Abstract
Archaea possess a basal transcriptional apparatus that resembles that of eukaryotes. Here we report the 2.1-Å crystal structure of the archaeal transcription factor complex formed by the TATA-box-binding protein (TBP), the transcription factor IIB homolog, and a DNA target, all from the hyperthermophile Pyrococcus woesei. The overall fold of these two basal transcription factors is essentially the same as that of their eukaryotic counterparts. However, in comparison with the eukaryotic complexes, the archaeal TBP–DNA interface is more symmetrical, and in this structure the orientation of the preinitiation complex assembly on the promoter is inverted with respect to that seen in all crystal structures of comparable eukaryotic systems. This study of the structural details of an archaeal transcription factor complex presents the opportunity to examine the evolution of the basal eukaryotic transcriptional apparatus from a stereochemical viewpoint and to extend our understanding of the physical biochemistry of transcriptional initiation.
The recent sequencing of the first archaeal genome has confirmed the designation of the archaea as a third kingdom of life, distinct from eubacteria and eukaryotes (1). Recent discoveries indicate that the basal components of transcription in archaea resemble those in the eukaryotic RNA polymerase II transcriptional system (2). Archaeal homologs to the transcription factors TATA-box binding protein (TBP) and transcription factor IIB (TFIIB) have been reported (3–6). As in eukaryotic polymerase II transcription, archaeal transcription is initiated by an A+T-rich TATA-like segment known as the “boxA” sequence, containing a consensus 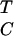 TTA
TTA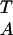 ANN (hereafter referred to as the boxA/TATA element) (7, 8). Interestingly, one of the common archaeal promoter sequences contains the boxA/TATA element, TTTATATA, which is an inverted canonical eukaryotic TATA box (8). Like eukaryotic TBP, archaeal TBP recognizes the boxA/TATA element, and transcription factor B (TFB) from Pyrococcus woesei (pwTFB) binds to the TBP–DNA complex (3). Homologs to other eukaryotic basal transcription factors, or TBP-associated factors (TAFs), however, are not present in the archaeal genome. In fact, promoter-specific transcription can occur in a purified reconstituted archaeal system, with only TBP, TFB, and polymerase (9).
ANN (hereafter referred to as the boxA/TATA element) (7, 8). Interestingly, one of the common archaeal promoter sequences contains the boxA/TATA element, TTTATATA, which is an inverted canonical eukaryotic TATA box (8). Like eukaryotic TBP, archaeal TBP recognizes the boxA/TATA element, and transcription factor B (TFB) from Pyrococcus woesei (pwTFB) binds to the TBP–DNA complex (3). Homologs to other eukaryotic basal transcription factors, or TBP-associated factors (TAFs), however, are not present in the archaeal genome. In fact, promoter-specific transcription can occur in a purified reconstituted archaeal system, with only TBP, TFB, and polymerase (9).
We present here the 2.1-Å crystal structure of the ternary archaeal transcription complex, illustrating the interaction of TBP, TFB, and a promoter fragment. The components are all from P. woesei, a hyperthermophilic archaeon which exhibits an optimal growth temperature of 105°C (10). The amino acid sequence of pwTBP is 36–41% identical to the conserved C-terminal core of its eukaryotic counterparts (3), and, as expected from this degree of homology, we have recently found that the general features of its three-dimensional structure are the same as those of eukaryotic TBP (11). pwTFB is 28–32% identical to full-length eukaryotic TFIIBs, and 25% identical to the C-terminal domain of the yeast RNA polymerase III transcription factor BRF (4). Of special relevance to this report is the fact that the C-terminal 200 residues of pwTFB (pwTFBc) correspond (32% identity) to the C-terminal protease-resistant core of TFIIB (TFIIBc), which interacts with TBP and whose structure has been determined both alone (12) and in a complex with TBP and the TATA element (13). In eukaryotes, TBP and TFIIBc each contain imperfect direct repeats, which may have diverged from a single domain. The P. woesei repeats are more exact than those of their eukaryotic counterparts; 28% and 39% identical for pwTFB and pwTBP, respectively, compared with 22% and 27% for human TFIIB and TBP (4, 5, 14). Thus, the archaeal proteins more closely resemble the presumed common ancestral transcription factors from which both archaeal and eukaryotic factors evolved (5). The structure of the ternary complex presented here should broaden our view of the evolution of transcriptional initiation and provide a stereochemical context for extending the thermodynamic and kinetic analysis of the transcription preinitiation complex to unusual solvent conditions and temperatures above the boiling point of water.
