with pNFAT-TA-luc and Renilla control plasmids for 16 h, 2 μg/ml of anti-CD3 and 1 μg/ml of anti-CD28 antibodies or 10 ng/ml PMA and 1 μM ionomycin were added to the medium, and the mixture was incubated for 8 h before harvesting for the luciferase assay.
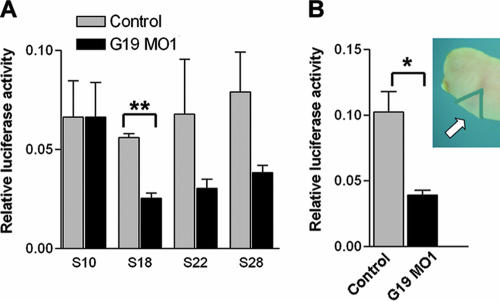
FIG. 5.
Depletion of XGRIM-19 inhibits NFAT activity. (A) Whole embryos injected with control or MO1 together with pNFAT-TA-luc reporter and control Renilla luciferase construct were harvested from different stages (as indicated), and a dual luciferase assay was performed. The average levels of luciferase activity are shown in the graph, with error bars representing the standard deviations of the means from three experiments. Activities in control and GRIM-19 KD show significant difference at stage 18 (**, P < 0.01), but not at stage 22 and 28, likely due to large variations. (B) The NFAT activity from the tissue of cardiac region (indicated by an arrow in the upper right panel) was measured at stage 28 using the same method as that described for panel A (P < 0.05).
siRNA.
KD of GRIM-19 and NDUFS3 by small interfering RNA (siRNA) in HeLa cells was performed using pSUPER.neo (OligoEngine) as described previously (19). The same methods were used to make KD cell lines in Jurkat cells except that 800 μg/ml of G418 was used for selection. Single clones were picked and expanded, and the KD efficiency was examined by Western blotting. Four clones displaying efficient KD of GRIM-19 or NDUFS3 were pooled and used for further experiments.
Intracellular calcium measurement.
HeLa cells (3 × 106) were washed and incubated in sample medium containing 2.5 μM Fura2-AM and 0.1% F-127 at 37°C for 30 min, followed by washing and deesterification in sample medium for 30 min. The cells were washed, resuspended in the sample medium, and transferred into a stirred, temperature-controlled (37°C) cuvette placed in a Shimadzu RF-5301PC spectrofluorophotometer (Shimadzu). Data were collected using dual-wavelength excitation at 340 and 380 nm and an emission wavelength of 510 nm at a frequency of 1 Hz. The sample media contained 25 mM HEPES (pH 7.4), 140 mM NaCl, 2 mM CaCl2, 5 mM KCl, and 1 mM MgCl2 in the presence of 5 mM d-glucose or 2 mM pyruvate.
RT-PCR.
RT-PCR was performed using a One-Step RT-PCR kit (QIAGEN). Primers for RT-PCR are listed in Table S1 in the supplemental material.
Mitochondrial complex I OXPHOS assay.
Embryos at stage 28 were collected and homogenized in a solution containing 750 mM 6-aminocarproic acid, 50 mM BisTris (pH 7.0), and 5 μl of 10% dodecylmaltoside. The lysate was centrifuged, and the supernatant was used for further assays. Separation of mitochondrial complexes was performed with 5% to 18% polyacrylamide gradient blue native polyacrylamide gel electrophoresis (PAGE). In-gel colorimetric reactions for OXPHOS of complex I and complex II were performed by blue native PAGE as described previously (20).
Transmission electron microscopy.
Hearts and somites were removed from Xenopus embryos at stage 45, fixed in 2.5% glutaraldehyde in 0.1 M phosphate-buffered saline overnight at 4°C, and further processed for staining, embedding, and sectioning as described previously (20). The ultrastructure of heart and skeletal muscle was examined with a Jeol JEM1010 transmission electron microscope.
Statistical analysis. The results were analyzed with the chi-square test or Mann-Whitney test and the Student t test. A P value of ≤0.05 was considered to be significant.
Nucleotide sequence accession number.
The sequence of X. laevis GRIM-19 was submitted to the GenBank database under accession number EF412965.
RESULTS
Cloning and expression pattern of XGRIM-19 in X. laevis.
Two clones showing identical nucleotide sequences were isolated from the X. laevis oocyte library and named XGRIM-19. Similar to the other species, XGRIM-19 encodes an open reading frame containing 144 amino acids and displays 92%, 72%, and 72% amino acid identity with GRIM-19 from X. tropicalis, mouse, and human, respectively (Fig. 1A). The expression pattern of XGRIM-19 during embryogenesis was examined by whole-mount in situ hybridization. The XGRIM-19 expression was detected in the animal cap and marginal zone of embryos, representing the future ectoderm and mesoderm, respectively, at the gastrulation stage (stage 10). During organogenesis, XGRIM-19 is specifically expressed in tissues with a high energy expenditure, such as neural, eye, skeletal muscle, kidney, and heart tissues (Fig. 1B). This observation is consistent with the function of GRIM-19 in MRC. The XGRIM-19 mRNA level was very low in the blastula stage (stages 5 to 9) and increased gradually after gastrulation (stages 11 to 40), with high levels at stages 30 and 40 (Fig. 1C). However, the XGRIM-19 protein, probably representing the maternal XGRIM-19 protein, can be detected from stages 5 to 15. The levels of XGRIM-19 protein also increased from stages 20 to 40 (Fig. 1D). These results may reflect a minimum requirement for the mitochondrial OXPHOS at the early stage of embryogenesis and an increased demand for it at the later stages.
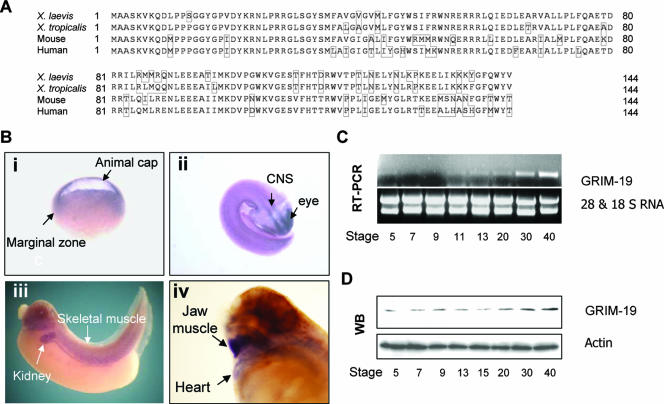
FIG. 1.
Cloning and expression pattern of XGRIM-19 in X. laevis. (A) Comparison of GRIM-19 amino acid sequences between X. laevis, X. tropicalis, mouse, and human. Amino acid numbers are indicated, and the nonconserved amino acids are highlighted with boxes. (B) Whole-mount in situ hybridization of XGRIM-19 in Xenopus embryos. XGRIM-19 mRNA was detected in the animal cap and the marginal zone of embryos at stage 10, as indicated in panel i. During organogenesis, XGRIM-19 mRNA was strongly expressed in the central nervous system (CNS) and eyes at stage 25 (ii), skeletal muscle and kidney at stage 35 (iii), and jaw muscle and heart (iv) at stage 42, as indicated. (C) XGRIM-19 mRNA in the different developmental stages was detected by RT-PCR with 28S and 18S RNA as loading controls. (D) XGRIM-19 protein level was detected by Western blot analysis (WB) using anti-mouse GRIM-19 antibody. The blot was reprobed with antiactin as a control.
