Abstract
The hGH cluster contains a single human pituitary growth hormone gene (hGH-N) and four placenta-specific paralogs. Activation of the cluster in both tissues depends on 5′ remote regulatory elements. The pituitary-specific locus control elements DNase I-hypersensitive site I (HSI) and HSII, located 14.5 kb 5′ of the cluster (position −14.5), establish a continuous domain of histone acetylation that extends to and activates hGH-N in the pituitary gland. In contrast, histone modifications in placental chromatin are restricted to the more 5′-remote HSV-HSIII region (kb −28 to −32) and to the placentally expressed genes in the cluster, with minimal modification between these two regions. These data predict distinct modes of hGH cluster gene activation in the pituitary and placenta. Here we used cell culture models to track structural changes at the hGH locus through placental-gene activation. The data revealed that this process was initiated in primary cytotrophoblasts by histone H3K4 di- and trimethylation and H4 acetylation restricted to HSV and to the individual placental-gene repeat (PGR) units within the cluster. Later stages of transcriptional induction were accompanied by enhancement and extension of these modifications and by robust H3 acetylation at HSV, at HSIII, and throughout the placental-gene regions. These data suggested that elements restricted to HSIII-HSV regions and each individual PGR might be sufficient for activation of the hCS genes. This model was tested by comparing hCS transgene expression in the placentas of mouse embryos carrying a full hGH cluster to that in placentas in which the HSIII-HSV region was directly linked to the individual hCS-A PGR unit. The findings indicate that the HSIII-HSV region and the PGR units, although targeted for initial chromatin structural modifications, are insufficient to activate gene expression and that this process is dependent on additional, as-yet-unidentified chromatin determinants.
Mammalian development is initiated by differentiation of the primordial blastocyst into the trophectoderm and inner cell mass (12, 54, 60). The inner cell mass gives rise to all somatic tissues, whereas the trophectoderm generates extraembryonic tissues that will constitute the fetal placenta. Early in development, the trophoblast stem cells of human extraembryonic structures diverge into two distinct lineages: villous and extravillous cytotrophoblasts (CTBs) (13, 53, 54, 67). The extravillous CTBs differentiate into invasive trophoblasts that migrate into the uterine wall in the process of embryo implantation. The villous CTBs proliferate and fuse to form a layer of syncytial cells that extend over the surface of the placental villi (24, 34, 47, 51). This multinucleate syncytiotrophoblast (STB) layer constitutes the interface between fetal and maternal circulation, mediating essential gas and nutrient exchange. In addition to having these exchange functions, CTBs and STBs synthesize gestational hormones that are secreted directly into the maternal circulation.
The morphological and functional transition from a CTB to a multinucleate STB has been the focus of intense study (24, 38, 50, 55, 60). Although expression profiling has identified an array of proteins associated with this process (5), the molecular mechanisms that drive trophoblast differentiation and STB formation remain to be defined (13, 14, 38, 53, 54). The critical roles of the STB in multiple aspects of human embryonic development and maternal-fetal communication and symbiosis make it likely that defects in this process underlie pathological abnormalities in placentation affecting fetal growth and development (67).
A number of gene induction processes are tightly linked to the formation and function of human STBs. Among the most robust of these processes is the induction of the human growth hormone (hGH) gene cluster (Fig. 1A). This cluster, located on chromosome 17q22-24, contains five genes generated by a series of segmental duplications in the primate genome (7, 11). The most 5′ gene in the cluster is the hGH-N gene. hGH-N is a major hormonal determinant of postnatal growth of bone and soft tissues (48). The remaining four genes in the cluster, from 5′ to 3′, are the human chorionic somatomammotropin-like (hCS-L), hCS-A, hGH variant (hGH-V), and hCS-B genes (11, 40). Expression of these four genes is restricted to the STB layer of placental villi. hCS-L is considered a pseudogene; its expression is blocked by multiple splice site mutations (45). hGH-V encodes fetal GH, which is expressed exclusively in STBs and secreted into maternal serum beginning early in the second trimester. hGH-V has growth-promoting, lactogenic, and other activities similar to those of hGH-N (2, 22, 39); its robust expression in the second and third trimesters represses maternal pituitary hGH-N expression, thus replacing much of the hGH-N in maternal serum (18, 36). The two hCS genes, hCS-A and hCS-B, are coexpressed and encode identical placental chorionic somatomammotropins, also known as placental lactogen. hCS constitutes the most abundant maternal serum protein (23, 61). Thus, the hGH-V, hCS-A, and hCS-B genes undergo a selective induction during the CTB-to-STB transition in the developing placenta. Of these three, hCS-A is by far the most robustly expressed gene in the term placenta (40). Defining the molecular events that underlie this induction should identify molecular mechanisms involved in STB formation and function.
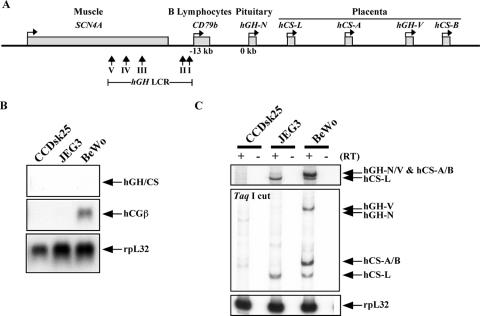
FIG. 1.
Expression of the hGH-hCS cluster genes in human placental cell lines. (A) Structure of the hGH gene cluster and its LCR. The hGH cluster is composed of five conserved genes, including the pituitary-specific hGH-N gene and placenta-specific hCS-L, hCS-A, hGH-V, and hCS-B genes. The expression of the hGH cluster genes is regulated by its LCR, which is far upstream of the cluster and overlaps two other tissue-specific genes, the B-lymphocyte-specific CD79b gene and the muscle-specific SCN4A gene. The LCR includes five DNase I-HSs, indicated as vertical arrows. The bent arrows above each gene indicate the transcriptional direction of the gene. (B) Expression of the early placental marker gene hCGβ was detected in BeWo cells by Northern analysis. Total RNAs from CCDsk-25, JEG3, and BeWo cell lines were fractionated in a formaldehyde-agarose gel and transferred to a nitrocellulose membrane. The blot was hybridized with a 32P-labeled hGH-hCS probe that detects all five genes. The blot was also hybridized with a probe for the early placental hCGβ mRNA. The constitutive rpL32 gene was used as a loading control. The signals were visualized by autoradiography. (C) Expression of the placental hGH-hCS genes is activated in BeWo cells. Total RNAs were purified and amplified by PCR. These starting PCR products are shown (top panel). The products were next digested with TaqI to distinguish the mRNAs corresponding to each of the hGH cluster genes (middle panel). Amplification of rpL32 mRNA was used as a positive control (bottom panel).
In previous studies, we demonstrated that activation of the hGH cluster in both the pituitary and the placenta is dependent on a set of remote regulatory elements (Fig. 1A) (8, 31, 58, 59, 62). These elements, located between 14.5 and 32 kb 5′ of the cluster, were identified initially in chromatin preparations by their sensitivity to DNase I (DNase I-hypersensitive site I [HSI] to HSV [31]). HSV and HSIII, at kb −32 and kb −28, respectively, are detected in pituitary somatotrope and placental STB chromatin; HSIV, at kb −30, is specific to STB chromatin; and HSI and HSII, at kb −14.5 to kb −15.5, are specific to somatotrope chromatin. In addition to the specificity of HS formation, the activation of the hGH cluster in pituitary and placenta is distinguished by observed patterns of histone modifications. Histone acetyltransferase activity recruited to HSI establishes a continuous 32-kb domain of histone acetylation connecting the locus control region (LCR) and the hGH-N promoter (16, 26). This acetylated domain facilitates trans-factor binding at the hGH-N promoter and the transcriptional activation of hGH-N (26, 27). Activation of the placental genes in term placental STBs is marked by activating histone modifications that are restricted to the HSV-HSIII region and to the placental genes (32); the extensive regions between HSIII and the cluster remain unmodified. These differences in HS formation and histone modifications predict two distinct pathways of long-range control by the LCR, a “spreading” or “tracking” mechanism in the pituitary (16, 17, 26) and a “looping” mechanism in the placenta (32). Based on the present knowledge of cellular differentiation and epigenetic alterations, it seems reasonable to propose that chromatin structures in the placenta are altered during the terminal transition of CTBs to STBs and the corresponding robust induction of gene expression from the hGH cluster. Analysis of this process of epigenetic modifications should address molecular mechanisms at critical stages of human placental development and further clarify the corresponding long-range interactions. Here we explore this process utilizing ex vivo cell culture models of STB differentiation. The data support a simplified model of placental-gene activation, and that model is tested by in vivo transgenic analysis.
MATERIALS AND METHODS
Cell culture.
The human choriocarcinoma cell lines BeWo and JEG3 and a human skin fibroblast line, CCDsk-25, were purchased from the American Type Culture Collection. JEG3 and CCDsk-25 cells were cultured at 37°C with 5% CO2 in minimum essential medium (Invitrogen Life Technologies, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen Life Technologies). BeWo cells were maintained at 37°C with 5% CO2 in F12 medium (Invitrogen Life Technologies) supplemented with 15% fetal bovine serum (HyClone), 0.2% glucose (Sigma-Aldrich, St. Louis, MO), 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen Life Technologies). To induce hGH/CS expression, BeWo cells were treated with either 80 μM forskolin (FSK) or 10 to 100 nM trichostatin A (TSA) and with the carrier dimethyl sulfoxide or ethanol in parallel controls. For long-term culture, 0.7 × 106 cells were spread on a 60-mm dish with or without a Matrigel coating (BD Biosciences, Palo Alto, CA). These were incubated overnight to allow cell attachment. Additional surface treatments that were assessed included collagen type I, collagen type IV, fibronectin, laminin, and poly(D) lysine (BD Biosciences). The day after plating was designated day 0, and the medium was subsequently changed every 2 days for the remainder of the culture period.
Isolation and culture of primary CTBs.
Purification of CTBs from human term placentas and their primary culture was conducted as previously described (5, 33). Third-trimester placentas were obtained from women with normal pregnancies and deliveries, and CTBs were isolated by enzymatic disaggregation and cultured as described previously (52). The CTBs were purified to >98% homogeneity by negative CD9 selection. The protocol for obtaining placentas was approved by the Human Investigation Committees of the University of Cincinnati and the Children's Hospital Medical Center. Second-trimester maternal serum was added to the culture medium to maximize hCS gene induction (52).