MATERIALS AND METHODS
C-terminally truncated (Δ182–191) pwTBP was overproduced and purified as described for full-length pwTBP, with the addition of a Superdex-75 gel filtration column (Pharmacia) as a final step (11). We were unable to grow x-ray-quality crystals in the presence of the acidic C-terminal cluster of 10 residues found only in some archaeal pwTBPs. The cluster was therefore deleted, leaving residues 1–181, which correspond to the conserved C-terminal functional core of TBP found in eukaryotes. pwTFBc was overproduced in a T7 RNA polymerase-based expression system (Novagen) in Escherichia coli. The pwTFBc backbone hydrolyzes spontaneously over a wide range of salt and pH conditions. Potassium acetate (200 mM) and Tris, pH 8.3, were necessary to prevent degradation. Cells were lysed by sonication for 3 min in 200 mM potassium acetate/10 mM Tris, pH 8.3. The supernatant was incubated for 10 min at 90°C, causing extensive precipitation of E. coli proteins. pwTFB in the remaining soluble fraction was purified to homogeneity over three chromatographic columns: Fast-S (Pharmacia) in 10 mM Tris, pH 8.3, 100–500 mM potassium acetate gradient; SOURCE 15S (Pharmacia) in 10 mM Tris, pH 8.3, 200–800 mM potassium acetate gradient; and a Superdex-75 gel-filtration (Pharmacia) column in 1 M potassium acetate, 10 mM Tris, pH 8.3, as the final step. Oligonucleotides were synthesized by the phosphoramidite method (Keck facility, Yale University) and purified by anion-exchange chromatography on a SOURCE 15Q column (Pharmacia) at pH 11.8 at room temperature. The DNA target was chosen from the promoter for the ef1α gene of P. woesei. The best crystals were grown with a 17-base-pair duplex containing the boxA/TATA element flanked by 2 base pairs upstream and 7 base pairs downstream (Fig. 1).
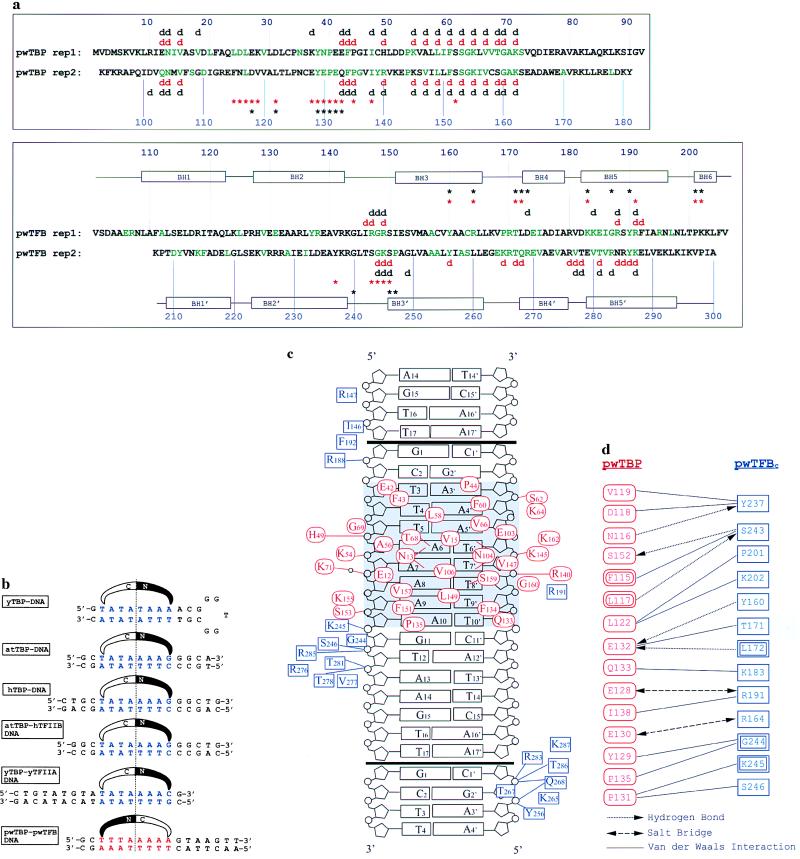
Figure 1.