KD of XGRIM-19 impairs MRC complex I activity in Xenopus embryos.
To investigate the role of mitochondrial OXPHOS in early embryonic development, we knocked down XGRIM-19 with two specific antisense MOs to block its translation in Xenopus embryos. MO1 targets the 5′ untranslated region of XGRIM-19 mRNA, and MO2 targets the sequence from the start codon (first ATG). Both MO1 and MO2 inhibited translation of XGRIM-19 mRNA in the in vitro translation experiment, whereas the control MO (designated C), consisting of a nucleotide sequence similar to that of MO1 but with four point mutations, had no effect (Fig. 2A). MO1 and MO2 showed similar phenotypes in vivo, but MO1 worked more efficiently than MO2 (data not shown) and was used in most of our experiments. When MO1 was injected into Xenopus embryos at stage 1, a decrease in GRIM-19 protein was observed at stage 15 (data not shown), and GRIM-19 was almost completely knocked down at stages 28 and 45 (Fig. 2B, lanes 2 and 5). The effect of MO1 on the complex I enzymatic activity was also evaluated with the blue native PAGE assay described previously (20). At stage 28, ∼80% of the complex I activity was inhibited in the XGRIM-19 KD embryos in comparison with the control embryos. However, the complex II activity was largely unaffected, and it was used as a control (Fig. 2C).
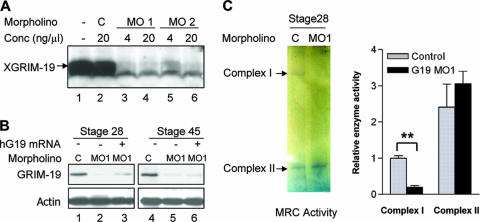
FIG. 2.
KD efficiency of XGRIM-19 and its effect on complex I activity. (A) XGRIM-19 MOs inhibit translation of XGRIM-19 mRNA in vitro. The pcDNA3-XGRIM-19 construct was transcribed and translated in vitro in the presence of XGRIM-19 MO1, MO2, or the control MO (C) at the indicated concentrations (Conc). (B) XGRIM-19 MO1 inhibits XGRIM-19 protein expression in vivo. Xenopus embryos were injected with control MO (lanes 1 and 4) or XGRIM-19 MO1 (lanes 2 and 5) at stage 1. Embryos were harvested at the indicated stages, and total lysates were prepared for Western blot analysis with anti-GRIM-19 antibody. For the rescue experiment, the hG19 mRNA was injected into two dorsal cells at stage 3 after injection with MO1 at stage 1 (lanes 3 and 6). (C) KD of GRIM-19 impairs complex I activity. The representative activities of complex I and complex II were assayed in the control (C) and XGRIM-19 KD (MO1) embryos at stage 28 by in-gel OXPHOS assay (left panel). Enzymatic activities of complex I and II in the in-gel assay were quantified by densitometry. The graph depicts the average of complex I or II activity from three independent experiments with standard deviations (right panel). **, statistical significance at a P value of <0.01, as determined by paired t test analysis.
KD of XGRIM-19 causes a heart defect in Xenopus embryos.
After injection with MO1, the embryos exhibited retarded growth, indicated by their smaller size (data not shown). The hindbrain, eyes, and skeletal muscles were developed with abnormal morphologies, which are very similar to the multisystem syndromes of human mitochondrial disease (see Fig. S1 in the supplemental material). Strikingly, the most severe defect occurred in heart development. In comparison with the normal heart shown in the control embryos (Fig. 3A) (see Video S2 in the supplemental material), no heart formation was observed for more than 70% of the XGRIM-19 KD embryos (Fig. 3A, yellow bar), although sometimes a beating nub was visible in the pericardial cavity (see Video S1 in the supplemental material). Only 20% of the XGRIM-19 KD embryos formed hearts, but with an abnormal morphology and a much smaller size than those of the control embryos (Fig. 3A, pink bar). There were a few or no circulating blood cells in heart chambers or blood vessels observed under the microscope in the GRIM-19 KD embryos. Histological study further revealed that in comparison to the well-developed hearts with intact ventricles and atria in the control embryos at stage 45 (Fig. 3C), most hearts of the XGRIM-19 KD embryos failed to undergo looping and retained their linear conformation without atrial and ventricular chamber formation (Fig. 3F). This result indicates that these embryos failed to develop beyond a very early stage in heart formation. Although a small percentage of the embryos seemed to undergo heart looping, the hearts were smaller and no well-formed atria or ventricles were observed (Fig. 3E). To confirm that the phenotype is due to the KD of XGRIM-19, hG19 mRNA was injected into two dorsal cells of the XGRIM-19 KD embryos at stage 3. The Xenopus-specific MO1 should have no effect on the hG19 mRNA owing to the sequence divergence between these two species. Injection with hG19 mRNA rescued more than 50% of the heart-deficient embryos and also restored blood circulation. However, enlarged hearts were observed for most of the rescued embryos (Fig. 3A, light blue bar) (see Video S3 in the supplemental material). Histological analysis confirmed that hG19 restored the heart structure, which was accompanied by heart enlargement (Fig. 3D). This could be due to dilation of the atria and a defect of the atrioventricular valves, in comparison to the control embryos that displayed intact atrioventricular valves (Fig. 3C). GRIM-19 protein levels at stage 28 were partially restored in the hG19-rescued group (Fig. 2B, lane 3), demonstrating the uptake and expression of hG19 mRNA. However, the GRIM-19 protein level did not show an increase at stage 45 (Fig. 2B, lane 6), possibly due to the eventual degradation of the hG19 mRNA.
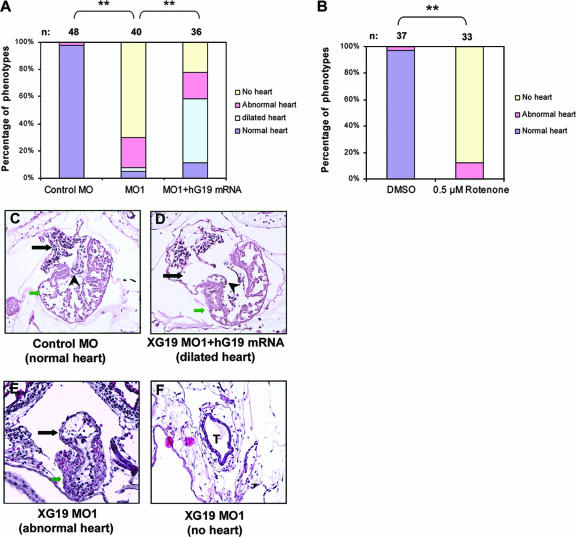
FIG. 3.