RT-PCR analyses.
Total RNA was extracted from cultured cells, term placentas, or mouse tissues using TRIzol reagent (Invitrogen Life Technologies) according to the manufacturer's instructions. The RNAs were treated with RNase-free DNase I (Invitrogen Life Technologies) at 37°C for 5 h and reextracted with TRIzol reagent. For the reverse transcriptase-PCR (RT-PCR) analysis of hGH gene cluster expression, the resulting RNAs were reverse transcribed with an oligo(dT) primer and Superscript III transcriptase (Invitrogen Life Technologies), and 2.5% of this RT reaction mixture was used for PCR. PCR was performed with a sense primer 32P labeled with T4 polynucleotide kinase (5′-GTCCCTGCTCCTGGCTTTTG-3′) and an antisense primer (5′-AGCAGCCCGTAGTTCTTGAG-3′). PCR was carried out for 25 to 35 cycles under the following conditions: 30 s at 94°C, 30 s at 57°C, and 2 min at 72°C. The amplified cDNA segments were 546 bp for hGH-N, hGH-V, hCS-A, and hCS-B and 492 bp for hCS-L. Digestion of the amplification products with TaqI generated an hGH-V fragment of 546 bp, an hGH-N fragment of 494 bp, an hCS-L fragment of 251 bp, and hCS-A and hCS-B fragments of 305 bp. The amplified and digested products were separated on 4% polyacrylamide gels, and the signals were visualized by autoradiography. As an internal control, a transcript of human ribosomal-protein large-subunit 32 (rpL32) mRNA was amplified using a specific primer pair (5′-GTGAAGCCCAAGATCGTCA-3′ and 5′-TGTTGCACATCAGCAGCAC-3′) (70). For the RT-PCR analysis of the relative levels of hCS-A and hCS-B gene expression, a primer pair of 5′-AGAACTACGGGCTGCTCT-3′ and 5′-AGGGCCAGGAGAGGCACT-3′ was used. Reverse transcription cDNA synthesis, with the reaction mixture primed with the antisense oligonucleotide, was followed by PCR amplification utilizing the same antisense primer together with the sense oligonucleotide labeled at its 5′ end with 32P (40). PCR products were then separated in 6% denaturing polyacrylamide gel, visualized by autoradiography, and subjected to PhosphorImager quantification. Expected sizes for the hCS-A and hCS-B RT-PCR-amplified fragments were 153 bp and 157 bp, respectively.
Northern blot analysis.
Total RNA was purified from cells or tissues using TRIzol reagent (Invitrogen Life Technologies) according to the manufacturer's protocol. The RNA was electrophoresed on a formaldehyde-agarose gel and transferred to a Zetabind membrane (Cuno Inc., Meriden, CT). The blot was hybridized for 18 h with a 32P-labeled probe at 42°C in 50% formamide, 5× Denhardt's solution, 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 1% sodium dodecyl sulfate, and 100 μg/ml salmon sperm DNA. The probes for hGH/hCS and rpL32 mRNAs were prepared by RT-PCR as described above. The probes for human chorionic gonadotropin β (hCGβ) and mouse placental lactogen II were also prepared by RT-PCR using specific primer pairs (5′-CATCACCGTCAACACCACCAT-3′ and 5′-TCACAGGTCAAGGGGTGGTC-3′ and 5′-ACTCCTCAGAGATGAAGCTG-3′ and 5′-ACATCACGACACTTCAGGAC-3′, respectively). All the probes were labeled with the Klenow fragment by using a random-priming labeling kit (Roche Diagnostics, Mannheim, Germany). The oligoprobe for 18S rRNA (5′-CGGCATGTATTAGCTCTAGAATTACCACAG-3′) (21) was 32P labeled with T4 polynucleotide kinase. After hybridization, the membrane was washed at 50°C in 0.1% sodium dodecyl sulfate-0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and exposed to Kodak Biomax film (Eastman Kodak, Rochester, NY).
HS region mapping.
Nuclei from cultured cells and from human term placentas were isolated and stored as described previously (32). HS regions were detected as described previously (17, 26, 31). Briefly, 300 to 500 μg of nuclei were digested with 45 units of RNase-free DNase I (Invitrogen Life Technologies) at 37°C in 1 ml of 50 mM Tris-HCl (pH 8.0), 0.1 M NaCl, 3 mM MgCl2, 1 mM CaCl2, and 1 mM phenylmethylsulfonyl fluoride. At the time points of 0, 2, 4, 8, and 15 min, 200-μl samples were transferred into new tubes containing 20 μl of 0.5 M EDTA. After the genomic DNA was purified, each sample was digested with EcoRI and the signals were detected by Southern blot hybridization.
ChIP assay of unfixed chromatin.
A chromatin immunoprecipitation (ChIP) assay with unfixed chromatin to determine the histone modification patterns was performed as previously described (32). In brief, 300 to 600 μg of nuclei was digested with micrococcal nuclease and soluble chromatin was collected. Immunoprecipitation was performed with antibodies for modified histones, and DNA was purified from the precipitated (bound) chromatin as well as from the fraction before the precipitation (input). Antibodies used were anti-acetyl histone H3, anti-acetyl histone H4, anti-dimethylated-histone H3 (Lys4) (Upstate Biotechnology, Inc., Lake Placid, NY), and anti-trimethylated-histone H3 (Lys4) (Abcam, Cambridge, United Kingdom). PCR was conducted with the serially diluted DNA samples from the bound and input fractions. The histone modification levels were calculated as ratios of the signal intensity of the bound fraction to that of the input fraction. Those ratios were further normalized to the corresponding ratio for the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter, considered 100. The amplimer of α-globin was assessed to determine the histone modification levels of an inactive gene. All the primers used in this study were described previously (32), except for the α-globin primers, whose sequences were 5′-ATGTTCCTGTCCTTCCCCACC-3′ and 5′-ATGGTGCTGTCTCCTGCCGA-3′. It should also be noted that the background in the day 4 CTB cultures, as monitored by modification at the α-globin control locus, was unusually and consistently high. We therefore further normalized the data set for this study to the day 4/day 0 modification ratio at the α-globin locus. For assessing the modification level of each gene in the cluster, we coamplified the 5′ region (amplimer set 233GH/CS) and digested the products with four restriction enzymes (HinfI, DraIII, PstI, and MscI) as described previously (32).
Transgenic constructs and generation of transgenic mouse lines.
A human bacterial artificial chromosome (BAC) genomic clone encompassing the hGH LCR and the entire hGH cluster was identified by a screen of a total genomic library (CITB Human B&C Library; Invitrogen Life Technologies) with probes corresponding to the TCAM1 and SCN4A genes (69). A 148.3-kb clone (clone 535D15) was identified, and a 123-kb insert was released by NotI digestion. To construct the HSIII-V/Pa(CSA)Ea transgene, the 11.5-kb ClaI-EcoRI fragment of cosmid clone GH5 (31) encompassing the placental LCR HSIII-HSV region was ligated to a 7.5-kb fragment containing the hCS-A locus, including its contiguous 5′ P element and contiguous 3′ putative enhancer. This hCS-A fragment was generated by PCR using a specific primer set (5′-GGTACCTGGTCCATGGTTGGCATGGTAACCCCTTAC-3′ and 5′-GTCGACGCGGCCGCACCACAACTGCCATCTCCTTTTTCTCC-3′). The long-distance PCR was performed with 50 μl that included 100 to 200 ng of the BAC DNA template, 1× LA PCR buffer (Takara Bio, Ohtsu, Japan), a 0.4 mM concentration of a deoxynucleoside triphosphate mixture, a 0.2 μM concentration of paired primers, 2.5 units of LA Taq (Takara Bio), and 1.3 units of Pfu Ultra DNA polymerase (Stratagene, La Jolla, CA) under the following conditions: 1 min at 94°C, 35 cycles of 20 s at 98°C and 10 min at 66°C, and a final extension for 10 min at 72°C.
The hGH/BAC and HSIII-HSV/Pa(CSA)Ea DNAs were adjusted to a concentration of 2 ng/μl in 10 mM Tris-HCl (pH 8.0), 1 mM EDTA and microinjected into the male pronuclei of fertilized mouse eggs. All procedures involved in generating the two sets of transgenic mouse lines were carried out by the University of Pennsylvania Transgenic and Chimeric Mouse Core Facility (http://www.med.upenn.edu/genetics/core-facs/tcmf/index.html). Positive founders were identified by dot blot analysis of tail DNA using the HSIII probe (32).
RESULTS
Activation of the hGH cluster in BeWo cells.
A number of human-placenta-derived cell lines have been used to model CTB function (63). Two of the most intensively studied are the choriocarcinoma lines JEG3 and BeWo. These two lines were studied with the aim of defining the structure of the hGH cluster prior to the CTB-to-STB transition. A human skin fibroblast line, CCDsk-25, was used as a nonexpressing control. Expression profiles of the JEG3, BeWo, and CCDsk-25 cell lines were initially assessed by Northern blotting. hCGβ mRNA served as a marker of CTB gene expression. The hCGβ gene is not linked to the hGH cluster, and its expression precedes activation of the hCS genes during gestation and during the CTB-to-STB differentiation (40). A map of the hGH-hCS multigene cluster, closely linked genes, and the hGH LCR region is shown (Fig. 1A). A single hybridization probe that recognizes all five of the evolutionarily related hGH-hCS mRNAs was used to monitor total output from the hGH cluster (hGH/CS mRNA). The Northern blot revealed an hCGβ mRNA signal in the BeWo cells, with no evidence of coexisting hCS expression (Fig. 1B). The same three mRNA samples were reassessed at higher levels of sensitivity by RT-PCR (35 cycles) with primers that coamplify all five of the hGH and hCS mRNAs (Fig. 1C, top panel). hGH/hCS mRNAs were detected in BeWo cells and at lower levels in JEG3 cells. These transcripts were absent in the CCDsk-25 fibroblasts. The output from the hGH cluster was further defined using a validated RT-PCR/endonuclease cleavage assay that distinguishes the individual transcripts (see Materials and Methods). Expression in JEG3 cells was limited to hCS-L. In contrast, BeWo cells expressed hGH-V, hCS-A, hCS-B, and hCS-L but no hGH-N mRNA. The expression of hCS mRNA in the BeWo cells, which could be detected only by RT-PCR at high cycle numbers, constituted less than 0.01% of that in STBs (see below). These trace levels of hCS and hGH-V mRNAs in the BeWo cells might represent expression from a small number of the cells that have spontaneously initiated STB differentiation or may represent trace expression from the BeWo population as a whole. In either case, analysis of hGH cluster chromatin in the BeWo cell model should reveal the epigenetic structure at the hGH-hCS locus prior to the robust activation characteristic of STB differentiation.