(a) Amino acid sequences for pwTBP and pwTFBc. The direct repeats of pwTBP and pwTFB are aligned. pwTBP amino acids in green are conserved with Arabidopsis thaliana TBP (atTBP), and pwTFBc residues in green are conserved with human TFIIBc. Red symbols above and below the sequence indicate interactions seen in this structure (d for DNA contacts and ∗ for protein contacts). Black symbols indicate the corresponding interactions in the eukaryotic ternary structure (11). Black rectangles indicate α-helices in pwTFBc. Contacting residues are defined as having a distance < 4.0 Å. (b) Comparison of the promoter fragment crystallized in this study to five different TATA boxes seen in other TBP complex structures: yeast TBP (yTBP)–DNA (15), A. thaliana TBP (atTBP)–DNA (16, 17), human TBP (hTBP)–DNA (18), atTBP–human TFIIB–DNA (11), and yeast TBP–TFIIA–DNA (19, 20). Sequences are centered on the pseudodyad axis of the TATA box, and the orientation is shown with a schematic representation of the N-terminal and C-terminal domains of TBP. (c) Protein–DNA interactions in this structure. pwTBP residues are red ovals, and pwTFBc residues are green rectangles. The boxA/TATA element is shaded. Black bars indicate the end of the crystallization oligonucleotide. Crystal packing contacts between DNA molecules simulate a contiguous B form helix with a smaller than normal twist of 9° at the junction. This allows pwTFB to interact with phosphates of neighboring molecules in both directions. Residues involved in van der Waals interactions are shown near the region of DNA they contact. Lines indicate hydrogen bonds or salt bridge interactions. (d) Residues interacting at the pwTBP–pwTFBc interface. pwTBP residues are red and pwTFBc residues, blue. Double rectangles or ovals indicate a main-chain interaction.
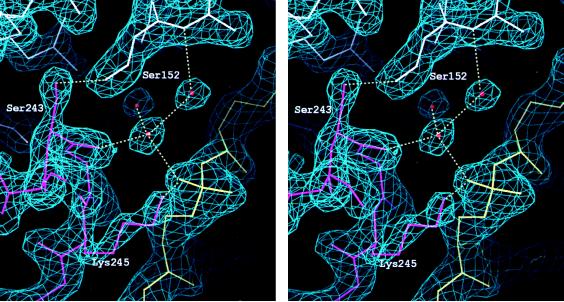
For selenomethionine-substituted crystals, selenomethionine was incorporated into both proteins as described (11). To form a ternary complex, both proteins were mixed in stoichiometric amounts with DNA, each at a concentration of 0.3 mM (18 mg/ml). Monoclinic crystals (C2, a = 125.7 Å, b = 91.2 Å, c = 74.2 Å, β = 122.7°, one complex per asymmetric unit) grew in 10–20 days at 18°C (typically 0.3 mm × 0.3 mm × 0.1 mm) from 4-μl hanging drops containing 2 μl of 8% (wt/vol) PEG 8000 and 200 mM potassium phosphate, pH 7.4, mixed with 2 μl of 0.3 mM pwTBP/pwTFBc/DNA complex in 150 mM potassium acetate and 10 mM Tris, pH 8.3, equilibrated over a reservoir containing 8% PEG 8000 and 200 mM potassium phosphate, pH 7.4. Crystals were briefly washed in cryoprotectant [20% PEG 8000, 20% (vol/vol) glycerol, and 200 mM potassium phosphate at pH 7.4], before freezing in the nitrogen cooling stream at 100 K. Highest resolution data were collected to 2.1-Å resolution at the Cornell High Energy Synchrotron Source (CHESS) beamline A1. All data sets were processed with denzo (21) [except Xentronics data sets, which were processed with xds (22)]. All data sets were scaled with scalepack (21). Using the uncomplexed pwTBP as a search model, an unambiguous molecular replacement solution, generated by x-plor (23) provided phases to 3 Å which were used to locate iodine and selenium sites by difference Fourier maps. Iodine and selenium sites were refined and phases were computed with ml-phare (24). Phases were improved by solvent flattening with dphase (G. Van Duyne, Yale) and combined with partial model phases by using ml-phare (24). The model was built with the program o (25), and positional and simulated annealing refinements were carried out with x-plor (23). The model was rebuilt periodically using sigma A maps (Fig. 2). Flat bulk solvent was modeled using x-plor (23, 27). The current model includes all 181 TBP residues, all 34 nucleotides, and all but the 7 N-terminal pwTFBc residues. The structure has been refined to an R factor of 21.2% and free R factor of 26.8%, containing 288 water molecules with refined temperature factors less than 55 Å2 at unit occupancy. No residues other than glycine have ϕ, ψ angles in disallowed regions. The crystallographic analysis is summarized in Table 1.
Figure 2.
Stereo pair of electron density of a sigma A weighted (26) 2|Fo| − |Fc| map, contoured at 2σ. pwTFBc residues are magenta, pwTBP residues are white, DNA is yellow, and water molecules are orange spheres.
Table 1.
Summary of crystallographic analysis: Data quality and refinement
| Type | Resolution limit, Å | λ, Å | Rsym | (last shell) | Reflections | % complete | Detector (source) | Phasing power, Å |
|---|---|---|---|---|---|---|---|---|
| Native | 3.0 | 1.54 | 5.3 | (14.4) | 12,942 | 91 | Xentronics (Yale) | |
| Se-Met | 2.4 | 0.9794 | 4.8 | (18.0) | 10,852 | 82 | Mar image plate (ESRF) | 0.13 |
| Anomalous | 0.32 | |||||||
| Iodo-1* | 3.0 | 1.54 | 6.4 | (30.1) | 8,749 | 61 | Xentronics (Yale) | 0.75 |
| Iodo-2* | 2.9 | 1.54 | 10.0 | (39.0) | 13,234 | 92 | Raxis (Yale) | 0.65 |
| Iodo-3* | 3.0 | 1.54 | 6.4 | (24.2) | 12,986 | 90 | Xentronics (Yale) | 1.03 |
| Iodo-4* | 2.1 | 0.920 | 8.8 | (39.6) | 40,619 | 92 | CCD (CHESS A1) | 0.75 |
| Avg. figure of merit: 0.5 | ||||||||
| Resolution, Å | R factor | Free R factor | ||||||
| 40–2.1 (2σ data) | 21.2 | 26.8 | ||||||
| 40–2.1 (all data) | 22.8 | 28.2 | ||||||
| rms deviations | bond lengths 0.009 Å; bond angles 1.55° | |||||||
Rsym = Σ|Ih − 〈Ih〉|/ΣIh, where 〈Ih〉 is the average intensity over Friedel and symmetry equivalents. Phasing power = Σ|FH|/Σ∥FPHobs| − |FPHcalc∥. Anomalous phasing power = Σ|F"|/Σ∥ADobs| − |ADcalc∥, where AD is Bijvoet difference. CHESS, Cornell High Energy Synchrotron Source; CCD, charge-coupled device; ESRF, European Synchrotron Radiation Facility.
Crystals were grown with 5-iodouracil replacing thymine in the DNA at the following positions (Fig. 1 shows the numbering scheme). Iodo-1: 3, 4, and 5; Iodo-2: 9′ and 10′; Iodo-3: 6′, 7′, and 8′; and Iodo-4: 6′ and 7′.
RESULTS AND DISCUSSION
TBP–DNA Interface.