KD of XGRIM-19 causes heart defect in Xenopus embryos. (A and B) Statistical data showing heart defects in control, XGRIM-19 MO1, or XGRIM-19 MO1 with hG19 mRNA rescue (A) and 0.5 μM rotenone or DMSO-treated WT Xenopus embryos (B). Numbers (n) of treated embryos are indicated. Heart development was examined under the microscope at stage 45 and grouped as “normal,” “abnormal,” “no heart,” or “dilated,” as indicated by different colors. Percentages of embryos displaying different phenotypes are presented as bars (**, P < 0.01). (C to F) Histological analysis of heart morphology in control (C), XGRIM-19 MO1-injected (E and F), and human GRIM-19 mRNA-rescued (D) embryos at stage 45. The numbers of embryos examined in each group were 4, 4, and 3, respectively. Transverse sections of the heart were examined, and only one set of embryos is represented. Black arrows indicate the atria, green arrows indicate ventricles, “T” indicates the heart tube, and the arrowhead indicates heart valves.
To further confirm that the heart deficiency in XGRIM-19 KD embryos was due to a depletion of MRC complex I, an inhibitor of complex I, rotenone, was incubated with the WT Xenopus embryos from stage 12, a point after gastrulation and the onset of organogenesis. This treatment led to a heart deficiency almost identical to that seen with the GRIM-19 KD embryos. These embryos exhibited phenotypes of either an abnormal heart (16%) or no heart (84%). In comparison, DMSO alone, a solvent of rotenone, did not result in heart deficiency (Fig. 3B). These results confirm that the heart defects had resulted from complex I deficiency.
KD of XGRIM-19 downregulates cardiac gene expression and NFAT activity.
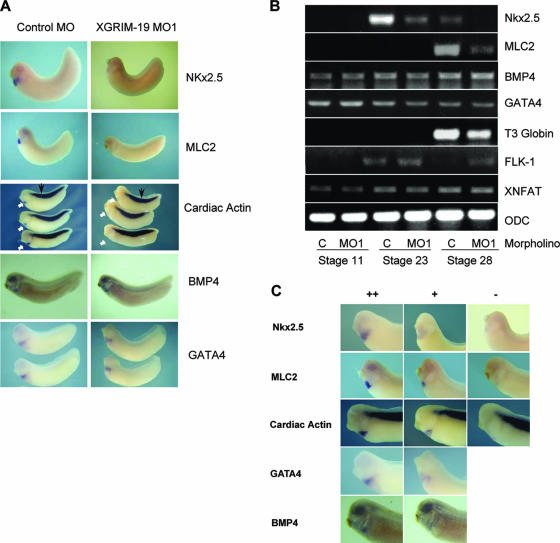
The effect of XGRIM-19 in cardiogenesis was further examined by expression of several cardiac differentiation markers. As shown in Fig. 4A, expression levels of Nkx2.5, myosin light chain 2 (MLC2) and cardiac actin were decreased in cardiac tissue of the XGRIM-19 KD embryos at stage 28. However, the cardiac actin expressed in somites was unaffected by KD of XGRIM-19. Expression of bone morphogenetic protein 4 (BMP4) and GATA binding protein 4 (GATA4), which are also implicated in the initiation of heart development, remained unaffected in the cardiac region (Fig. 4A). Consistent with the in situ hybridization results, RT-PCR analysis also showed decreased expression of Nkx2.5 at stages 23 and 28 and of MLC2 at stage 28 in XGRIM-19 KD embryos (Fig. 4B). The blood cell marker T3 globin was also decreased. Expression of BMP4 and GATA4 was not affected. FLK-1, a blood vessel marker, was slightly increased in the XGRIM-19 KD embryos at stage 28. As a control, the expression of ornithine decarboxylase remained the same in both control and XGRIM-19 KD embryos (Fig. 4B). Three sets of images representing normal, reduced, or the absence of gene expression detected by in situ hybridization are shown in Fig. 4C, and the statistical data for the gene expression in the control and GRIM-19 KD embryos based on this classification are given in Table 1. Together, these results suggest that specific pathway(s) and genes are affected by GRIM-19 KD during cardiomyogenesis.
FIG. 4.
Depletion of XGRIM-19 downregulates expression of several cardiac genes and NFAT activity. (A) Whole-mount in situ hybridization showing the expression of Nkx2.5, MLC2, cardiac actin, BMP4, and GATA4 in the control (left panels) or the XGRIM-19 MO1 (right panels) embryos at stages 28 (lateral views). The cardiac actin expression in the heart region is indicated by white arrows and in the somite by black arrows. (B) Various gene expression levels in the embryos injected with control MO (C) or XGRIM-19 MO1 at different stages, as detected by RT-PCR. ODC, ornithine decarboxylase. (C) Three sets of images representing normal (++), reduced (+), or absence of (−) gene expression. Statistical data on heart-specific gene expression in control and XGRIM-19 KD embryos are summarized in Table 1.
TABLE 1.
Specific gene expression in control and XGRIM-19 KD embryosa
| mRNA (embryo stage) | Control MO
|
XGRIM19 MO1
|
||||||
|---|---|---|---|---|---|---|---|---|
| − (%) | + (%) | ++ (%) | n | − (%) | + (%) | ++ (%) | n | |
| Nkx2.5* (22) | 0 | 10 | 90 | 10 | 0 | 75 | 25 | 8 |
| Nkx2.5** (28) | 0 | 4 | 96 | 57 | 21 | 68 | 11 | 47 |
| MLC2** (28) | 0 | 0 | 100 | 12 | 19 | 75 | 6 | 16 |
| Cardiac actin** (28) | 0 | 4 | 96 | 26 | 20 | 65 | 15 | 20 |
| BMP4 (28) | 0 | 23 | 77 | 22 | 0 | 14 | 86 | 21 |
| GATA4 (28) | 0 | 7 | 93 | 14 | 0 | 26 | 74 | 19 |
Summary of statistical data on heart-specific gene expression (Nkx2.5, MLC2, cardiac actin, BMP4, and GATA4) at different stages in embryos injected with either control MO or XGRIM-19 MO1. −, no expression; +, reduced expression; ++, normal expression (as indicated by the images shown in Fig. 4); n, total number of embryos used in whole-mount in situ hybridization; ** and *, P < 0.01 and P < 0.05, respectively, for the differences in gene expression between control and XGRIM-19 KD embryos.
The transcription factor NFAT family contributes to the development of the immune, heart, vasculature, and nervous systems. Mouse NFATc1 to NFATc4 homologues are found in the GenBank database of X. tropicalis, but only one that is homologous to human NFATc3 has been cloned for X. laevis (37). We examined the expression level of XNFAT and did not find any obvious change in the XGRIM-19 KD embryos (Fig. 4B). In a search of the NCBI X. laevis database, an unknown protein (MGC82799) was found to be highly homologous to the X. tropicalis NFATc1. The mRNA expression of this protein also did not show any obvious change in the XGRIM-19 KD embryos (see Fig. S2 in the supplemental material).
We then tested the activity of endogenous XNFAT(s) in both whole embryos and the cardiac region by injecting a reporter plasmid, pNFAT-TA-luc, which contains an interleukin 2 promoter, into the Xenopus embryos. The results showed that in comparison to the control embryos, the NFAT activity in the GRIM-19 KD embryos was significantly reduced in both the whole embryos at stage 18 (Fig. 5A) and the cardiac region at stage 28 (Fig. 5B).