The “competence” of the hGH cluster to be induced in the BeWo cells was next assessed. Such an induction would attest to the “poised” state of the cluster and possibly allow us to follow the chromatin transition from CTB to STB. Attempts to induce expression in the BeWo cells by treatment with the histone deacetylase inhibitor TSA or with the adenylyl cyclase activator FSK (see Materials and Methods) (65, 68) were ineffective (Fig. 2A and B and data not shown). However, in agreement with prior studies (68), FSK triggered a striking enhancement of hCGβ expression (compare Fig. 2A and B). A third approach was based on the model that the transition from CTB to STB is enhanced by cell-cell interactions. The BeWo cells were grown to high density over a 32-day period to allow the cells to become tightly packed and multiply layered. As assessed by Northern blotting, the hCGβ mRNA was rapidly induced early in the study and reached a robust plateau between days 4 and 8 (Fig. 2C). This hCGβ mRNA induction was followed by a steady increase in expression from the hGH cluster (hGH/CS mRNA) (Fig. 2C). Beyond 32 days, the high cell density resulted in dislodgment from the plastic surface. Analysis of the expression profile by the RT-PCR/endonuclease cleavage assay confirmed that the activation of the hGH cluster in the BeWo cells paralleled that seen in native STBs, with selective induction of the hCS-A, hCS-B, and hGH-V mRNAs (Fig. 2D). Consistent with this native pattern of induction, hGH-N and hCS-L mRNAs were not detected. To further optimize this approach, the long-term culture was repeated using a variety of cell adhesion substrates (see Materials and Methods). The only significant improvement was obtained with Matrigel. The cells grown on this matrix activated the hGH cluster more rapidly and robustly than those grown on noncoated plates (Fig. 2E). Importantly, the placenta-specific profile of mRNA accumulation was the same as on the noncoated dishes; i.e., the induction of hGH-V, hCS-A, and hCS-B mRNAs was selective (data not shown). These studies demonstrated that hGH cluster gene expression can be induced by growing BeWo cells to high density.
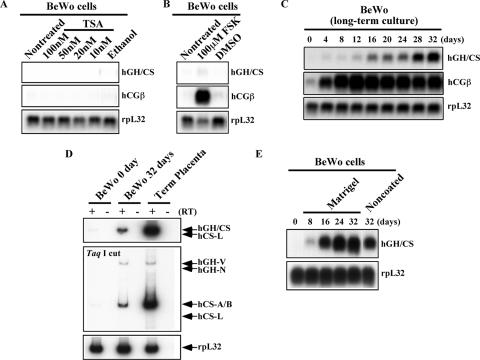
FIG. 2.
Induction kinetics of the placental hGH-hCS genes in BeWo cells. (A) Histone deacetylase inhibition with TSA failed to induce BeWo cell gene expression. The cells were treated with the indicated concentrations of TSA for 12 h. Total RNA was analyzed by Northern blotting using ethanol as a vehicle control. Hybridization probes used are indicated to the right of each panel. (B) FSK-induced BeWo cell syncytialization resulted in only a moderate increase in hGH-hCS expression. BeWo cells were induced to syncytialize by culturing them in the presence of 80 μM FSK for 4 days. Total RNA from the treated cells was used for Northern blot analyses. The syncytialization was confirmed by hCGβ induction. Dimethyl sulfoxide (DMSO) was the vehicle control. (C) Placental hGH-hCS expression was induced by long-term culture of BeWo cells. The cells were plated at a high density and incubated without chemical additives for 32 days. Total RNA (20 μg) was collected every 4 days for analysis by Northern blotting. (D) The BeWo hGH-hCS transcript profile after 32 day of culture was qualitatively similar to that in term placentas. RT-PCR analysis was performed as described for Fig. 1C but with total RNA (5 μg) from BeWo cells at days 0 and 32 of culture and from human term placental villi. hCS-A, hCS-B, and hGH-V mRNAs were detected in all three samples; no hGH-N or hCS-L signals were observed. (E) Matrigel coating of the culture plates enhanced hGH-hCS activation in BeWo cells. The BeWo cells were cultured on Matrigel-coated dishes for 32 days, and total RNA was collected every 8 days, as described for panel C above. The Northern blot analysis showed higher and faster induction of hGH-hCS genes than that shown in panel C when cells were grown on Matrigel.
Activation of the hGH cluster in primary CTBs.
A second model for analysis of the CTB-to-STB transition is ex vivo differentiation of primary placental CTBs. CTBs can be isolated in a highly enriched state from normal human term placentas (33). These can be induced to differentiate into STBs by culturing them over a 4- to 6-day period in media supplemented with second-trimester maternal serum (5, 52). Northern blot analysis of freshly prepared primary CTBs revealed the presence of hCGβ mRNA in the absence of mRNAs originating from the hGH cluster (Fig. 3A, day 0 lane). The primary CTBs were then incubated in media containing second-trimester maternal serum (52), and the cells were assessed on a daily basis for gene expression. hCGβ mRNA levels remained fairly stable over the first 4 days of culture and then demonstrated a low-level induction on days 5 and 6. In contrast, there was a steady induction of expression from the hGH cluster throughout the 6-day culture period. This induction is consistent with the results of prior reports using this same protocol (5, 52). While days 0 and 1 were negative for hGH-hCS mRNAs by Northern analysis and day 2 showed only trace levels, the more sensitive RT-PCR/endonuclease assay of day 0 revealed trace levels of hCS mRNA. Furthermore, day 2 mRNA samples revealed trace levels of hCS and hGH-V mRNAs in the absence of hGH-N and hCS-L (Fig. 3B). The trace expression of these placental mRNAs from the hGH cluster in the day 0 CTBs could reflect low-level contamination of the CTB preparation with STBs or trace expression of the cluster in the CTB population as a whole.
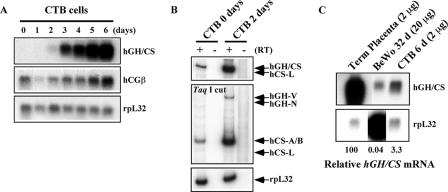
FIG. 3.
hGH-hCS genes were induced in ex vivo cultures of primary human placental CTBs. (A) Expression of the hGH cluster genes in primary placental CTB cell culture. The CTBs were cultured in the presence of second-trimester maternal serum for 6 days to induce spontaneous differentiation into syncytial cells capable of expressing the hGH-hCS genes. Total RNA was prepared from daily aliquots and analyzed by Northern blotting. Probes used are indicated to the right of the autoradiographs. (B) hCS-A, hCS-B, and hGH-V are expressed by the induced CTBs. RT-PCR analysis was conducted with RNA from CTBs at days 0 and 2 of culture. The PCR products were digested with TaqI and analyzed as described for Fig. 1C. The hCS-A and hCS-B mRNAs were detected at day 0, and their expression and that of hGH-V were induced by day 2. (C) Quantitative comparison of hGH-hCS expression levels among the placental model systems used in this study. Northern blot analysis was performed on total RNA from human term placenta, cultured primary CTBs at day 6 (CTB 6 d), and day 32 BeWo cells (BeWo 32 d). Relative hGH-hCS mRNA levels, estimated using the rpL32 signals as a loading control, are shown at the bottom.
The preceding studies demonstrated that the hGH cluster is minimally active in both BeWo cells and freshly harvested CTBs. In both models, the cluster could be induced in culture and the profile of mRNA expression from the induced cluster corresponded to that of primary STBs. As a final assessment of these two models, we compared the maximal levels of hCS mRNA in the induced BeWo cells (at day 32 of cell culture) and CTBs (at day 6 of culture) to that in a preparation of primary term placental villi (Fig. 3C). In this comparison, the level of expression from the hGH cluster in both models was significantly below that in the term placenta. When normalized to a ribosomal-protein mRNA (rpL32), the levels of hCS mRNA in day 32 BeWo cells and day 6 CTBs were 0.04% and 3.3% of those in primary placental villous tissue. Although this may be an underestimation because the comparison is based on steady-state mRNA levels and the hCS genes were in the process of induction, it is clear that the numbers of cells generated in both induction models fell far below that seen in the native STB population. This may limit the utility of these cell culture models, once cells are induced, to accurately reflect the final STB chromatin state. However, defining the structure of the hGH chromatin locus in BeWo cells and CTBs in their “poised” state, prior to induction (day 0 BeWo cells and day 0 CTBs), and comparing the epigenetic profiles with those of the fully induced cluster in the primary STBs should establish the endpoints of the CTB-to-STB differentiation process.
DNase I-HSs that mark the hGH LCR in STB chromatin are present in uninduced BeWo cells and in primary CTBs.
Of the five DNase I-HSs that mark proven and/or potential elements of the hGH LCR, HSIII and HSV are constitutive, HSI and HSII assemble selectively in pituitary somatotrope cells, and HSIV is STB specific. To determine whether the chromatin structures characteristic of STB chromatin are already established at the CTB stage of differentiation, nuclear chromatin samples from BeWo cells and primary CTBs were DNase I mapped for HSIII, HSIV, and HSV (Fig. 4). Parallel HS mapping studies were also carried out on day 32 BeWo cells to determine whether the culture-based induction altered this conformation. Analysis of day 6 CTBs could not be performed because the amount of material after 6 days in culture was inadequate for reliable DNase I/Southern blot analysis. These DNase I mapping studies revealed that HSIII, HSIV, and HSV were already present in BeWo cell and in day 0 CTB chromatin. Of particular note, placenta-specific HSIV was present in BeWo chromatin at both days 0 and 32, in CTB chromatin at day 0, and in primary STB but not fibroblast chromatin. Constitutive HSIII was present in all samples. HSV could not be well visualized in this study and was not further pursued (17, 31). These results indicated that the placental (STB) chromatin configuration, as reflected by the presence of HSIV, is established during the CTB stage prior to transcriptional induction.
