Abstract
The nicotinic acetylcholine receptor (AcChoR) is a ligand-gated ion channel that is activated upon binding of acetylcholine. α-Neurotoxins, in particular α-bungarotoxin (α-BTX), bind specifically and with high affinity to the AcChoR and compete with binding of the natural ligand. We employed a 15-mer phage-display peptide library to select epitopes reacting with α-BTX. Phages bearing the motif YYXSSL as a consensus sequence were found to bind with high affinity to α-BTX. The library-derived peptide (MRYYESSLKSYPD) bears amino acid sequence similarities to a region of the α-subunit of the Torpedo muscle AcChoR, as well as of other muscle and neuronal AcChoRs that bind α-BTX. The library-derived peptide and the corresponding peptides containing residues 187–199 of the Torpedo AcChoR α-subunit (WVYYTCCPDTPYL), as well as peptides analogous to the above region in the neuronal AcChoR (e.g., human α7; ERFYECCKEPYPD) that binds α-BTX, inhibit the binding of α-BTX to the intact Torpedo AcChoR with IC50 values of 10−6 M. A synthetic peptide from a neuronal AcChoR that does not bind α-BTX (e.g., human α2; ERKYECCKEPYPD) which differs by just one amino acid from the homologous peptide from the α-BTX-binding protein (α7)—i.e., Lys in α2 and Tyr in α7—does not inhibit the binding of α-BTX to Torpedo AcChoR. These results indicate the requirement for two adjacent aromatic amino acid residues for binding to α-BTX.
The nicotinic acetylcholine receptor (AcChoR) is a ligand-gated ion channel that is activated by binding of acetylcholine (AcCho). In muscle, the functional AcChoR molecule is a pentameric complex of α2βγδ subunits, or α2βɛδ subunits. The ligand-binding site of the AcChoR has been studied extensively and found to be located on the α-subunit, in the vicinity of cysteines 192 and 193. It was further shown that the ligand-binding domains are formed at the interfaces between the α-γ and α-δ subunits (for reviews, see 1–3). Affinity-labeling experiments indicated that within the α-subunit, cysteines 192 and 193, which form an intrachain disulfide in the intact receptor (4), and the conserved aromatic residues, Tyr-93, Trp-149, Tyr-190, and Tyr-198, are labeled by derivatives of agonists or antagonists (4–9) and are thus within the binding site, or very close to it.
The curarimetic snake α-neurotoxins, such as α-bungarotoxin (α-BTX), are potent competitive inhibitors of AcChoR function and are highly toxic due to functional blockade of AcChoRs at the neuromuscular junction. Because of the high affinity and specificity of α-BTX for the AcChoR, many studies of the AcChoR ligand-binding site have focused on the binding site for α-BTX. Though the binding sites for AcCho and α-BTX probably overlap, structural requirements for their binding may differ, and amino acid residues critical for binding of the toxin may not be the same as those critical for binding of AcCho (10, 11). Previous studies have demonstrated that short synthetic peptides containing cysteines 192 and 193 bind α-BTX specifically, though with moderate affinity (12, 13). Major components of this binding were shown to be contained within a synthetic dodecapeptide corresponding to residues 185–196 of the Torpedo AcChoR α-subunit (13, 14). NMR analysis indicated that five residues in this dodecapeptide (HWVYY, residues 186–190) are in contact with α-BTX (15).
Additional evidence for localization of the binding site for α-BTX within this particular region of the α-subunit came from recent analysis of the presumed ligand-binding site of AcChoRs from animals that are resistant to α-BTX (10, 16–18). This analysis indicated unique sequence differences between toxin-resistant and toxin-sensitive receptors, concentrated within a very small region of the α-subunit, close to cysteines 192 and 193.
Another approach in searching for a peptide that interacts specifically with α-BTX is to employ random peptide libraries (19, 20). One advantage of such an approach is that the selector molecule is not “committed” to any target molecule known to interact with it and is allowed to select peptides from a completely random collection of peptide-presenting phages. We have previously employed phage–epitope libraries to study the AcChoR and have identified peptide epitopes for two anti-AcChoR monoclonal antibodies (21, 22).