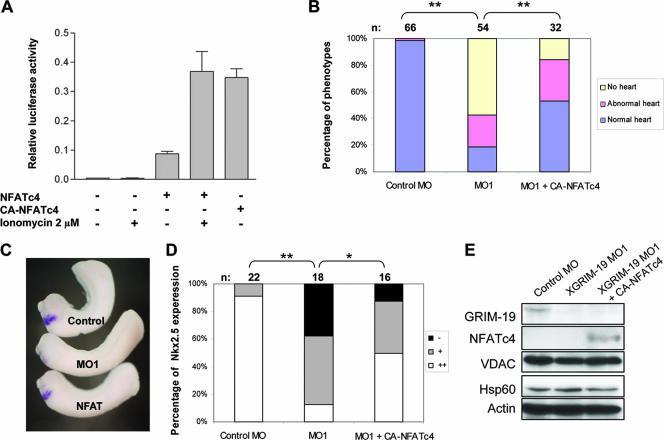
CA-NFATc4 rescues the heart defect in XGRIM-19 KD embryos.
Our results suggested that the NFAT activity is affected by downregulation of XGRIM-19. We next examined whether an active form of NFAT could rescue the heart defects caused by XGRIM-19 KD. One NFAT family member, NFATc4, is reported to regulate myocardium formation in mice (4), and transgenic mice expressing a constitutive active form of NFATc4 exhibit heart hypertrophy (30). We constructed a CA-NFATc4 and measured its transcriptional activity in mammalian cells by luciferase assays. Activity of the WT NFATc4 was low in unstimulated cells and could be increased by ionomycin, a calcium ionophore that triggers an increase in intracellular Ca2+. In contrast, CA-NFATc4 showed constitutive activity in the absence of ionomycin, and the level was comparable to the that of the stimulated WT NFATc4 (Fig. 6A). We subsequently injected CA-NFATc4 mRNA into two dorsal cells of Xenopus embryos at stage 3, after injecting MOs at stage 1. Less than 20% of the XGRIM-19 KD embryos displayed normal heart development, but injection with CA-NFATc4 mRNA increased this number to more than 50% (Fig. 6B). Conversely, embryos with severe heart defects decreased from 60% to 18% (Fig. 6B). In addition, the development of atria, ventricles, and functional blood vasculature was visible in most of the CA-NFATc4-rescued embryos (see Video S4 in the supplemental material). Furthermore, the active NFATc4 could also restore Nkx2.5 expression in the GRIM-19 KD embryos, as shown by in situ hybridization (Fig. 6C). The statistical data indicated that 91% of the control embryos expressed normal level of Nkx2.5, whereas only 11% of GRIM-19 KD embryos expressed Nkx2.5 normally; the rest of the embryos displayed either low or no expression. Introducing CA-NFATc4 mRNA to the GRIM-19 KD embryos increased the percentage of normal-level-expressing embryos from 11% to 50% and decreased the percentage of nonexpressing embryos from 33% to 13% (Fig. 6D). In Fig. 6E, the XGRIM-19 KD efficiency and the expression of CA-NFATc4 protein are shown. Other mitochondrial proteins used as controls, such as VDAC and heat shock protein Hsp60, remained unaffected by either KD of XGRIM-19 or expression of CA-NFATc4.
FIG. 6.
Rescue of heart deficiency by CA-NFATc4 in XGRIM-19 KD Xenopus embryos. (A) Comparison of NFATc4 and CA-NFATc4 activity in MCF-7 cells. MCF-7 cells were cotransfected with the pNFAT-TA-luc construct and either pcDNA3-NFATc4 or pcDNA3-CA-NFATc4. Cells were either left untreated or were treated with ionomycin to increase intracellular calcium levels. Dual luciferase assays were performed in triplicate, and the average luciferase activity from three independent experiments is shown, with error bars representing the standard deviations of the means. (B) CA-NFATc4 partially rescues the heart defect in XGRIM-19 KD embryos. Embryos were injected with control MO, XGRIM-19 MO1, or XGRIM-19 MO1 and CA-NFATc4 mRNA, as indicated. The percentages of embryos displaying heart phenotypes of normal, abnormal, and no heart are indicated as bars, and the number of embryos is shown on top (n) (**, P < 0.01). (C) Embryos were injected with control MO, XGRIM-19 MO1, or XGRIM-19 MO1 and CA-NFATc4 mRNA. Nkx2.5 mRNA was detected by in situ hybridization in these embryos at stage 28. (D) The percentages of embryos displaying an absence of (−), weak (+), or normal (++) Nkx2.5 expression are indicated as bars, and the number of embryos is shown on top (n) (**, P < 0.01; *, P < 0.05). (E) Gene expression in embryos injected with control MO, XGRIM-19 MO1, or XGRIM-19 MO1 and CA-NFATc4 mRNA. Total embryo lysate at stage 28 was harvested and analyzed by Western blotting with antibodies against GRIM-19, NFATc4, VDAC, Hsp60, and actin, as indicated on the left.
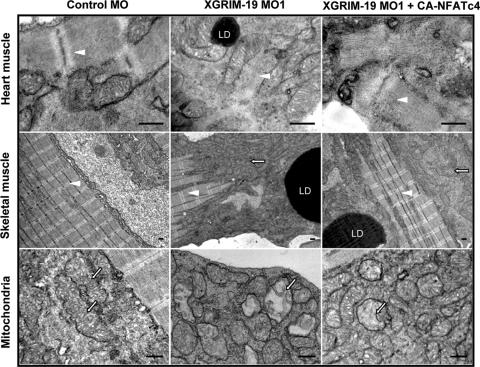
NFATc4 rescues the defects of sarcomere formation in the heart muscles.
NFAT was reported to regulate myocardium formation by affecting sarcomeric protein expression (38). We checked the ultrastructure of heart and skeletal muscles in the control, XGRIM-19 KD, and CA-NFATc4-rescued embryos by electron microscopy. Heart muscles from control embryos exhibited normal sarcomere morphology with alternating rows of M and Z lines. In contrast, the sarcomere structure was disorganized and lacked clear M and Z lines in XGRIM-19 KD embryos and, in most cases, only scattered individual filaments were observed. However, injection with CA-NFATc4 mRNA partially rescued the sarcomere defect in the heart muscle of XGRIM-19 KD embryos by showing restoration of M and Z lines (Fig. 7, top panels). Statistic data suggested that differences between the sarcomere structure of the XGRIM-19 KD and that of the CA-NFATc4-rescued embryos were significant (see Fig. S3 in the supplemental material). In the skeletal muscle of the GRIM-19 KD embryos, however, the sarcomere structure was retained, although its size was decreased (Fig. 7, middle panels). On the other hand, a dramatic increase of mitochondrial number—a hallmark of mitochondrial myopathy (10)—was observed for both XGRIM-19 KD and CA-NFATc4-rescued embryos. The mitochondrial morphology was also abnormal. Some of them were rounded and swollen, with disorganized cristae inside the matrix (bottom panels). In addition, we also observed formation of lipid droplets, which is a common phenotype occurring in human mitochondrial diseases, in both heart and skeletal muscles of the XGRIM-19 KD and CA-NFATc4-rescued embryos. Taken together, these results indicate that CA-NFATc4 is able to rescue the defects in sarcomere formation in the heart muscles, but not the defects in mitochondria.