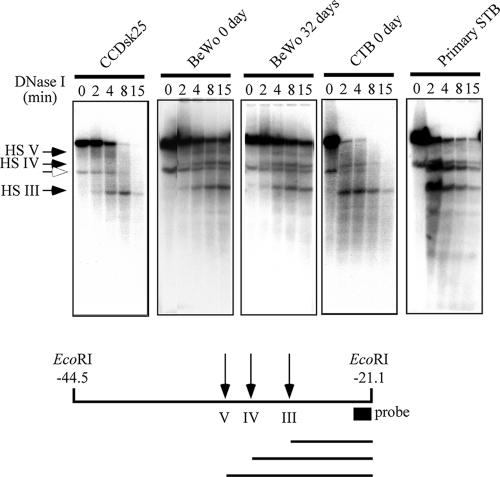
FIG. 4.
DNase I-HS mapping in the BeWo, CTB, and STB chromatins. Nuclei from the indicated cells were digested with DNase I, genomic DNA was purified from each sample, and the DNAs were digested with EcoRI and subjected to Southern blot hybridization analysis. The positions corresponding to HSIII, HSIV, and HSV are indicated by arrows at the left. An open arrowhead points to a nonspecific band that was detected before the DNase I treatment (time zero). At the bottom is a diagram of the EcoRI fragment showing the probe position and the sizes of each fragment after DNase I digestion. The results reveal the presence of HSIV and the placenta-specific HS in day 0 and 32 BeWo cells as well as in primary CTBs and STBs. HSIV is absent from the parallel analysis of the human fibroblast line CCDsk-25, which does not express any of the hGH cluster genes. A diagram of the relative fragment sizes is at the bottom.
Histone modification patterns at the hGH locus in BeWo cells before and after transcriptional activation.
The formation of placenta-specific HSIV in uninduced BeWo cells and freshly isolated CTBs suggests that chromatin structures at the hGH cluster in these cells have been “primed” for subsequent induction (Fig. 4). The structure of the hGH locus at the preinduction stage was further assessed by ChIP analysis (16, 32). Four modifications of core histones H3 and H4 were chosen for study based on their consistent linkage to gene activation: histone H3 and H4 acetylation and histone H3K4 di- and trimethylation (41, 49). H3K4 methylation has been linked to gene activation in a number of systems, and the addition of two rather than three methyl groups at the critical lysine may play distinct roles in activation pathways (35, 43, 56). Chromatin containing each of these modifications was enriched by immunoprecipitation (see Materials and Methods), and DNA sequences in these samples were assayed for enrichment over the starting material (“input”) at 14 sites along the cluster. These sites span 98 kb and encompass the entire hGH LCR and hGH cluster, along with flanking regions (Fig. 5A). The analysis takes into account the fact that the five genes within the cluster were generated via segmental duplications of an ancestral hGH-hCS precursor along with its flanking sequences (7, 11). Due to this evolutionary history, several amplimer sets detect three or four copies of the repeated regions within the cluster. A core placental-gene repeat (PGR) unit, encompassing each placentally expressed gene along with 3 kb of 5′-end-flanking regions, was evaluated at high resolution with a set of four additional amplimers (Pe0, Pe2, Pe6, and Pe9). This PGR includes the “P-element,” which has been postulated to contribute to the specificity of placental expression for these genes (17, 46). An element that enhances hCS expression in cell transfection studies (“Enhancer”) is located 3′ of the three hCS genes (but not hGH-V) and is separately detected by a specific amplimer (En) (66).
FIG. 5.
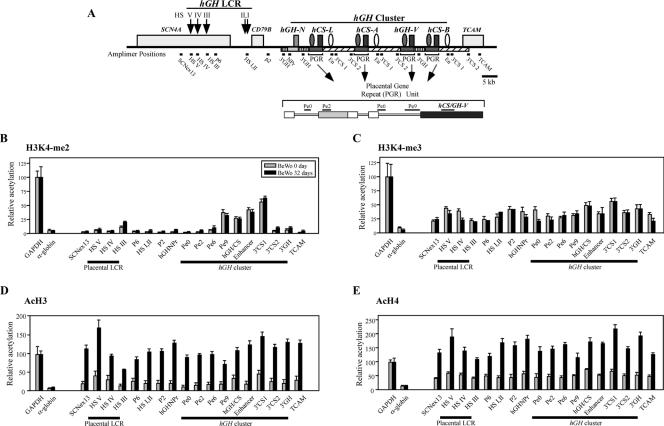
Histone modifications at the hGH cluster and its LCR in BeWo cells during hGH-hCS activation. (A) Positions of amplimers used in the ChIP assays. The structure of the hGH cluster, linked genes, and the hGH LCR are depicted. P-elements and the putative hCS enhancers are indicated (shaded and open ovals, respectively). Shaded and striped rectangles indicate extensive conserved segments among the PGR units and the hGH-N gene. The amplimer names and their positions are shown below the map. The PGR unit is also shown in an expanded format. (B) The histone H3K4 dimethylation pattern in BeWo cells was established prior to gene activation and did not change after long-term culture. Soluble chromatin was collected from BeWo cells at days 0 and 32 of culture by digesting nuclei with micrococcal nuclease. The ChIP analysis was conducted with anti-dimethylated-histone H3K4 antibody, and the resulting input and bound fractions were subjected to PCR amplification. The bound/input signal ratio at each amplicon (A) was normalized to that at the GAPDH promoter, which was considered 100. The relative histone modification levels in day 0 (shaded bars) and day 32 (black rectangles) BeWo cells are shown. Standard error values are indicated. The α-globin promoter was used as a negative control. (C) Histone H3K4 trimethylation was detected at low levels across the entire locus in BeWo cells at both day 0 and day 32. Histone H3K4 trimethylation patterns, examined by ChIP analysis with anti-trimethylated-H3K4 antibody, were analyzed and plotted as described for panel B. (D) Histone H3 acetylation was significantly induced across the locus during the 32 days of BeWo cell culture. Histone H3 acetylation patterns, using anti-acetylated-histone H3 antibody, were studied as described for panel B. (E) Histone H4 acetylation was significantly induced across the locus during BeWo cell culture. The ChIP assay was performed, analyzed, and plotted as described for panel B but with anti-acetyl histone H4 antibody.
The day 0 and day 32 BeWo cells were studied for enrichment of H3K4 methylation across the hGH locus (Fig. 5B and C). The controls for both the H3K4-me2 and H3K4-me3 ChIP studies revealed a 20-fold difference between the expressed GAPDH locus and the repressed α-globin locus. The H3K4-me2 analysis in the BeWo day 0 cells revealed a well-defined island of H3K4-me2 modification that extended from the placental-gene promoters through the hCS enhancer element (amplimers Pe9 through 3′CS1). The hGH-N promoter and its flanking regions remained unmodified. This pattern of H3K4-me2 modification was stable during the induction period in that the same pattern was observed in the day 32 BeWo cells. H3K4 trimethylation in the day 0 BeWo cells showed minimal variation throughout the region, with the exception of a slight relative enrichment at the 5′ boundary of the LCR (HSV) and over the level in structural genes. As in the case of the H3K4-me2 analysis, the pattern and level of H3K4-me3 remained stable over the induction period (day 32).
The BeWo cells were next analyzed for histone H3 and H4 acetylation. In the uninduced, day 0 BeWo cells, the levels of H3 acetylation across the entire hGH locus were quite low, with marginal elevations at HSV and in the immediate proximity of the structural genes (Fig. 5D, gray bars). ChIP analysis of H4 acetylation of the BeWo day 0 chromatin revealed slightly higher levels but a similar flat profile (Fig. 5E, gray bars). Analysis of the induced day 32 BeWo cells revealed a robust enrichment in H3 and H4 acetylation across the entire region (Fig. 5D and E, black bars). These levels of H3 and H4 acetylation equaled or exceeded those of the GAPDH-positive control. The enrichment for H3 acetylation was highest at the 5′ boundary of the LCR (HSV), with a progressive decrease at HSIV and HSIII. Otherwise, the levels across the locus differed by less than twofold. The H4 acetylation pattern similarly showed a robust increase of acetylation across the entire region compared to that in the day 0 samples with evidence for selective enrichment at HSV compared to that at HSIV and HSIII.
In summary, at day 0 of BeWo culture, we observed a well-defined island of H3K4-me2 modification at the PGR units that extended from the placental-gene promoters to their 3′ enhancer regions. During the subsequent induction period, this segment of H3K4-me2 modification remained unaltered, while there was a robust increase in H3 and H4 acetylation across the entire locus. These data suggested that H3K4 dimethylation and H3/H4 acetylation play distinct roles in the process of placental-gene activation within the hGH cluster, with methylation corresponding to the “primed” state and acetylation tracking with the subsequent transcriptional induction.
Histone modification patterns in primary CTBs.
We next applied ChIP analysis to primary CTBs. Chromatin was isolated from freshly prepared CTB nuclei (day 0 CTBs) or CTB nuclei following 4 days in STB differentiation media (day 4 CTBs). Day 4 cells rather than day 6 cells were used for ChIP analysis due to the more reliable and consistent harvest of chromatin at this time point. The results of these assays are displayed along with the previously reported profiles for primary STB chromatin (open bars) (32) to facilitate a direct comparison (Fig. 6). Levels of H3K4-me2 are exceedingly low in the primary CTBs; the most prominent modification was noted at HSV, with more-moderate levels of modification immediately 5′ and 3′ of the hGH/CS genes (Pe9 and Enhancer, respectively) (Fig. 6A). In the transition to day 4, the levels of H3K4-me2 modification at HSV show a sharp decrease, while modification at the PGR units extends to encompass a continuous domain extending from amplimers Pe6 to 3′CS1. Primary STBs show a further decrease in HSV to background levels, while modification within the PGR continues to demonstrate an overall increase within the PGR (except at the 3′ enhancer). The analysis of H3K4-me3 in day 0 CTB chromatin revealed a discrete and prominent peak at HSV and enrichment over that in the PGR unit (Fig. 6B). In contrast to the H3K4-me2 data, the substantial modifications at HSV are fully sustained in day 4 CTBs and STBs. The only further change in H3K4-me3 modification that appeared to take place during the CTB-to-STB differentiation process was a twofold increase in modification 3′ of the hCS enhancer (amplimer 3′CS1) and 3′ of the two GH genes (amplimer 3′GH). Thus, H3K4 di- and trimethylation was limited to HSV and the PGR units and is established early in the differentiation process.