Like its eukaryotic counterparts, pwTBP binds in the minor groove of an eight-base-pair boxA/TATA element, imposing a similar severe distortion (Fig. 3). Two pairs of pwTBP phenylalanine side chains penetrate between the first two and last two base pairs, which form two kinks, as in the eukaryotic complexes (15–19, 29). When viewed in the projection of maximum bend, the trajectory of the promoter is bent by approximately 65°, compared with 70° for the eukaryotic ternary complex (13) and 80° for eukaryotic binary complexes (15, 16). pwTBP in the complex is essentially unchanged compared with uncomplexed pwTBP (11). At the center of the boxA/TATA element there are six hydrogen bonds between TBP side chains and the N3 of adenine and the O2 of thymine; the remaining hydrogen bond acceptors of the minor groove are buried in a hydrophobic van der Waals contact surface (Fig. 1). There are no T-A steps present at the kinks in this complex, therefore there is no possibility of intrastrand hydrogen bonding of the type seen in the eukaryotic complexes (15–18).
Figure 3.
(a) Ribbons (28) drawing of the overall structure of the ternary complex. pwTBP is in red, the N-terminal domain of pwTFBc is green, the C-terminal domain is yellow, and DNA is blue. (b) Stereo drawing of pwTFBc, with the same orientation as in a.
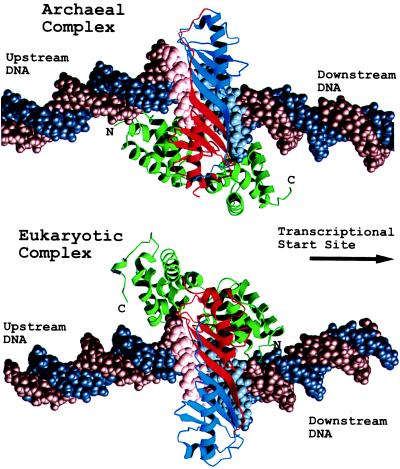
Compared with the orientation of eukaryotic TBP, pwTBP binds to the promoter DNA in an inverted orientation with respect to the start site of transcription (Fig. 4). This places the C-terminal stirrup on the “downstream” end of the boxA/TATA element rather than the “upstream” end as seen in the crystal structure of all eukaryotic TBP-containing complexes (13, 15–20). Since the pwTFBc binds to the C-terminal stirrup of pwTBP in a manner similar to its eukaryotic homologs, the orientation of pwTFBc on the boxA/TATA element is also inverted with regard to the start site. The issue of polarity is discussed below.
Figure 4.
Ribbons (28) drawing of the archaeal and eukaryotic ternary complexes, with B-form DNA extended by modeling in both directions outside of the oligonucleotides used in crystallizations. DNA is depicted as a space-filling model. Proteins are depicted as ribbons. The N-terminal sequence repeat of TBP is red and the C-terminal repeat is blue, and TFBc and TFIIBc are green.
Comparison of the Archaeal and Eukaryotic Ternary Complexes.
Like TFIIBc, pwTFBc is composed of two cyclin A-like domains, indicating that the cyclin fold predates the evolution of the eukaryotic nucleus. The two domains straddle the C-terminal stirrup of pwTBP, with stereochemistry similar, but not identical, to that of the eukaryotic complex (Fig. 1). pwTFBc interacts with the DNA backbone but not the bases, both upstream and downstream of the boxA/TATA-box sequence (Fig. 1). Like human TFIIBc, pwTFBc presents a positively charged surface to the TBP–DNA complex (Fig. 5). pwTFBc and human TFIIBc each contact DNA with 10 positively charged amino acids, 8 of which are located at equivalent positions in their respective amino acid sequence (Fig. 1). A total of 3,200 Å2 of solvent-exposed surface is buried upon binding of pwTFBc to pwTBP–DNA: 1,400 Å2 in the pwTBP interface and 1,800 Å2 in the DNA interface. These values are comparable to those of the corresponding eukaryotic complex (13). A superposition of the corresponding Cα and C1′ atoms of the eukaryotic and archaeal ternary complexes gives an rms deviation of 2.9 Å. As apparent from Fig. 5, the deviations are primarily due to en bloc domain shifts rather than changes in the tertiary structure within domains.