FIG. 7.
Ultrastructure of heart and skeletal muscle from Xenopus embryos at stage 45. Embryos injected by control MO, GRIM-19 MO1, or GRIM-19 MO1 plus CA-NFAT were examined with a transmission electron microscope. XGRIM-19 KD causes severely perturbed sarcomeres in heart muscle (white arrowhead, top middle panel). Mitochondrial proliferation (white arrow, middle panel in middle row) and morphological abnormalities in skeletal muscle were observed (white arrow, bottom middle panel). CA-NFATc4 can rescue the sarcomere formation in heart muscle (top right panel). Lipid droplets (LD) are commonly visible in XGRIM-19 KD muscles. Scale bars, 0.5 μM.
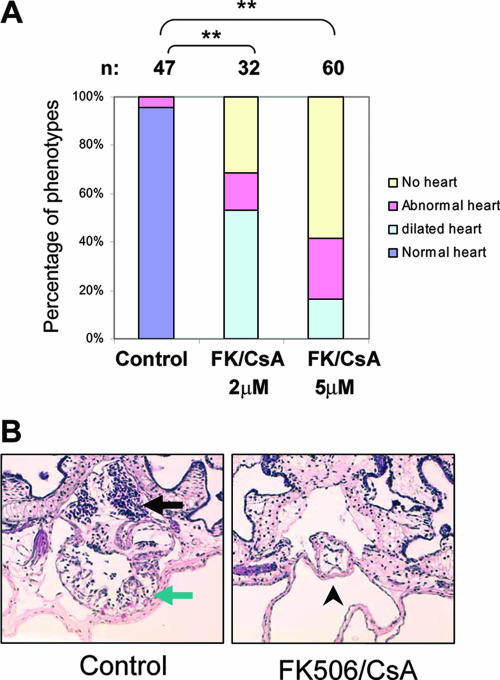
Inhibition of calcineurin also results in early heart defects similar to those of GRIM-19 KD embryos.
To further demonstrate that the function of MRC in the early heart development is through regulation of NFAT in Xenopus, we treated the embryo with calcineurin inhibitor FK506 and CsA from stage 15. The heart development was examined under a microscope at stage 42. As shown in Fig. 8A, treatment with 5 μM of FK506 and CsA caused heart formation to fail in 58% of the total embryos. The heart remained as a heart tube and failed to loop (Fig. 8B), which is identical to the phenotype seen with the GRIM19 KD and rotenone-treated embryos (Fig. 3). The effect of the inhibitors was dose dependent, as treatment with 2 μM of FK506 and CsA had a similar but less severe impact on heart development. In this group, in addition to the “no heart” and “abnormal heart” phenotypes, about 50% of embryos had a dilated heart. This could be due to the heart valve defects as seen with the GRIM-19 KD embryos partially rescued by hG19 mRNA (Fig. 3D). These data demonstrate the critical role of NFAT in mediating early heart development in Xenopus.
FIG. 8.
Inhibition of calcineurin-NFAT activity results in early heart defect in Xenopus embryos. (A) Statistical data showing heart defects in the Xenopus embryos treated with solvent (control) or 2 μM or 5 μM of FK506 and CsA. The number (n) of treated embryos is indicated. Heart development was examined under the microscope at stage 42 and grouped as “normal,” “abnormal,” “no heart,” or “dilated” heart, indicated by different colors. The percentages of embryos displaying different phenotype are presented as bars (**, P < 0.01). (B) Transverse sections represent the normal heart in control or the heart tube in the Fk506/CsA-treated embryos at stage 42. Black arrow, atrium; green arrow, ventricle; arrowhead, heat tube.
To further explore the possible effect of MRC/NFAT on later heart development in Xenopus, embryos from stage 37 to 40, a window for heart valve initiation, were incubated with a complex I inhibitor, rotenone. A deficiency in atrioventricular valve formation was detected (see Fig. S4 in the supplemental material). Interestingly, this defect was morphologically indistinguishable from that in the embryos incubated with the NFAT inhibitor FK506 at stages 37 to 43. The data therefore also reveal the role of MRC in the heart valve formation in Xenopus.
KD of XGRIM-19 or NDUFS3 impairs calcium mobilization and calcium-induced NFAT activity.
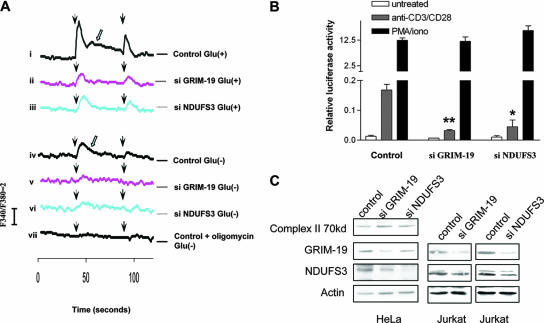
Intracellular calcium is the key regulator of NFAT. We examined whether depletion of MRC complex I disturbs calcium signaling. MRC-deficient stable cell lines were created by knocking down GRIM-19 or NDUFS3, another complex I subunit, with siRNA in HeLa cells, and their effect on calcium regulation was examined by measuring changes in the intracellular calcium concentration, [Ca2+]i. Both KD and control cells were grown in medium supplemented with glucose and treated with pulsatile stimulation with a 50-s application of an InsP3-generating agonist, His. The basic calcium levels in control and KD cells were not obviously different from one another (∼150 nM). However, the control cells responded to His with a rapid and transient increase in [Ca2+]i (Fig. 8A, panel i), which indicates amounts of Ca2+ being released from the intracellular stores, such as ER, via its receptors, InsP3Rs. Following the rapid and transient Ca2+ spike, elevated levels of [Ca2+]i were sustained, which probably represents the Ca2+ influx through CRAC on the PM, as triggered by ER Ca2+ depletion (panel i). This persistent Ca2+ signal is necessary to activate NFAT (41). A second application of His resulted in another spike in Ca2+ concentrations, at lower levels than the first, likely due to an intrinsic desensitization of the His receptor. In contrast to the control cells, both GRIM-19 and NDUFS3 KD cells responded to the His stimulation poorly, with a small [Ca2+]i elevation which was insufficient for stimulation of a subsequent calcium entry, as shown by lack of the sustained [Ca2+]i elevation (panels ii and iii). Although in most cells, ATP is generated mainly by MRC via OXPHOS, it can also be produced by glycolysis. To ascertain the specificity of MRC-produced ATP in the modulation of calcium signals, we checked the calcium mobilization without glucose but with pyruvate in the medium. The cells in such a medium can produce ATP only through OXPHOS. The results showed an increase of [Ca2+]i triggered by His in the control cells, but to a lesser extent than with the same cells grown in the presence of glucose. In addition, a lower induction of calcium influx was observed (panel iv). KD of GRIM-19 or NDUFS3 further decreased the calcium release triggered by His (panels v and vi). In comparison, the addition of oligomycin, which inhibits MRC complex V to prevent ATP synthesis, completely impaired the intracellular Ca2+ release and subsequent Ca2+ entry (panel vii). These results suggest that both OXPHOS and glycolysis are important for calcium mobilization. Since cellular ATP is produced mainly via mitochondrial OXPHOS in vivo, it is conceivable that MRC plays a more important role in the control of Ca2+ mobilization in vivo than in the cultured cells.