FIG. 6.
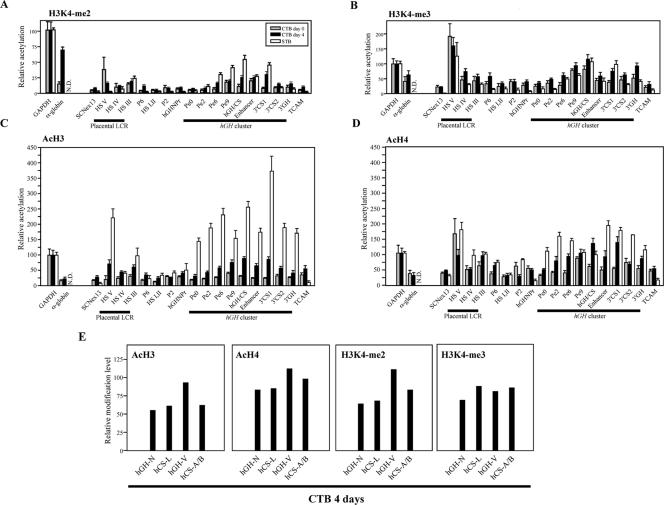
Histone modification at the hGH cluster and its LCR in primary CTBs before and after 4 days of differentiation. (A) Histone H3K4 dimethylation was established after day 4 of culture in primary CTBs. The amplimer positions are the same as those in Fig. 5A. Primary CTBs were isolated from human term placentas and cultured for 4 days in the presence of second-trimester maternal serum. The ChIP assay was performed with anti-dimethylated-histone H3K4 antibody as described for Fig. 5B on chromatin from freshly prepared CTBs (day 0) and after culture (day 4). For comparison, corresponding values for modifications of chromatin isolated from primary placental STBs, previously reported by us (32), are shown. (B) The histone H3K4 trimethylation pattern was largely preset in the freshly prepared, day 0 CTBs. The histone H3K4 trimethylation pattern was examined by ChIP with anti-trimethylated-H3K4 antibody, analyzed, and labeled. (C) Histone H3 acetylation was induced at the hGH locus during differentiation from CTBs to STBs. The histone H3 acetylation patterns were determined by ChIP with anti-acetylated-histone H3 antibody. (D) Histone H4 acetylation was induced at the hGH locus during differentiation of the CTBs. The histone H4 acetylation levels were investigated by ChIP with anti-acetylated-histone H4 antibody. (E) All genes in the hGH cluster were similarly modified in the day 4 CTBs. Histone modification levels at each cluster gene were examined by ChIP. The PCR analysis of the isolated DNA was performed with an amplimer set common to the five genes; the amplified products were digested with four restriction enzymes to distinguish each signal.
Assessment of H3 acetylation by ChIP revealed very low levels of modification across the hGH locus in the day 0 CTBs, with no evidence of enrichment at any position (Fig. 6C, shaded bars). The 4-day induction period boosted levels of the H3 acetylation modification by two- to threefold at HSV and HSIII as well as over that of the promoters in the PGR duplication unit and extending into the 3′-end-flanking enhancer region (Fig. 6C, black bars). The increase in H3 acetylation seen in CTBs from day 0 to day 4 was further exaggerated and broadened in the fully induced STB chromatin locus (open bars). The increase in H3 acetylation became more prominently marked over that of HSV and HSIII. Thus, the levels of H3 acetylation in the regions encompassing the placental genes were dramatically increased and spread to flanking sequences. H4 acetylation analysis (Fig. 6D) revealed that HSV was prominently modified in day 0 cells and that this level of modification remained stable in day 4 cells and in the STBs. Modification in day 0 cells at the hCS promoter (Pe9) remained constant in the day 4 CTBs and STBs, while levels of acetylation on both sides of this site increased incrementally in the day 4 and STB chromatin. As was the case with H3 acetylation, H4 acetylation in the STBs extended to encompass the entire cluster, with the specific exclusion of the hGH-N promoter.
Modifications of each of the individual genes in the cluster were analyzed in the day 4 CTBs using a previously described coamplification/restriction endonuclease assay that is specifically designed to distinguish the 5′ termini of the five genes (32; see also Materials and Methods). (Modifications in this region were insufficient for accurate analysis of the day 0 CTBs.) These modifications in the day 4 CTB samples, summarized in Fig. 6E, revealed that all genes in the cluster were equally modified within a twofold range of each other. These relatively equivalent levels of modification among the five paralogs agree with a similar analysis carried out with STBs (32) and highlight the lack of a direct correlation between modification at the individual paralogs in the cluster and their corresponding steady-state mRNA levels.
In summary, the ChIP studies revealed distinct distributions and dynamics of H3K4 methylation and H3/H4 acetylation during the CTB-to-STB transition. These modifications are summarized in Fig. 7. The initial H3K4 di- and trimethylation modifications in the day 0 and 4 CTBs were localized to the HSIII-HSV region and to the PGR units. The levels of H3K4-me2 increased during the STB transition but maintained the restricted pattern. The hGH-N gene appeared to be transiently modified at day 4 (Fig. 6E), and this modification was subsequently lost in the STBs, while the modifications over the PGR units were strengthened and extended. There appeared to be a complete absence of H3 acetylation at the beginning of the process (day 0 CTBs), and H4 acetylation at HSV and within the cluster was marginal. Remarkably, the levels of both H3 and H4 acetylation underwent a dramatic increase in parallel with the induction of hCS/hGH-V transcription in the day 4 CTB and primary STB populations. At the end of this differentiation process, the 5′ remote HS regions and the entire placentally expressed portion of the hGH cluster were encompassed within a continuous domain of H3/H4 acetylation. Of note, the region between HSIII and the cluster remained entirely excluded from these histone modifications.
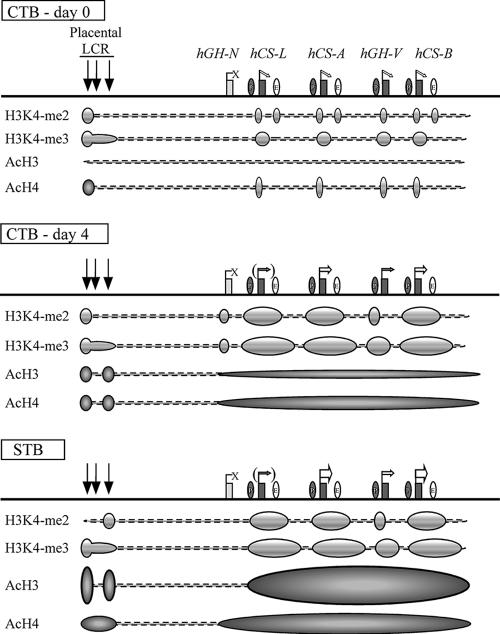
FIG. 7.
Summary of epigenetic modifications during the transition of human placental CTBs to STBs. (Top) The histone modification patterns in freshly prepared primary CTBs are diagrammed below the map. Prior to terminal STB differentiation, all the placental LCR HSs were already formed and the placental genes were expressed at trace levels, whereas hGH-N was totally inactive. Moderate levels of histone H3K4 dimethylation were observed at discrete regions, including HSV, the sites of the promoters of the placental genes, and the sites of the putative enhancers. Histone H3K4 trimethylation was also established at this point within the PGR regions. In contrast, only minimum levels of H3 acetylation were observed across the locus, and H4 acetylation was limited to HSV and at the placental-gene promoters. (Middle) After 4 days of culture, most of the CTBs fused to form a syncytium and the expression of the placental genes, especially hCS-A and hCS-B, was dramatically enhanced. Histone H3K4 di- and trimethylation patterns were well established, and the histone H3/H4 acetylation levels increased modestly at the LCR and at the cluster region in a block encompassing the four placental genes. (Bottom) In the full-term placental STBs, histone H3/H4 acetylation increased to maximum levels and the epigenetic patterns necessary for full activation of the placental genes were completed. The four placental genes are expressed at the highest levels, whereas hGH-N, lacking the epigenetic modifications, remains inactive in STBs.
Testing of the HSIII-HSV region and an individual PGR unit for sufficiency of gene activation.
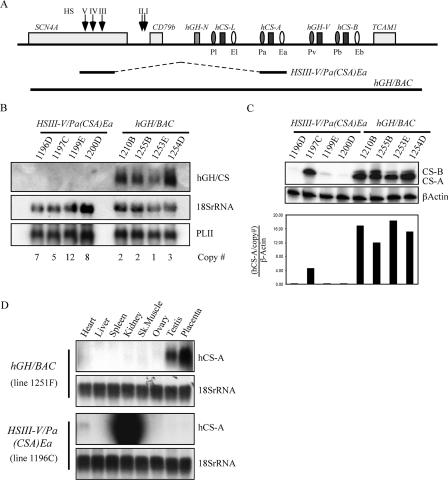
The initial modifications at the hGH locus appeared to be limited to the LCR determinants and to the individual placental-gene units (see above; summarized in Fig. 7). These data suggest that the modifications of the chromatin at each PGR unit, in conjunction with the remote 5′ HS elements, may suffice at the critical first step in the gene activation pathway. Whether these modifications are sufficient to trigger subsequent events involved in robust transcriptional activation is not clear. To test this possibility, we assembled a minimal expression unit in which an 11.5-kb genomic fragment encompassing HSIII to HSV was linked to a 7.5-kb genomic fragment encompassing hCS-A and its contiguous 5′ P-element and 3′ enhancer flanking sequences (Fig. 8A). Expression from this HSIII-V/Pa(CSA)Ea transgene was compared to that of the 123-kb BAC transgene encompassing the entire hGH locus and contiguous hGH LCR (hGH/BAC). Four mouse lines carrying each of these two transgenes were established. Transgenic males from each line were crossed with wild-type females. In the case of the 1254D line, the transgene had inserted into the X chromosome as determined by the inheritance pattern. For this reason, it was necessary to cross a transgenic female with a wild-type male in order to overcome the paternal imprinting that occurs in extraembryonic tissues in mice. Individual 18.5-day transgenic embryos generated by these crosses were identified, and hCS-A gene expression was assessed per transgene copy in the corresponding placentas (Fig. 8B and C). As is the case for expression from the hGH cluster in the human term placenta (40), the expression from hGH/BAC in the transgenic mouse placenta was predominantly hCS-A mRNA (Fig. 8C). The hCS-A expression in all four hGH/BAC lines was robust and copy number dependent, indicating strong site-of-integration independence. In contrast, expression from the HSIII-V/Pa(CSA)Ea transgene by Northern analysis was evident only in the placenta after prolonged exposure (data not shown). By the more sensitive RT-PCR (Fig. 8C), hCS-A expression from the HSIII-V/Pa(CSA)Ea transgene could be reliably detected in the placentas of one of the four lines; a second line expressed at a much lower level, and expression from the two remaining lines could be detected only at trace levels by high-cycle amplification (data not shown). Thus, the HSIII-V/Pa(CSA)Ea transgene was poorly expressed in the placenta and its expression failed to correlate with transgene copy number, indicating a marked sensitivity to the site of transgene integration. The specificity of expression from the two transgenes was further assessed in a panel of eight tissues (Fig. 8D). The expression of hCS from the hGH/BAC was appropriately confined to the placenta, with trace levels in the testes. In clear contrast, the expression of the HSIII-V/Pa(CSA)Ea transgene was strongly expressed in the kidney (Fig. 8D and data not shown).