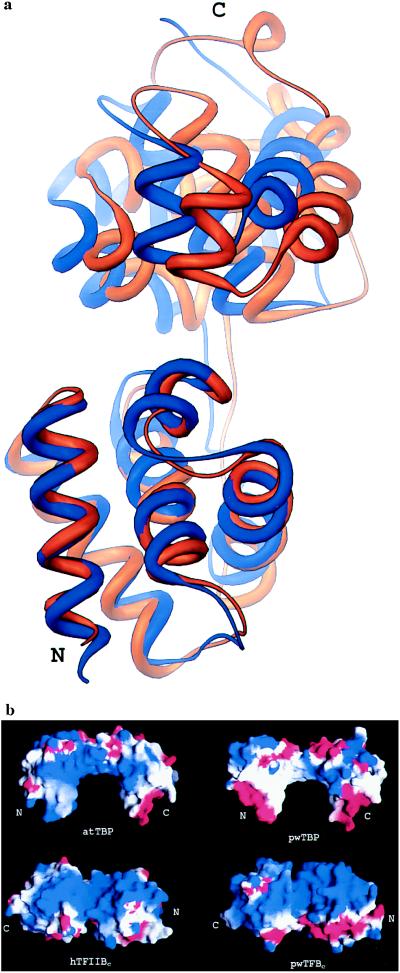
Figure 5.
(a) Superposition of the N-terminal cyclin A-like domains of pwTFBc (blue) and human TFIIBc (orange) in the eukaryotic ternary complex (14). pwTFBc residues 108–197 were aligned with human TFIIB residues 113–202, giving an rms deviation of 1.1 Å for corresponding Cα atoms. In a different superposition (not shown), pwTFB residues 211–300 can be aligned with human TFIIBc residues 214–303, giving an rms deviation of 1.2 Å for corresponding Cα atoms. (b) Comparison of the electrostatic potential surface of pwTBP with A. thaliana TBP (atTBP), and human TFIIBc (hTFIIBc) with pwTFBc. Molecular surfaces and electrostatic potentials were rendered with grasp (30), calculated using 0.1 M salt, and −2kT (red) to +2kT (blue). Coordinates for the eukaryotic ternary complex were provided by S. K. Burley (Rockefeller University, New York).
Fig. 5 compares the conformation of pwTFBc with that of human TFIIBc (13) in their respective ternary complexes. Each cyclin A-like domain of pwTFBc superimposes quite well individually, but there is a slight difference in the relative orientation of the two domains. There are three main structural differences between pwTFBc and human TFIIBc: (i) the linker between the two domains of pwTFBc has two additional residues and a short helical turn formed by residues 201–204; (ii) the last 13 residues at the C terminus of eukaryotic TFIIB are absent from pwTFBc; and (iii) the short helix BH6′ found in human TFIIBc (13) is absent from pwTFBc. None of these changes have an obvious effect on the architecture of the complex.
Orientation of the Preinitiation Complex.
All eukaryotic TBP/DNA complexes whose structures have been determined show the TBP bound to the TATA element in a common orientation—i.e., with the C-terminal stirrup contacting the upstream half of the boxA/TATA element, and the N-terminal stirrup contacting the downstream half. The inverted orientation in this structure and the prevalence of inverted eukaryotic TATA sequences in archaeal boxA/TATA box elements raise an interesting question: does pwTBP alone bind with a preferred orientation to the promoter and, if it does, is the orientation opposite to that of eukaryotes?
An inverted arrangement of pwTBP/pwTFB on an archaeal promoter would imply a different mode of interaction of the basal factors with RNA polymerase. Two observations are consistent with this possibility. First, in eukaryotes, the N terminus of TFIIB contacts RNA polymerase II through an interaction with TFIIF (31). While the N terminus of archaeal TFB is conserved with that of eukaryotic TFIIB, the absence of a TFIIF homolog in the archaeal genome suggests the possibility of a different arrangement between the general factors and the archaeal polymerase. Second, the DNase I footprint of the Pyrococcus TBP/TFB/DNA complex shows a clear difference in the pattern of protection in comparison to the eukaryotic complex (9).