To investigate whether MRC deficiency can also affect calcium-dependent NFAT activity, we knocked down either GRIM-19 or NDUFS3 in a human T-lymphocyte line, Jurkat, and tested the endogenous NFAT activity. Anti-CD3 and CD28 were used to activate T-cell receptor-NFAT signaling through the induction of intracellular calcium release and the subsequent activation of CRAC (23, 36). As shown in Fig. 8B, treatment of control Jurkat T cells with a combination of anti-CD3 and CD28 antibodies resulted in an obvious increase of NFAT activity. However, the same treatment failed to raise NFAT activity to similar levels in the GRIM-19 or NDUFS3 KD cells. A calcium ionophore, ionomycin, activated NFAT signaling by direct induction of Ca2+ influx via alteration of the PM permeability for calcium, without requiring any Ca2+ release or a subsequent influx in CRAC-dependent Ca2+ (23). Treatment with ionomycin and PMA drastically stimulated NFAT activity in both control and KD cell lines, indicating that the defect was not due to loss of NFAT function per se in the KD cells. The KD efficiency was confirmed in HeLa and Jurkat cells (Fig. 8C). While GRIM-19- and NDUFS3-specific siRNAs downregulated their respective protein levels, a complex II 70-kDa subunit was not affected, indicating the specificity of the KD. Together, these results reveal the roles of MRC in regulating intracellular Ca2+ dynamics and the Ca2+-dependent NFAT activity.
DISCUSSION
Xenopus as a model for studying MRC functions in early embryonic development.
The specific role of MRC in the control of embryonic development is difficult to dissect in mammalian systems, because embryonic lethality occurs as a result of the MRC dysfunction. The amphibian embryo provides a useful model for studying the early embryonic developmental events, especially those of heart development, since amphibians do not depend on cardiac function for much of their embryonic development (29). In X. laevis, despite severe defects in heart formation and development, the embryos with XGRIM-19 KD remained alive for at least 2 weeks. Furthermore, in contrast to the GRIM-19 knockout in mice, KD of XGRIM-19 in Xenopus did not lead to early embryonic lethality, perhaps due to the presence of maternal XGRIM-19, which is sufficient for the energy demand in the earlier stages of embryonic development. Therefore, this system allows us to study the MRC functions in the early embryonic development and establish the heart as one of the first major targets controlled by MRC in the organogenesis.
Specific functions of the MRC in the development of the heart.
In Xenopus embryos, the heart is the first organ to develop (32). Heart induction begins during gastrulation. In response to the signals from underlying endodermal cells, the precardiac mesoderm gives rise to bilaterally symmetrical heart primordia (14, 32). The two heart primordia migrate to fuse into a single heart tube that undergoes looping, segmentation, and valve formation. The cardiomyocytes undergo high levels of proliferation as the tube transforms into the three-chambered heart (33). The heart tube is formed at stage 32 and starts to loop at stage 33 (29). Most XGRIM-19 KD embryos remain at the heart tube stage, indicating that the defects occur from the heart tube looping. Interestingly, this phenotype is similar to that reported for Nkx2.5 knockout mice (27, 40). Nkx2.5 starts to express in the cardiac crescent from stage 13 and is the first marker expressed in the specified cardiomyocyte (7, 21, 24). We found that Nkx2.5 expression was inhibited in the XGRIM-19 KD embryos at stages 22, 23, and 28 (Fig. 4), suggesting that the MRC may be involved in the maintenance of Nkx2.5 expression. However, it is not clear whether the MRC plays any role in heart initiation and cardiomyocyte specification. We examined the sections crossing the cardiac region of the stage 28 embryos by in situ hybridization with Nkx2.5 as a marker. It seems that GRIM-19 KD does not affect the cardiac precursor cell specification, but the Nkx2.5 level in these cells was obviously reduced (see Fig. S5 in the supplemental material). Furthermore, the Nkx2.5 expression at stage 15 was not shown differently in the XGRIM-19 KD embryo compared to the control embryos. These observations seem to suggest that the MRC is not essential for heart initiation and cardiomyocyte specification. However, due to the presence of the maternal XGRIM-19 protein, which is not affected by GRIM-19 morpholino at the early stages, a function of the MRC in the early stage of heart development cannot be excluded. We have detected the expression of XGRIM-19 in the mesoderm, but it was barely detectable in the endoderm at the gastrulation stage of the WT embryos (Fig. 1B, i), a result which also provides support for the role of the MRC in the heart initiation. In addition to the early effect, complex I inhibitor exhibits an inhibitory effect on atrioventricular valve formation (see Fig. S4 in the supplemental material), suggesting that the MRC also has a function in the later stages of heart development.
In addition to the heart defect, knocking down XGRIM-19 causes less-severe defects in other tissues, such as the brain, eyes, and muscles, although embryonic convergence and extension and the general organogenesis in GRIM-19 KD embryos appear to be normal (data not shown). GRIM-19 KD embryos exhibit extensive apoptosis in neural tissues but not in the muscles (see Fig. S1B in the supplemental material), suggesting that mitochondrial ATP production is crucial for maintaining cell survival in neural tissues. As for the muscular tissues, XGRIM-19 KD affects cardiac muscles more severely than skeletal muscles (Fig. 7). In agreement with this result, GRIM-19 KD caused a decrease in the cardiac actin expression only in the heart area, not in the skeletal muscles (Fig. 4A). These results suggest that the MRC has profound effects on the early embryonic development by governing complex and specific signaling pathways.
NFAT is a key factor regulated by MRC-triggered Ca2+ signaling in Xenopus heart development.
In this study, we found that activation of XNFAT(s) was repressed in the XGRIM-19 KD embryos, and the expression of a CA mouse NFATc4 in these embryos allowed the MRC-mediated upstream regulatory signaling events to be bypassed and rescued the heart developmental defects (Fig. 6B) (see Video S4 in the supplemental material). Furthermore, inhibition of calcineurin with FK506 and CsA led to a heart deficiency similar to those seen with the GRIM-19 KD embryos (Fig. 8). These data demonstrate the essential role of NFAT, which functions downstream of MRC, in the early heart development in Xenopus. However, such early heart defects were not observed for the NFAT-deficient mice. This result suggests that NFAT may not be required for the early heart development in mouse, and mouse and Xenopus have fundamental differences in requirements for NFAT signaling in early cardiac development. In mouse, NFAT is crucial for heart valve development in two phases. In the first one, NFAT (c2, c3, and c4) in the myocardium is required for the initiation of heart valve morphogenesis. Later, the second wave of calcineurin/NFAT signaling is needed in the endocardium for valve remodeling (6). In our experiment, the inhibition of complex I from stage 37 to 40 (valve initiation stages) led to defects in atrioventricular valve formation (see Fig. S4 in the supplemental material). This phenotype is similar to that caused by the inhibition of calcineurin. These results indicate that the MRC also plays an important role in heart valve formation in Xenopus by regulating NFAT. However, inhibition of Xenopus embryos with the complex I inhibitor rotenone after stage 40 caused extensive death of embryos (data not shown). Thus, it is unknown whether the MRC also affects the second wave of NFAT activity during heart valve remodeling.