FIG. 8.
HSIII-HSV is insufficient to activate hCS-A expression in the transgenic placenta. (A) Transgene constructs. Each of the two transgene constructs is shown below the map of the hGH locus. Each of the four P-elements was named to correspond with its adjacent gene's name (Pl, Pa, Pv, and Pb), and the three enhancers were named similarly (El, Ea, and Eb). The hGH/BAC transgene encompasses the entire region. The HSIII-V/Pa(CSA)Ea transgene is composed of an 11.5-kb fragment encompassing HSV through HSIII ligated directly to a 7.5-kb hCS-A gene fragment that includes its contiguous 5′ and 3′ sequences, as shown. Sets of transgenic mouse lines were generated with each of these two constructs. (B) Expression of the hGH cluster genes in the transgenic placentas. Four lines of mice carrying the HSIII-V/Pa(CSA)Ea transgene, each with a unique transgene insertion site, were generated and studied (1196D, 1197C, 1199E, and 1200D). Similarly, four unique transgenic lines carrying hGH/BAC were established and studied (1210B, 1252B, 1253E, 1254D). Total RNAs were purified from the transgenic placentas of the indicated lines and subjected to Northern blot analysis. The transgene copy number in each line is indicated at the bottom of each lane. The probes for 18S rRNA and mouse placental lactogen II (PLII) were used as controls. Although robust and copy-number-dependent hGH-hCS expression was observed for all four hGH/BAC lines, no signals were detected for any of the HSIII-V/Pa(CSA)Ea lines at the level of Northern blot sensitivity. (C) Comparison of the hCS-A expression levels in transgenic placentas. RT-PCR analysis was performed to amplify the hCS-A and hCS-B mRNAs using total RNAs isolated from placentas of the indicated transgenic lines. The signal intensity for hCS-A was normalized to that of β-actin as well as to the transgene copy numbers noted in panel B. The calculated relative expression levels are plotted in the frame below. The hGH/BAC lines showed strong site-of-integration-independent and copy-number-dependent expression of hCS-A in placenta. In contrast, expression in the HSIII-V/Pa(CSA)Ea lines was quite low and showed marked position effects. (D) The HSIII-V/Pa(CSA) gene failed to retain placental specificity. Northern blot analysis was performed with RNAs prepared from the indicated tissues. The tissues were isolated from the lines with the highest transgene copy number for each construct. Line 1251F (hGH/BAC) had 35 copies of the transgene; line 1196C (HSIII-V/Pa(CSA)Ea) had 63 copies. The oligoprobe for 18S rRNA was used as a loading control. hCS-A expression is predominantly placenta specific in the hGH/BAC line but is ectopically expressed at high levels in kidneys of the HSIII-V/Pa(CSA)Ea mice.
In summary, the HSIII-V/Pa(CSA)Ea transgene was unable to establish an autonomous chromatin domain or selectively target gene expression to the placenta. The expression that did occur in the placentas of the HSIII-V/Pa(CSA)Ea transgenic embryos was well below that of the hGH/BAC transgene. Thus, although early histone modifications within the cluster were limited to the placental-gene units and the 5′, remote HS determinants, additional sequences and/or genomic organization are necessary for full and appropriate placental-gene activation.
DISCUSSION
The terminal differentiation of CTBs to STBs in placental villi is paralleled by a well-defined shift in gene expression (5, 24). A robust example of this shift is the induction of hCS expression from the hGH gene cluster. We report two dynamic cell culture models of placental-cell differentiation that recapitulate this shift in gene expression. These models reveal distinct patterns and timing for two categories of activating histone modifications within the locus—the methylation of H3K4 and the acetylation of H3 and H4. The initial modifications that define the activated hGH cluster in both models are localized to each of the conserved PGR units and to the 5′, remote HS determinants. In vivo functional testing revealed that these sites of initial chromatin modification are not in themselves sufficient for full and appropriate activation of the hGH cluster in the placenta. These data suggested that additional elements situated between the cluster and the remote 5′ HS determinants or involving the native overall configuration of the multigene cluster are involved in this process.
Differentiation of the placental cells.
Two cell culture models were found to reliably recapitulate transcriptional induction of the placental genes from the hGH cluster. Clearly defined increases in expression from the placental genes of the hGH cluster were observed in BeWo cells during long-term culture. However, the level of gene expression in this system, even at its peak, was well below that of the primary placental STBs (Fig. 3C). This is not entirely surprising given that the BeWo cells, even at high density, fail to show clear evidence for the syncytialization characteristic of STB differentiation in vivo (unpublished observations). The relevance of the BeWo cells to the in vivo situation is, however, supported by molecular characterizations of this cell culture model. Expression of a number of genes playing important roles in STB formation, such as TEF-5 (28, 29, 30), PPARγ (6), Gcm-1 (4, 57), and HERV-W/syncytin (9, 19, 44), can be detected, and syncytin mRNA levels are induced threefold by day 16 of culture (our unpublished data). Although it is not clear why growing the cells to high density induced the placental-gene expression in the absence of syncytialization, the high-density culture of BeWo cells reported here represents a useful, although imperfect, model for placental-gene regulation.
The induction of gene expression from the hGH cluster in the explanted CTBs was substantially more robust than that observed in the BeWo cells. This model is also closer to the “physiologic” system; primary CTBs freshly explanted from term placenta form syncytia and recapitulate on a global scale the STB gene expression profile (24). A weakness of this model is the difficulty in obtaining sufficient numbers of cells for detailed study. Importantly, we demonstrated in the present study that both the day 0 BeWo cells and day 0 CTBs are “poised” to express the placental genes. Thus, analysis of their chromatin structure can be informative in defining the initial stage of this process.
Histone acetylation and methylation play different roles in placental-gene activation within the hGH cluster.
In a previous report, we observed distinct patterns of histone acetylation and methylation at the hGH locus in primary STBs (32). These distinct patterns suggested that these modifications may be independently targeted and may play different roles in the control of gene expression. Our current data extend this observation by suggesting that localized histone H3K4 methylation and H4 acetylation precede global and robust H3 and H4 histone acetylation of the locus in both BeWo cell and primary CTB cultures. In the BeWo cells, the histone H3K4 dimethylation pattern is present at day 0 and the subsequent induction of placental-gene transcription is paralleled by generalized and robust acetylation throughout the locus. Similarly, in the primary CTBs at day 0, the histone H3K4 di- and trimethylation within the LCR and within the placental-gene duplication units appears to define the “poised” state of the cluster prior to transcriptional activation. Significant levels of histone H4 acetylation were also detected in the day 0 CTBs, primarily at HSV and at the placental-gene promoters. By day 4 of culture, the levels of methylation and acetylation increased and extended. The patterns then became remarkably similar to, although not as great as, those seen in primary STBs. Levels of histone acetylation across the cluster appeared to track with levels of gene transcription both in the BeWo cells and in the CTB culture model. These results point to distinct roles for histone H3K4 methylation in early gene activation and for H3 and H4 acetylation in subsequent transcriptional enhancement.
Distinct roles for histone acetylation and methylation during cell differentiation or development in a number of systems have been described. At the murine IgH locus, the HS4 regulatory determinant is H3K4 dimethylated in pro-B cells, whereas histone acetylation is observed at later stages of B-cell differentiation and during IgH induction (20). The mouse Hoxb9 gene also undergoes H3K4 methylation prior to histone H3 acetylation as embryonic stem cells are induced to differentiate by retinoic acid (10). A similar schedule of modifications is observed at the human HNF4α promoter (25), the human collagenase promoter (42), and the mouse IL-2 gene regulatory region (1). These results are consistent with our present data, suggesting that histone H3K4 methylation precedes acetylation at many gene loci and may serve specific mechanistic functions in the early stages of gene activation. However, this pattern is not absolute. Several reports have indicated that histone acetylation and methylation may occur concurrently or in the inverse temporal order (3, 37, 41, 64). Thus, the epigenetic regulation of subgroups of genes can reflect different temporal orders and functions of histone modifications.
Activation of placental genes in the hGH cluster.
The “poised” configuration of the hGH cluster in day 0 BeWo cells and CTBs is accompanied by the formation of placenta-specific HSIV as well as constitutive HSIII and HSV (Fig. 4). Therefore, the full sets of HSs that define the hGH LCR in the placenta are already assembled at a time when the CS genes were barely active. This finding is consistent with observations at other loci that DNase I-HSs can form in differentiating cells prior to transcriptional activation (3, 37, 64). The subsequent acetylation of H3 and H4 at the placental LCR elements and within the hGH cluster tracks with transcriptional induction (Fig. 7). The coordinated increase in histone H3 acetylation at the 5′ remote HS and at the gene repeat units seen as CTBs transit to STBs (day 4 CTBs to STBs) is consistent with a direct mechanistic and possibly physical communication. A “looping” model, previously proposed to explain the long-range interaction of the LCR with the placental genes over a distance exceeding 40 kb, is consistent with such interactions (15, 32).