To the extent that pwTBP has a preferred orientation on the boxA/TATA element it is not clear what specifies it. The pwTBP/DNA interface is even more symmetrical than that of the eukaryotic complexes (Fig. 1). The distribution of side-chain contacts in the archaeal system is exactly symmetrical. Of the four asymmetrically positioned side-chain contacts present in eukaryotic TBPs, the most significant is Pro-135 (archaeal numbering), which has been suggested to play a major role in determining the polarity by which eukaryotic TBPs bind to the promoter (29). In eukaryotic TBPs the corresponding position in the first sequence repeat is occupied by an alanine, which Juo et al. (29) argue is sterically more acceptable to the functional groups of the base pair at the first position of the TATA box. In the case of pwTBP, proline is present at this position in both repeats, and both prolines contact DNA in a similar way. Thus the proline-clash model (29) for directionality is not applicable to pwTBP. As pointed out by DeDecker et al. (11), the striking asymmetry of electrostatic charge potential seen in eukaryotic TBP is not present in pwTBP (Fig. 5).
Under the solvent conditions and temperature of crystallization, pwTBP requires bound pwTFBc to adhere effectively to its promoter (3). If pwTFBc were to bind preferentially to the flanking DNA on one side of the boxA/TATA element it could influence the polarity of pwTFBc/pwTBP/promoter assembly. Due to the absence of pwTFBc–base interactions, however, a mechanism by which pwTFBc would bind preferentially to one side of the boxA/TATA element and, thereby, specify the orientation of the ternary complex is not obvious. Unfortunately, the DNA flanking the boxA/TATA element in the duplex used in these crystals is not long enough to provide all of the contacts with pwTFBc, rather pwTFBc makes contacts with neighboring DNA fragments in the lattice which are packed in near-continuous helical register. Thus any orienting effect that pwTFBc might have would not be accurately represented in this crystal.
In summary, there are four considerations consistent with TBP and TFB in an inverted orientation during the assembly of the archaeal preinitiation complex: (i) the inverted orientation seen in the crystal structure of this archaeal ternary complex, (ii) the need for a different linkage between the basal factors and polymerase implied by the absence of an archaeal TFIIF equivalent, (iii) the inverted sequence of many boxA/TATA sequences, and (iv) the distinctive pattern of protection in the DNase I footprint of the archaeal complex (9).
On the other hand, the stereochemical basis for an orienting preference is not clear, given the high degree of structural and electrostatic symmetry of the archaeal TBP/promoter interface, and the absence of base-specific contacts between the promoter and pwTFBc. In view of the fact that, in archaea, TBP, TFB, and polymerase are sufficient for supporting promoter-specific transcription, further experiments will be required to establish the orientation preference of preinitiation assembly on archaeal promoters, the degree to which transcription initiation is unidirectional and the molecular mechanism by which these polarities are achieved.
Acknowledgments
We thank Drs. Andrew Thompson of ESRF (Grenoble, France) and Steven Ealick (CHESS, Ithaca, NY) for access to their synchrotron sources, Drs. Yong Wang (Yale) and Serge Pares (Yale) for help with data collection, and Drs. Ron Albright (Yale), James Geiger (Yale), Sean Juo (Yale), Steven Hahn (Fred Hutchinson Cancer Research Center, Seattle), and Steve Jackson (Wellcome Cancer Research Campaign, Cambridge, U.K.), for helpful discussions. Work was supported in part by a grant from the National Institutes of Health (GM15225). P.F.K. and B.S.D. were National Institutes of Health Predoctoral Trainees, and G.G. was a Fellow of the Irvington Institute.
ABBREVIATIONS
- TBP
TATA-box-binding protein
- TFIIB
transcription factor IIB
- TFB
transcription factor B
- pwTFB and pwTBP
Pyrococcus woesei TFB and TBP
- pwTFBc
C-terminal 200 residues of pwTFB
- TFIIBc
C-terminal protease-resistant core of TFIIB
- CHESS
Cornell High Energy Synchrotron Source
- ESRF
European Synchrotron Radiation Facility
Footnotes
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, Brookhaven National Laboratory, Upton, NY 11973 (reference 1ais).