In the present study we utilized a 15-mer phage–epitope library to select a peptide that interacts specifically with α-BTX and blocks its interaction with the AcChoR. The library-derived peptide bears a similarity to the ligand-binding region in the α-subunit of both muscle and neuronal α-BTX-binding AcChoRs. Identification of this peptide confirms the localization of the binding site within the receptor molecule and allows a more precise determination of the boundaries of the binding site. This peptide could also be employed as a lead compound in the search for effective antidotes against toxin intoxication. In this connection, the constrained conformation attained by the peptide in a complex with α-BTX as revealed by NMR studies (see the companion article, ref. 23), should obviously be taken into consideration.
MATERIALS AND METHODS
Epitope Library.
The 15-mer phage–peptide library employed in this study was provided by James J. Devlin (Cetus) and was constructed by use of phage M13 as described (20). The library consists of about 2 × 107 different 15-residue peptide-presenting phages.
Biotinylation of α-Bungarotoxin.
HPLC-purified α-BTX from the snake venom of Bungarus multicinctus was purchased from Sigma. For biotinylation, 100 μg of α-BTX in 100 μl of 0.1 M NaHCO3 (pH 8.6) were incubated for 2 h at room temperature with 5 μg of biotin amidocaproate N-hydroxysuccinimide ester (Sigma) from a stock solution of 1 mg/ml in dimethylformamide, and then dialyzed at 4°C against PBS (0.14 M NaCl/0.01 M phosphate buffer, pH 7.4).
Isolation of Peptide-Presenting Phages from the Phage–Peptide Library.
A sample of the amplified library, containing 1011 infectious phage particles, was subjected to three rounds of selection (panning) and amplification. In the first round, 1011 phage particles were incubated overnight at 4°C with 20 ng/ml biotinylated-α-BTX (bio-α-BTX) in 40 μl of 0.5% BSA in PBS. The phages were diluted with 1 ml 0.5% Tween 20 in PBS (T/P) and allowed to react for 30 min at room temperature with streptavidin (10 μg/ml)-coated Petri dishes (60 mm) with gentle shaking. Unbound phages were removed by extensive washing (10 times, 10 min each) in T/P and the remaining phages were eluted with 0.8 ml 0.1 M HCl, titrated to pH 2.2 with glycine. The eluate was neutralized with 48 μl of 2 M Tris base and used to infect Escherichia coli K91 cells. The phage eluted in the first round were amplified and subjected to a second round of affinity purification with the bio-α-BTX at 2 ng/ml. Phage from the second round were amplified and subjected to a third round with 0.2 ng/ml bio-α-BTX. The progressively decreasing α-BTX concentrations used in the second and third rounds, and the use of excess phages, were intended to facilitate selection for high-affinity epitopes. After three pannings, individual bacterial colonies containing amplified phage clones were grown in a microtiter plate and screened by ELISA for their ability to specifically bind bio-α-BTX, essentially as described (21). Phage DNAs were sequenced to determine the amino acid sequences of their inserted peptides.
α-BTX Binding to Isolated Phages.
Binding of α-BTX to phages was analyzed by ELISA. Wells of microtiter plates (Maxisorb; Nunc) were coated with 100 μl of rabbit anti-phage M13 serum at a dilution of 1:1000 (in 0.1 M NaHCO3, pH 8.6) and incubated overnight at 4°C. Coated plates were washed three times with T/P and 100 μl of enriched phage clones, each containing 109 phage particles, were then added to the wells and incubated for 1 h at 37°C. Wells were blocked with 1.5% BSA and 1.5% hemoglobin (in PBS) for 1–2 h at room temperature, washed, and incubated with bio-α-BTX (1 μg/ml, or as otherwise specified) overnight at 4°C. For inhibition experiments, peptides were preincubated with bio-α-BTX for 30 min at 37°C prior to their addition to phage-coated wells. Following washings, bound bio-α-BTX was detected after incubation for 1 h at room temperature with alkaline phosphatase-conjugated extravidin (BioMakor, Rehovot, Israel), diluted 1:2,000. α-BTX was estimated by the enzymatic activity of alkaline phosphatase, with N-p-nitrophenyl-phosphate as a substrate, and the color developed after 50 min was determined using a microtiter plate reader at 405 nm. Positive phage clones were propagated, and the DNA in the inserted epitope region was sequenced at the Biological Services Unit of the Weizmann Institute using an automated 373A DNA sequencer and the Taq DyeDeoxy Terminator Cycle Sequencing Kit (Applied Biosystems).