The gene units in the hGH cluster are highly conserved in structure, and the placental units appear to be individually targeted by the initial H3K4 methylation activities. These observations suggest that the LCR and an individual PGR unit might be sufficient for activating placental-gene expression. To test this model, we compared the expression from the cluster contained in a BAC transgene encompassing the entire cluster and LCR with that in a more limited transgene containing the HSIII-HSV region and the single most active PGR unit (hCS-A). The BAC transgene demonstrated robust, placenta-specific, site-of-integration-independent, and copy-number-dependent expression, indicating that the 123-kb sequence included all the components needed to fully activate the placental genes in vivo (Fig. 8B and C). This is consistent with the results of an analysis of mice carrying a somewhat shorter hGH/P1 transgene (62). In contrast, expression of the HSIII-V/Pa(CSA)Ea transgene was quite low in the placenta (Fig. 8B and C), was sensitive to the site of integration, lacked placenta specificity, and demonstrated marked ectopic expression in the kidney (Fig. 8D). This inability of a transgene that links the HSIII-HSV components directly to hCS-A to activate placental expression contrasts markedly with the robust and pituitary-specific expression of a parallel short transgene that links the hGH-N gene directly to the pituitary-specific LCR unit (HSI, HSII) (8, 26, 31, 58, 59). Thus, the current data highlight substantial differences between the pathways of gene activation of the pituitary and placental genes from the hGH cluster. These data are in agreement with our prior model indicating that activation of the hGH-N gene is mediated by direct extension of activating signals from its LCR elements, although the active placental genes communicate with the corresponding 5′ remote determinants in a more complex manner (17, 26, 27, 32).
The present studies revealed that the HSIII-HSV region is not sufficient to activate the hCS-A gene. It is useful in this context to ask whether these remote 5′ HSs that form in placental chromatin are in fact an LCR for placental-gene expression. Our studies have consistently demonstrated that the HSIII-HSV region undergoes chromatin alterations that are specific to the placenta; these sites form in placental chromatin and include the specific formation of DNase I-HSIV (reference 31 and the present study). Furthermore, localized enrichment for histone modifications is specific to the HSIII-HSV region in the placenta (reference 32 and the present study). In functional studies, we have shown that transgenes that retain the HSIII-HSV region in its native spacing express hCS specifically and robustly in mouse placentas (62). Transgenes that do not include the HSIII-HSV region do not express hCS in mouse placentas (31). In addition, multiple transgenic lines in which the HSIII-HSV region is intact but in which deletions have removed limited determinants between HSIII and the gene cluster failed to decrease or alter hCS transgene expression in mouse placentas (26). These combined data support a model in which the HSIII-HSV region is a critical remote regulatory region for placental-gene expression. However, the present data demonstrating that the HSIII-HSV region is not sufficient for hCS activation in the placenta differ from similar studies that demonstrated the sufficiency of HSI and HSII LCR elements to activate the hGH-N gene in the pituitary. Thus, the question of whether the HSIII-HSV region is necessary for this activity needs to be further addressed.
The present data lead us to conclude that the activation of placental hGH-hCS genes involves the sequential establishment of chromatin restructuring at the hGH LCR, discrete histone H3K4 methylation targeted to the individual placental-gene units, and subsequent and expansive histone H3 and H4 acetylation during placental differentiation. The functional comparison of the hGH/BAC and HSIII-V/Pa(CSA)Ea transgenes points to the existence of elements in addition to the HSIII-HSV region and the individual PGR unit that are indispensable for appropriate site-of-integration-independent and copy-number-dependent expression of the hGH-hCS genes in placentas. These missing components may be responsible for the higher-order organization of this chromatin region and a corresponding looping configuration that has been postulated to occur in the activation process (32). The identification of such elements and structures, as well as the reconstruction of the entire process of placental hGH-hCS gene activation, remains an important challenge for future studies.
Acknowledgments
We thank Michael Hubert for harvesting and culturing primary CTBs. We thank Jean Richa, Pei Fu He, and Kathleen Moosbrugger of the University of Pennsylvania Transgenic and Chimeric Mouse Core Facility for their help in generating transgenic mice.
This work was supported by R01s HD46737, HD25147 (N.E.C. and S.A.L.), and HD07447 (S.H.).
Footnotes
Published ahead of print on 16 July 2007.
REFERENCES
- 1.Adachi, S., and E. V. Rothenberg. 2005. Cell-type-specific epigenetic marking of the IL2 gene at a distal cis-regulatory region in component, nontranscribing T-cells. Nucleic Acids Res. 33:3200-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alsat, E., J. Guibourdenche, D. Luton, F. Frankenne, and D. Evain-Brion. 1997. Human placental growth hormone. Am. J. Obstet. Gynecol. 177:1526-1534. [DOI] [PubMed] [Google Scholar]
- 3.Anguita, E., J. Hughes, C. Heyworth, G. A. Blobel, W. G. Wood, and D. R. Higgs. 2004. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 23:2841-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anson-Cartwright, L., K. Dawson, D. Holmyard, S. J. Fisher, R. A. Lazzarini, and J. C. Cross. 2000. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 25:311-314. [DOI] [PubMed] [Google Scholar]
- 5.Aronow, B. J., B. D. Richardson, and S. Handwerger. 2001. Microarray analysis of trophoblast differentiation: gene expression reprogramming in key function categories. Physiol. Genomics 6:105-116. [DOI] [PubMed] [Google Scholar]
- 6.Barak, Y., M. C. Nelson, E. S. Ong, Y. Z. Jones, P. Ruiz-Lozano, K. R. Chien, A. Koder, and R. M. Evans. 1999. PPARγ is required for placental, cardiac, and adipose tissue development. Mol. Cell 4:585-595. [DOI] [PubMed] [Google Scholar]
- 7.Barrera-Saldaña, H. A. 1998. Growth hormone and placental lactogen: biology, medicine and biotechnology. Gene 211:11-18. [DOI] [PubMed] [Google Scholar]
- 8.Bennani-Baiti, I. M., S. L. Asa, D. Song, R. Iratni, S. A. Liebhaber, and N. E. Cooke. 1998. DNase I-hypersensitive sites I and II of the human growth hormone locus control region are a major developmental activator of somatotrope gene expression. Proc. Natl. Acad. Sci. USA 95:10655-10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blond, J.-L., F. Besème, L. Duret, O. Bouton, F. Bedin, H. Perron, B. Mandrand, and F. Mallet. 1999. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virol. 73:1175-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambeyron, S., and W. A. Bickmore. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18:1119-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, E. Y., Y.-C. Liao, D. H. Smith, H. A. Barrera-Saldaña, R. E. Gelinas, and P. H. Seeburg. 1989. The human growth hormone locus: nucleotide sequence, biology, and evolution. Genomics 4:479-497. [DOI] [PubMed] [Google Scholar]
- 12.Cross, J. C., Z. Werb, and S. J. Fisher. 1994. Implantation and the placenta: key pieces of the development puzzle. Science 266:1508-1518. [DOI] [PubMed] [Google Scholar]
- 13.Cross, J. C., D. Baczyk, N. Dobric, M. Hemberger, M. Hughes, D. G. Simmons, H. Yamamoto, and J. C. P. Kingdom. 2003. Genes, development and evolution of the placenta. Placenta 24:123-130. [DOI] [PubMed] [Google Scholar]
- 14.Cross, J. C. 2005. How to make a placenta: mechanisms of trophoblast cell differentiation in mice. Placenta 26(Suppl. A):S3-S9. [DOI] [PubMed] [Google Scholar]
- 15.Dean, A. 2006. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 22:38-45. [DOI] [PubMed] [Google Scholar]
- 16.Elefant, F., N. E. Cooke, and S. A. Liebhaber. 2000. Targeted recruitment of histone acetyltransferase activity to a locus control region. J. Biol. Chem. 275:13827-13834. [DOI] [PubMed] [Google Scholar]
- 17.Elefant, F., Y. Su, S. A. Liebhaber, and N. E. Cooke. 2000. Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region. EMBO J. 19:6814-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankenne, F., J. Closset, F. Gomez, M. L. Scippo, J. Smal, and G. Hennen. 1988. The physiology of growth hormones (GHs) in pregnant women and partial characterization of the placental GH variant. J. Clin. Endocrinol. Metab. 66:1171-1180. [DOI] [PubMed] [Google Scholar]
- 19.Frendo, J.-L., D. Olivier, V. Cheynet, J.-L. Blond, O. Bouton, M. Vidaud, M. Rabreau, D. Evain-Brion, and F. Mallet. 2003. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell. Biol. 23:3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrett, F. E., A. V. Emelyanov, M. A. Sepulveda, P. Flanagan, S. Volpi, F. Li, D. Loukinov, L. A. Eckhardt, V. V. Lobanenkov, and B. K. Birshtein. 2005. Chromatin architecture near a potential 3′ end of the Igh locus involves modular regulation of histone modifications during B-cell development and in vivo occupancy at CTCF sites. Mol. Cell. Biol. 25:1511-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman, W. E., G. Goldberg, L. H. Bowman, D. Steinmetz, and D. Schlessinger. 1983. Mouse rDNA: sequences and evolutionary analysis of spacer and mature RNA regions. Mol. Cell. Biol. 3:1488-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodman, H. M., L.-R. Tai, J. Ray, N. E. Cooke, and S. A. Liebhaber. 1991. Human growth hormone variant produces insulin-like and lipolytic responses in rat adipose tissue. Endocrinology 129:1779-1783. [DOI] [PubMed] [Google Scholar]