We found that Nkx2.5 expression was suppressed in the XGRIM-19 KD embryos but was rescued by injecting active NFATc4 (Fig. 4 and Fig. 6C and D), suggesting a possible regulatory role of NFAT on Nkx2.5. Our preliminary search has revealed six potential NFAT binding sites in the reported cis-regulatory region of the mouse Nkx2.5. Investigation of whether NFAT directly regulates Nkx2.5 expression is ongoing.
Since NFAT activity is controlled via dephosphorylation by calcium-dependent phosphatase calcineurin (8), it is possible that calcium signaling could have been perturbed by KD of GRIM-19. Because of technical difficulties, this could not be tested directly in whole embryos. To overcome this obstacle, GRIM-19- and NDUFS3-deficient cell lines were produced and used to measure intracellular calcium responses. The Ca2+ concentration stimulated by treatment of His was indeed diminished in these two cell lines. Furthermore, the endogenous NFAT activity in response to T-cell receptor-mediated Ca2+ elevation was also abrogated (Fig. 9). These results demonstrate a crucial regulatory effect of MRC complex I in calcium signaling, which is consistent with previous reports showing that MRC function and ATP production are essential for the sperm-triggered calcium oscillation and homeostasis (12, 25).
FIG. 9.
GRIM-19 KD compromises intracellular calcium mobilization and NFAT activity. (A) GRIM-19 and NDUFS3 KD HeLa cells and control cells expressing nonspecific siRNA (si) were stimulated with 2.5 μM His (arrowhead) in sample medium containing 5 mM glucose (i to iii) or 2 mM pyruvate (iv to vii). As a control, 25 μM oligomycin was added to medium to inhibit complex V activity (vii). Calcium mobilization was measured using a luminescence spectrophotometer. The Fura 340/380 ratio indicates the change in [Ca2+]i. The open arrow indicates Ca2+ influx. (B) Control, GRIM-19, and NDUFS3 KD Jurkat cells were transiently transfected with pNFAT-TA-luc and stimulated with either anti-CD3/CD28 or ionomycin/PMA. Endogenous NFAT activity was analyzed by luciferase assay. The difference between NFAT activity in the KD cell lines in comparison to that in the control is statistically significant (**, P < 0.01; *, P < 0.05). (C) Western blots showing the KD efficiency of GRIM-19 and NDUFS3 in HeLa and Jurkat cells.
Mechanism of MRC's regulatory role in heart development.
Based on the current data, we propose a model for the role of the MRC in the regulation of cardiogenesis. Activation of the calcium-calcineurin-NFAT pathway is initiated by hormones and growth factors binding to their specific receptors on the PM that leads to the activation of phospholipase C and catalysis of InsP3 production. InsP3 binds to the receptors on the ER membrane and opens the Ca2+ channels to release the ER-stored Ca2+ into the cytoplasm. The resultant ER calcium depletion leads to a more prolonged cytosolic calcium plateau by triggering the opening of CRAC on the PM. This calcium plateau in turn activates calcium/calmodulin-dependent phosphatase calcineurin and its downstream target NFAT. Activated NFAT translocates to the nucleus, where it stimulates target genes, such as Tnl and TnT (38), which are involved in the contraction of heart muscle. Altogether, our findings reveal a crucial role of mitochondrial MRC in cardiogenesis through the regulation of cytosolic calcium signals and the subsequent calcium-calcineurin-dependent NFAT pathway. This could be achieved by modulating the intracellular calcium release from the ER. ATP production by mitochondrial MRC increases the storage of calcium in the ER via Ca2+ ATPases (SERCA). It may also regulate the sensitivity of InsP3 receptors on the ER (3). Conversely, calcium release from the ER leads to a higher calcium level in mitochondria, which stimulates the Krebs cycle and MRC enzymatic activity and augments ATP production (28). MRC dysfunction perturbs Ca2+ homeostasis, which impairs NFAT activation and causes a heart defect in Xenopus embryos.
Supplementary Material
Acknowledgments
We thank K. Guo and J. Li for histological analysis and C. P. Ng and X. H. Lim for electron microscopy. We are grateful for A. L. Miller's help with calcium signaling. We also thank Y.-J. Jiang and C. P. Lim for critical readings of the manuscript.
This work was supported by the Agency for Science, Technology and Research of Singapore. X. Cao is an adjunct staff member in the Department of Biochemistry, National University of Singapore.
Footnotes
Published ahead of print on 16 July 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Angell, J. E., D. J. Lindner, P. S. Shapiro, E. R. Hofmann, and D. V. Kalvakolanu. 2000. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J. Biol. Chem. 275:33416-33426. [DOI] [PubMed] [Google Scholar]
- 2.Berridge, M. J., M. D. Bootman, and H. L. Roderick. 2003. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4:517-529. [DOI] [PubMed] [Google Scholar]
- 3.Bezprozvanny, I., and B. E. Ehrlich. 1993. ATP modulates the function of inositol 1,4,5-trisphosphate-gated channels at two sites. Neuron 10:1175-1184. [DOI] [PubMed] [Google Scholar]
- 4.Bushdid, P. B., H. Osinska, R. R. Waclaw, J. D. Molkentin, and K. E. Yutzey. 2003. NFATc3 and NFATc4 are required for cardiac development and mitochondrial function. Circ. Res. 92:1305-1313. [DOI] [PubMed] [Google Scholar]
- 5.Carafoli, E. 2002. Calcium signaling: a tale for all seasons. Proc. Natl. Acad. Sci. USA 99:1115-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, C. P., J. R. Neilson, J. H. Bayle, J. E. Gestwicki, A. Kuo, K. Stankunas, I. A. Graef, and G. R. Crabtree. 2004. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell 118:649-663. [DOI] [PubMed] [Google Scholar]
- 7.Cleaver, O. B., K. D. Patterson, and P. A. Krieg. 1996. Overexpression of the tinman-related genes XNkx-2.5 and XNkx-2.3 in Xenopus embryos results in myocardial hyperplasia. Development 122:3549-3556. [DOI] [PubMed] [Google Scholar]
- 8.Crabtree, G. R., and E. N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109(Suppl.):S67-S79. [DOI] [PubMed] [Google Scholar]
- 9.de la Pompa, J. L., L. A. Timmerman, H. Takimoto, H. Yoshida, A. J. Elia, E. Samper, J. Potter, A. Wakeham, L. Marengere, B. L. Langille, G. R. Crabtree, and T. W. Mak. 1998. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 392:182-186. [DOI] [PubMed] [Google Scholar]