Inhibition of α-BTX Binding to AcChoR by Synthetic Peptides.
AcChoR was extracted from the electric organ of Torpedo californica by Triton X-100 and purified as described (24). Purified Torpedo AcChoR (5 μg/ml) was used to coat ELISA plates as described (21). Inhibition of the binding of α-BTX to AcChoR by synthetic peptides was carried out as described above for the inhibition of α-BTX binding to phages.
Synthetic Peptides.
Synthetic peptides were prepared in the Chemical Services Unit at the Weizmann Institute by the solid-phage automated method of Merrifield (25). The synthetic peptides employed in this study are listed in Table 1.
Table 1.
Origin and sequence of synthetic peptides studied
| Peptide | Sequence |
|---|---|
| Library-derived peptides | |
| 15-mer | YMRYYESSLKSYPDW |
| 17-mer | KKYMRYYESSLKSYPDW |
| 14-mer | MRYYESSKLSYPDW |
| 13-mer* | MRYYESSLKSYPD |
| 12-mer | MRYYESSLKSYP |
| 11-mer | MRYYESSLKSY |
| Peptides derived from sequences of Torpedo AcChoR | |
| α1 187-199 | WVYYTCCPDTPYL |
| α1 185-196 | KHWVYYTCCPDT |
| Peptides derived from sequences of neuronal α-BTX-binding AcChoRs | |
| Rat α7 | EKFYECCKEPYPD |
| Chick α8 | ELYYECCKEPYPD |
| Human α7 | ERFYECCKEPYPD |
| Peptide derived from the sequence of a neuronal AcChoR that does not bind α-BTX | |
| Human α2 | ERKYECCKEPYPD |
Library-derived peptide.
RESULTS
Selection of Peptide-Presenting Phages by Bio-α-BTX.
To isolate peptides representing the α-BTX binding site within AcChoRs or other proteins that may interact specifically with α-BTX, we screened a 15-mer phage–peptide library with bio-α-BTX. After three cycles of panning and phage amplification, 90 individually isolated bacterial colonies were grown in microtiter plates and their phages were assayed for toxin binding. ELISA analysis revealed that 60 of these clones exhibited specific binding to α-BTX. DNA from 23 phage clones, selected arbitrarily from the positive clones, was sequenced; the deduced peptide sequences are shown in Table 2. The phage expressing the sequence YMRYYESSLKSYPDW appeared to be dominant among the 23 analyzed phages and was represented in 16 phage clones that displayed high affinity for α-BTX. Half maximal binding (HMB) of α-BTX to the phage was obtained at 5 × 10−8 M. Three phage clones expressed the sequence FTYYQSSLEPLSPFY and exhibited similar affinity (HMB of 5 × 10−8 M) to α-BTX. These two sequences contain the consensus motif YYXSSL. In the additional phage clones sequenced, the inserted peptide sequences were TMTFPENYYSERPYH (in two clones), PPPIFRYYEYWPTSY and HDKLFTFYQNSXSSY. The three latter clones had a lower binding affinity for α-BTX (HMB of 5 × 10−7 M) and contained a consensus motif of two adjacent aromatic residues (YY or FY). Most of the following analysis was performed with the phage expressing YMRYYESSLKSYPDW—i.e., the one represented with the highest frequency (16 of 23).
Table 2.
Peptides selected from a 15-mer phage-peptide library by α-BTX
| Peptide sequence* | Frequency | Binding,† M |
|---|---|---|
| YMRYYESSLKSYPDW | 16 | 5 × 10−8 |
| FTYYQSSLEPLSPFY | 3 | 5 × 10−8 |
| TMTFPENYYSERPYH | 2 | 5 × 10−7 |
| PPPIFRYYEYWPTSY | 1 | 5 × 10−7 |
| HDKLFTFYQNSXSSY | 1 | 5 × 10−7 |
Amino acid sequences (derived from DNA) of the inserted 15-mer peptide within the minor coat protein pIII.