- 23.Handwerger, S. 1991. Clinical counterpoint: the physiology of placental lactogen in human pregnancy. Endocr. Rev. 12:329-336. [DOI] [PubMed] [Google Scholar]
- 24.Handwerger, S., and B. Aronow. 2003. Dynamic changes in gene expression during human trophoblast differentiation. Recent Prog. Horm. Res. 58:263-281. [DOI] [PubMed] [Google Scholar]
- 25.Hatzis, P., and I. Talianidis. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 10:1467-1477. [DOI] [PubMed] [Google Scholar]
- 26.Ho, Y., F. Elefant, N. Cooke, and S. Liebhaber. 2002. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell 9:291-302. [DOI] [PubMed] [Google Scholar]
- 27.Ho, Y., F. Elefant, S. A. Liebhaber, and N. E. Cooke. 2006. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell 23:365-375. [DOI] [PubMed] [Google Scholar]
- 28.Jacquemin, P., J. A. Martial, and I. Davidson. 1997. Human TEF-5 is preferentially expressed in placenta and binds to multiple functional elements of the human chorionic somatomammotropin-B gene enhancer. J. Biol. Chem. 272:12928-12937. [DOI] [PubMed] [Google Scholar]
- 29.Jacquemin, P., V. Sapin, E. Alsat, D. Evain-Brion, P. Dolle, and I. Davidson. 1998. Differential expression of the TEF family of transcription factors in the murine placenta and during differentiation of primary human trophoblasts in vitro. Dev. Dyn. 212:423-436. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, S.-W., K. Wu, and N. L. Eberhardt. 1999. Human placental TEF-5 transactivates the human chorionic somatomammotropin gene enhancer. Mol. Endocrinol. 13:879-889. [DOI] [PubMed] [Google Scholar]
- 31.Jones, B. M., B. R. Monks, S. A. Liebhaber, and N. E. Cooke. 1995. The human growth hormone gene is regulated by a multicomponent locus control region. Mol. Cell. Biol. 15:7010-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura, A. P., S. A. Liebhaber, and N. E. Cooke. 2004. Epigenetic modifications at the human growth hormone locus predict distinct roles for histone acetylation and methylation in placental gene activation. Mol. Endocrinol. 18:1018-1032. [DOI] [PubMed] [Google Scholar]
- 33.Kliman, H. J., J. E. Nestler, E. Sermasi, J. M. Sanger, and J. F. Strauss III. 1986. Purification, characterization, and in vitro differentiation of cytotrophoblast from human term placentae. Endocrinology 118:1567-1582. [DOI] [PubMed] [Google Scholar]
- 34.Knöfler, M., R. Vasicek, and M. Schreiber. 2001. Key regulatory transcription factors involved in placental trophoblast development. Placenta 22(Suppl. A):S83-S92. [DOI] [PubMed] [Google Scholar]
- 35.Lachner, M., R. J. O'Sullivan, and T. Jenuwein. 2003. An epigenetic road map for histone lysine methylation. J. Cell Sci. 116:2117-2124. [DOI] [PubMed] [Google Scholar]
- 36.Lacroix, M. C., J. Guibourdenche, J. L. Frendo, F. Muller, and D. Evain-Brion. 2002. Human placental growth hormone. Placenta 23(Suppl. A):S87-S94. [DOI] [PubMed] [Google Scholar]
- 37.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 38.Loregger, T., J. Pollheimer, and M. Knöfler. 2003. Regulatory transcription factors controlling function and differentiation of human trophoblast—a review. Placenta 24(Suppl. A):S104-S110. [DOI] [PubMed] [Google Scholar]
- 39.MacLeod, J. N., I. Worsley, J. Ray, H. G. Friesen, S. A. Liebhaber, and N. E. Cooke. 1991. Human growth hormone-variant is a biologically active somatogen and lactogen. Endocrinology 128:1298-1302. [DOI] [PubMed] [Google Scholar]
- 40.MacLeod, J. N., A. K. Lee, S. A. Liebhaber, and N. E. Cooke. 1992. Developmental control and alternative splicing of the placentally expressed transcripts from the human growth hormone gene cluster. J. Biol. Chem. 267:14219-14226. [PubMed] [Google Scholar]
- 41.Margueron, R., P. Trojer, and D. Reinberg. 2005. The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 15:163-176. [DOI] [PubMed] [Google Scholar]
- 42.Martens, J. H. A., M. Verlaan, E. Kalkhoven, and A. Zantema. 2003. Cascade of distinct histone modifications during collagenase gene activation. Mol. Cell. Biol. 23:1808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6:838-849. [DOI] [PubMed] [Google Scholar]
- 44.Mi, S., X. Lee, X.-P. Li, G. M. Veldman, H. Finnerty, L. Racie, E. LaVallie, X.-Y. Tang, P. Edouard, S. Howes, J. C. Keith, Jr., and J. M. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785-789. [DOI] [PubMed] [Google Scholar]
- 45.Misra-Press, A., N. E. Cooke, and S. A. Liebhaber. 1994. Complex alternative splicing partially inactivates the human chorionic somatomammotropin-like (hCS-L) gene. J. Biol. Chem. 269:23220-23229. [PubMed] [Google Scholar]
- 46.Nachtigal, M. W., B. E. Nickel, and P. A. Cattini. 1993. Pituitary-specific repression of placental members of the human growth hormone gene family: a possible mechanism for locus regulation. J. Biol. Chem. 268:8473-8479. [PubMed] [Google Scholar]
- 47.Norwitz, E. R., D. J. Schust, and S. J. Fisher. 2001. Implantation and the survival of early pregnancy. N. Engl. J. Med. 345:1400-1408. [DOI] [PubMed] [Google Scholar]
- 48.Ohlsson, C., B. A. Bengtsson, O. G. Isaksson, T. T. Andreassen, and M. C. Slootweg. 1998. Growth hormone and bone. Endocr. Rev. 19:55-79. [DOI] [PubMed] [Google Scholar]
- 49.Peterson, C. L., and M.-A. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14:R546-R551. [DOI] [PubMed] [Google Scholar]
- 50.Pötgens, A. J. G., U. Schmitz, P. Bose, A. Versmold, P. Kaufmann, and H.-G. Frank. 2002. Mechanisms of syncytial fusion. Placenta 23(Suppl. A):S107-S113. [DOI] [PubMed] [Google Scholar]
- 51.Red-Horse, K., Y. Zhou, O. Genbacev, A. Prakobphol, R. Foulk, M. McMaster, and S. J. Fisher. 2004. Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J. Clin. Investig. 114:744-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richards, R. G., S. M. Hartman, and S. Handwerger. 1994. Human cytotrophoblast cells cultured in maternal serum progress to a differentiated syncytial phenotype expressing both human chorionic gonadotropin and human placental lactogen. Endocrinology 135:321-329. [DOI] [PubMed] [Google Scholar]
- 53.Roberts, R. M., T. Ezashi, and P. Das. 2004. Trophoblast gene expression: transcription factors in the specification of early trophoblast. Reprod. Biol. Endocrinol. 2:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossant, J., and J. C. Cross. 2001. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2:538-548. [DOI] [PubMed] [Google Scholar]
- 55.Rote, N. S., S. Chakrabarti, and B. P. Stetzer. 2004. The role of human endogenous retroviruses in trophoblast differentiation and placental development. Placenta 25:673-683. [DOI] [PubMed] [Google Scholar]
- 56.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. T. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 57.Schreiber, J., E. Riethmacher-Sonnenberg, D. Riethmacher, E. E. Tuerk, J. Enderich, M. R. Bösl, and M. Wegner. 2000. Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol. Cell. Biol. 20:2466-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shewchuk, B. M., S. L. Asa, N. E. Cooke, and S. A. Liebhaber. 1999. Pit-1 binding sites at the somatotrope-specific DNase I hypersensitive sites I, II of the human growth hormone locus control region are essential for in vivo hGH-N gene activation. J. Biol. Chem. 274:35725-35733. [DOI] [PubMed] [Google Scholar]
- 59.Shewchuk, B. M., S. A. Liebhaber, and N. E. Cooke. 2002. Specification of unique Pit-1 activity in the hGH locus control region. Proc. Natl. Acad. Sci. USA 99:11784-11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simmons, D. G., and J. C. Cross. 2005. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev. Biol. 284:12-24. [DOI] [PubMed] [Google Scholar]
- 61.Simpson, E. R., and P. C. MacDonald. 1981. Endocrine physiology of the placenta. Annu. Rev. Physiol. 43:163-188. [DOI] [PubMed] [Google Scholar]
- 62.Su, Y., S. A. Liebhaber, and N. E. Cooke. 2000. The human growth hormone gene cluster locus control region supports position-independent pituitary- and placenta-specific expression in the transgenic mouse. J. Biol. Chem. 275:7902-7909. [DOI] [PubMed] [Google Scholar]
- 63.Sullivan, M. H. F. 2004. Endocrine cell lines from the placenta. Mol. Cell. Endocrinol. 228:103-119. [DOI] [PubMed] [Google Scholar]
- 64.Szutorisz, H., C. Canzonetta, A. Georgiou, C.-M.Chow, L. Tora, and N. Dillon. 2005. Formation of an active tissue-specific chromatin domain initiated by epigenetic marking at the embryonic stem cell stage. Mol. Cell. Biol. 25:1804-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor, R. N., E. D. Newman, and S. Chen. 1991. Forskolin and methotrexate induce an intermediate trophoblast phenotype in cultured human choriocarcinoma cells. Am. J. Obstet. Gynecol. 164:204-210. [DOI] [PubMed] [Google Scholar]
- 66.Walker, W. H., S. L. Fitzpatrick, and G. F. Saunders. 1990. Human placental lactogen transcriptional enhancer: tissue specificity and binding with specific proteins. J. Biol. Chem. 265:12940-12948. [PubMed] [Google Scholar]
- 67.Watson, E. D., and J. C. Cross. 2005. Development of structures and transport functions in the mouse placenta. Physiology 20:180-193. [DOI] [PubMed] [Google Scholar]
- 68.Xu, B., L. Lin, and N. S. Rote. 1999. Identification of a stress-induced protein during human trophoblast differentiation by differential display analysis. Biol. Reprod. 61:681-686. [DOI] [PubMed] [Google Scholar]
- 69.Yoo, E. J., I. Cajiao, J.-S. Kim, A. P. Kimura, A. Zhang, N. E. Cooke, and S. A. Liebhaber. 2006. Tissue-specific chromatin modifications at a multigene locus generate asymmetric transcriptional interactions. Mol. Cell. Biol. 26:5569-5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young, J. A. T., and J. Trowsdale. 1985. A processed pseudogene in an intron of the HLA-DPβ1 chain gene is a member of the ribosomal protein L32 gene family. Nucleic Acids Res. 13:8883-8891. [DOI] [PMC free article] [PubMed] [Google Scholar]