- 10.DiMauro, S., E. Bonilla, M. Zeviani, M. Nakagawa, and D. C. DeVivo. 1985. Mitochondrial myopathies. Ann. Neurol. 17:521-538. [DOI] [PubMed] [Google Scholar]
- 11.Duchen, M. R. 2000. Mitochondria and calcium: from cell signalling to cell death. J. Physiol. 529(Pt. 1):57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dumollard, R., P. Marangos, G. Fitzharris, K. Swann, M. Duchen, and J. Carroll. 2004. Sperm-triggered [Ca2+] oscillations and Ca2+ homeostasis in the mouse egg have an absolute requirement for mitochondrial ATP production. Development 131:3057-3067. [DOI] [PubMed] [Google Scholar]
- 13.Fearnley, I. M., J. Carroll, R. J. Shannon, M. J. Runswick, J. E. Walker, and J. Hirst. 2001. GRIM-19, a cell death regulatory gene product, is a subunit of bovine mitochondrial NADH:ubiquinone oxidoreductase (complex I). J. Biol. Chem. 276:38345-38348. [DOI] [PubMed] [Google Scholar]
- 14.Fishman, M. C., and K. R. Chien. 1997. Fashioning the vertebrate heart: earliest embryonic decisions. Development 124:2099-2117. [DOI] [PubMed] [Google Scholar]
- 15.Graef, I. A., F. Chen, L. Chen, A. Kuo, and G. R. Crabtree. 2001. Signals transduced by Ca2+/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell 105:863-875. [DOI] [PubMed] [Google Scholar]
- 16.Hajnoczky, G., L. D. Robb-Gaspers, M. B. Seitz, and A. P. Thomas. 1995. Decoding of cytosolic calcium oscillations in the mitochondria. Cell 82:415-424. [DOI] [PubMed] [Google Scholar]
- 17.Harland, R. M. 1991. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 36:685-695. [DOI] [PubMed] [Google Scholar]
- 18.Hirst, J., J. Carroll, I. M. Fearnley, R. J. Shannon, and J. E. Walker. 2003. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta. 1604:135-150. [DOI] [PubMed] [Google Scholar]
- 19.Huang, G., Y. Chen, H. Lu, and X. Cao. 2007. Coupling mitochondrial respiratory chain to cell death: an essential role of mitochondrial complex I in the interferon-beta and retinoic acid-induced cancer cell death. Cell Death Differ. 14:327-337. [DOI] [PubMed] [Google Scholar]
- 20.Huang, G., H. Lu, A. Hao, D. C. Ng, S. Ponniah, K. Guo, C. Lufei, Q. Zeng, and X. Cao. 2004. GRIM-19, a cell death regulatory protein, is essential for assembly and function of mitochondrial complex I. Mol. Cell. Biol. 24:8447-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komuro, I., and S. Izumo. 1993. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc. Natl. Acad. Sci. USA 90:8145-8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsson, N. G., J. Wang, H. Wilhelmsson, A. Oldfors, P. Rustin, M. Lewandoski, G. S. Barsh, and D. A. Clayton. 1998. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18:231-236. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, R. S. 2001. Calcium signaling mechanisms in T lymphocytes. Annu. Rev. Immunol. 19:497-521. [DOI] [PubMed] [Google Scholar]
- 24.Lints, T. J., L. M. Parsons, L. Hartley, I. Lyons, and R. P. Harvey. 1993. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119:419-431. [DOI] [PubMed] [Google Scholar]
- 25.Liu, L., K. Hammar, P. J. Smith, S. Inoue, and D. L. Keefe. 2001. Mitochondrial modulation of calcium signaling at the initiation of development. Cell Calcium 30:423-433. [DOI] [PubMed] [Google Scholar]
- 26.Lufei, C., J. Ma, G. Huang, T. Zhang, V. Novotny-Diermayr, C. T. Ong, and X. Cao. 2003. GRIM-19, a death-regulatory gene product, suppresses Stat3 activity via functional interaction. EMBO J. 22:1325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyons, I., L. M. Parsons, L. Hartley, R. Li, J. E. Andrews, L. Robb, and R. P. Harvey. 1995. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 9:1654-1666. [DOI] [PubMed] [Google Scholar]
- 28.McCormack, J. G., and R. M. Denton. 1993. The role of intramitochondrial Ca2+ in the regulation of oxidative phosphorylation in mammalian tissues. Biochem. Soc. Trans. 21(Pt. 3):793-799. [DOI] [PubMed] [Google Scholar]
- 29.Mohun, T. J., L. M. Leong, W. J. Weninger, and D. B. Sparrow. 2000. The morphology of heart development in Xenopus laevis. Dev. Biol. 218:74-88. [DOI] [PubMed] [Google Scholar]
- 30.Molkentin, J. D., J. R. Lu, C. L. Antos, B. Markham, J. Richardson, J. Robbins, S. R. Grant, and E. N. Olson. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93:215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nieuwkoop, P. D., and J. Faber. 1994. Normal table of Xenopus laevis. Garland Publishing, Inc., New York, NY.
- 32.Olson, E. N., and M. D. Schneider. 2003. Sizing up the heart: development redux in disease. Genes Dev. 17:1937-1956. [DOI] [PubMed] [Google Scholar]
- 33.Pasumarthi, K. B., and L. J. Field. 2002. Cardiomyocyte cell cycle regulation. Circ. Res. 90:1044-1054. [DOI] [PubMed] [Google Scholar]
- 34.Putney, J. W., Jr., and G. S. Bird. 1993. The signal for capacitative calcium entry. Cell 75:199-201. [DOI] [PubMed] [Google Scholar]
- 35.Rizzuto, R., P. Bernardi, and T. Pozzan. 2000. Mitochondria as all-round players of the calcium game. J. Physiol. 529(Pt. 1):37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salomon, B., and J. A. Bluestone. 2001. Complexities of CD28/B7: CTLA-4 costimulatory pathways in autoimmunity and transplantation. Annu. Rev. Immunol. 19:225-252. [DOI] [PubMed] [Google Scholar]
- 37.Saneyoshi, T., S. Kume, Y. Amasaki, and K. Mikoshiba. 2002. The Wnt/calcium pathway activates NF-AT and promotes ventral cell fate in Xenopus embryos. Nature 417:295-299. [DOI] [PubMed] [Google Scholar]
- 38.Schubert, W., X. Y. Yang, T. T. Yang, S. M. Factor, M. P. Lisanti, J. D. Molkentin, M. Rincon, and C. W. Chow. 2003. Requirement of transcription factor NFAT in developing atrial myocardium. J. Cell Biol. 161:861-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schultz, B. E., and S. I. Chan. 2001. Structures and proton-pumping strategies of mitochondrial respiratory enzymes. Annu. Rev. Biophys. Biomol. Struct. 30:23-65. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka, M., Z. Chen, S. Bartunkova, N. Yamasaki, and S. Izumo. 1999. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 126:1269-1280. [DOI] [PubMed] [Google Scholar]
- 41.Timmerman, L. A., N. A. Clipstone, S. N. Ho, J. P. Northrop, and G. R. Crabtree. 1996. Rapid shuttling of NF-AT in discrimination of Ca2+ signals and immunosuppression. Nature 383:837-840. [DOI] [PubMed] [Google Scholar]
- 42.Wallace, D. C. 1999. Mitochondrial diseases in man and mouse. Science 283:1482-1488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.