Binding represents the concentration resulting in half-maximal binding to the isolated phage. Binding data were obtained by ELISA.
Specificity of the Interaction of α-BTX with the Library-Selected Peptide.
The specificity of the interaction of α-BTX with the YMRYYESSLKSYPDW phage was further assessed by inhibition experiments with the corresponding synthetic peptide. Because the synthetic peptide YMRYYESSLKSYPDW was rather insoluble, two basic amino acid residues (lysines) were added to its N-terminal end to increase its solubility. As shown in Fig. 1, the synthetic peptide KKYMRYYESSLKSYPDW, but not the nonrelevant control peptide (PMTLPENYFSERPYHK), inhibited the binding of α-BTX to the phage with an IC50 of 1 × 10−8 M.
Figure 1.
Inhibition of α-BTX binding to YMRYYESSLKSYPDW phage by its corresponding synthetic peptide.
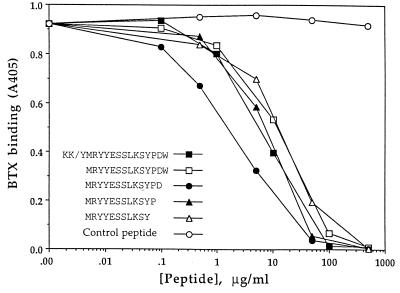
To determine whether the library-selected synthetic peptide bears relevance to α-BTX binding sites on the nicotinic AcChoR, and as such may be an AcChoR-specific mimotope, we tested the ability of the synthetic peptide to inhibit the binding of α-BTX to Torpedo AcChoR. As shown in Fig. 2, the peptide KKYMRYYESSLKSYPDW inhibits the binding of α-BTX to the AcChoR in a concentration-dependent manner with an IC50 of 1.7 × 10−5 M, which is about two orders of magnitude lower than the IC50 obtained upon inhibition of the binding of α-BTX to the peptide-presenting phage (see Fig. 1).
Figure 2.
Inhibition of α-BTX binding to Torpedo AcChoR by the library-selected synthetic peptide and its derivatives.
The consensus motif (YYXSSL) present in the high-affinity phages encompasses only part of the peptide selected from the library. We therefore shortened this peptide at both ends to find an optimal peptide in terms of its affinity for the α-BTX. As depicted in Fig. 2, the peptides MRYYESSLKSYPDW, MRYYESSLKSYPD, MRYYESSLKSYP, and MRYYESSLKSY inhibited interaction of α-BTX with AcChoR with IC50 values of 1.7 × 10−5 M, 2.8 × 10−6 M, 1.1 × 10−5 M, and 1.3 × 10−5 M, respectively. The best inhibition was obtained with the 13-mer MRYYESSLKSYPD peptide, which was designated the library-derived peptide and employed for further analysis.
Relevance of the Library-Derived Peptide to Sequences in Muscle and Neuronal AcChoR.
As the library-derived peptide MTYYESSLKYPD inhibited strongly the interaction between α-BTX and AcChoR (IC50 of 2.8 × 10−6 M), we searched for structural homologies between this peptide and sequences on the α-subunit of AcChoR, and in particular in the vicinity of the tandem cysteines (192 and 193) present within the AcChoR-binding site. Since all the library-selected peptides had two adjacent aromatic residues, we searched for similar residues in Torpedo AcChoR. Tyrosines 189 and 190 are the only two adjacent aromatic amino acid residues present in the extracellular domain of the α-subunit. These are separated by one residue from the tandem cysteines (192 and 193), which may bear structural similarity to the two serines present at this position in the library-consensus motif (YYXSSL). We therefore examined the reactivity of the synthetic peptide WVYYTCCPDTPYL, which corresponds to residues 187–199 of the Torpedo AcChoR α-subunit (Torpedo 187–199) and has the same number of residues on both ends of the consensus motif as the library-derived peptide. As seen in Fig. 3, the capacity of this synthetic peptide to inhibit the binding of α-BTX to Torpedo AcChoR is similar to that of the library-selected peptide. The Torpedo 187–199 peptide is a better inhibitor, by almost two orders of magnitude, than the synthetic Torpedo 185–196 peptide, which we previously found to contain the major determinants of the α-BTX binding site (13).