References
- 1.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, et al. Science. 1996;273:1017–1045. [Google Scholar]
- 2.Langer D, Hain J, Thuriaux P, Zillig W. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowlands T, Baumann P, Jackson S P. Science. 1994;264:1326–1328. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- 4.Qureshi S A, Baumann P, Rowlands T, Khoo B, Jackson S P. Nucleic Acids Res. 1995;23:1775–1781. doi: 10.1093/nar/23.10.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qureshi S A, Khoo B, Baumann P, Jackson S P. Proc Nat Acad Sci USA. 1995;92:6077–6081. doi: 10.1073/pnas.92.13.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creti R, Londei P, Cammarano P. Nucleic Acids Res. 1993;21:2942. doi: 10.1093/nar/21.12.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hain J, Reiter W-D, Hudepohl U, Zillig W. Nucleic Acids Res. 1992;20:5423–5428. doi: 10.1093/nar/20.20.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer J R, Daniels C J. J Bacteriol. 1995;177:1844–1849. doi: 10.1128/jb.177.7.1844-1849.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hausner W, Wettach J, Hethke C, Thomm M. J Biol Chem. 1996;271:30144–30148. doi: 10.1074/jbc.271.47.30144. [DOI] [PubMed] [Google Scholar]
- 10.Adams M W W. Annu Rev Microbiol. 1993;47:627–658. doi: 10.1146/annurev.mi.47.100193.003211. [DOI] [PubMed] [Google Scholar]
- 11.DeDecker B S, O’Brien R, Fleming P, Geiger J H, Jackson S P, Sigler P B. J Mol Biol. 1996;264:1072–1084. doi: 10.1006/jmbi.1996.0697. [DOI] [PubMed] [Google Scholar]
- 12.Bagby S, Kim S, Maldonado E, Tong K I, Reinberg D, Ikura M. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 13.Nikolov D B, Chen H, Halay E D, Usheva A A, Hisatake K, Lee D K, Roeder R G, Burley S K. Nature (London) 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 14.Marsh T L, Reich C I, Whitelock R B, Olsen G J. Proc Natl Acad Sci USA. 1994;91:4180–4184. doi: 10.1073/pnas.91.10.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim Y, Geiger J H, Hahn S, Sigler P B. Nature (London) 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 16.Kim J L, Nikolov D B, Burley S K. Nature (London) 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 17.Kim J L, Burley S K. Nat Struct Biol. 1994;1:638–653. doi: 10.1038/nsb0994-638. [DOI] [PubMed] [Google Scholar]
- 18.Nikolov D B, Chen H, Halay E D, Hoffmann A, Roeder R G, Burley S K. Proc Natl Acad Sci USA. 1996;93:4862–4867. doi: 10.1073/pnas.93.10.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geiger J H, Hahn S, Lee S, Sigler P B. Science. 1996;272:830–836. doi: 10.1126/science.272.5263.830. [DOI] [PubMed] [Google Scholar]
- 20.Tan S, Hunziker Y, Sargent D F, Richmond T J. Nature (London) 1996;381:127–134. doi: 10.1038/381127a0. [DOI] [PubMed] [Google Scholar]
- 21.Otwininski Z. In: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S W, editors. Warrington, U.K.: Science and Engineering Council/Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 22.Kabsh W. J Appl Crystallogr. 1988;21:916–924. [Google Scholar]
- 23.Brunger A T. x-plor Version 3.1, A System for X-ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1993. [Google Scholar]
- 24.Collaborative Computing Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 25.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Reed R. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 27.Jiang J-S, Brunger A T. J Mol Biol. 1994;243:100–115. doi: 10.1006/jmbi.1994.1633. [DOI] [PubMed] [Google Scholar]
- 28.Carson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 29.Juo Z S, Chiu K T, Leiberman P M, Baikalov I, Berk A J, Dickerson R E. J Mol Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 30.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 31.Ha I, Roberts S, Maldonado E, Sun X, Kim X-S, Green M, Reinberg D. Genes Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]