Figure 3.
Inhibition of α-BTX binding to AcChoR by the library-derived peptide and the corresponding peptides derived from the α-subunit of Torpedo. AcChoR. The peptide ERKYECCKEPYPD was used as a control.
Interestingly, we noticed that sequences of the α-subunits of the neuronal α-BTX-binding AcChoRs (α7, α8, and α9), at the region corresponding in position to the Torpedo-derived peptide 187–199, bear an even more striking similarity to the library-derived peptide than the Torpedo α-subunit sequences (see Table 1). As demonstrated in Fig. 4, inhibition of the binding of α-BTX to Torpedo AcChoR by synthetic peptides containing the amino acid sequence of the neuronal (α7 and α8) AcChoR is similar to its inhibition by the library-derived peptide.
Figure 4.
Inhibition of α-BTX binding to Torpedo AcChoR by peptides derived from neuronal α-BTX-binding AcChoR at positions corresponding to residues 187–199 of the Torpedo α-subunit.
An aromatic residue is present at position 189 in all AcChoRs that bind α-BTX (muscle and electric organ α1 or neuronal α7, α8, and α9), and is absent in muscle AcChoRs that are resistant to α-BTX (e.g., from snake, mongoose, or hedgehog) and in neuronal nicotinic AcChoRs which do not bind α-BTX (α2 to α6). It was therefore of interest to find out whether the binding properties of these peptides are affected by replacing the aromatic residue at position 189 in the synthetic peptides by a nonaromatic amino acid. Indeed, as shown in Fig. 5A, substitution of Tyr-189 in the Torpedo receptor peptide 187–199, or phenylalanine in the corresponding human α7 receptor peptide, by a lysine, abolished binding to α-BTX. Replacing this tyrosine in the library peptide by either lysine or serine also resulted in a marked decrease in the inhibition of α-BTX binding to AcChoR (Fig. 5B). In the library peptide, replacing the arginine present one amino acid upstream of that tyrosine by serine, resulted in only a small decrease in the inhibition of α-BTX binding. These experiments indicate that an aromatic residue at position 189 of the AcChoR α-subunit is necessary for α-BTX binding.
Figure 5.
Replacement of the aromatic residue at position 189 in the AcChoR-derived peptides (A) and in the library-derived peptide (B) by lysine abolishes the inhibition of α-BTX binding to Torpedo AcChoR.
DISCUSSION
The availability of short synthetic peptides that interact with biologically active molecules forms the basis for the development of specific reagents (drugs) that can interfere with the function of such molecules. In the present study, we employed a phage–epitope library to identify peptides that interact specifically with α-BTX and interfere with its binding to AcChoR. This is a random strategy for which the selector molecule (i.e., α-BTX) does not require any information concerning the structure of the target molecule AcChoR. Theoretically, by employing this random selection, α-BTX could have selected peptides reacting with any site on the toxin and not necessarily with the site that interacts with AcChoR. Interestingly, we found that the library-selected peptides bear remarkable similarities to a region in the AcChoR α-subunit, which is known to be within the AcChoR binding site. This may not be surprising, as the association of α-BTX with the AcChoR (IC50 = 10−11 M) is stronger than the association of α-BTX with other putative molecules (e.g., antibodies).
The most striking similarity between the library-selected peptides and the binding site of the AcChoR is the presence of two adjacent tyrosines (YY or in some cases FY) in all the library-selected peptides (see Table 2) as well as in the binding sites of all the α-BTX-binding AcChoRs (muscle and neuronal) at positions 189 and 190 in the α-subunit. Tyr-190 is highly conserved in all AcChoRs, whether they bind α-BTX or not. Affinity-labeling experiments have localized this tyrosine at the agonist binding site (2, 3, 5). Replacement of this tyrosine by phenylalanine had a dramatic effect on ligand binding and channel activation, and increased the IC50 for α-BTX by two orders of magnitude (26). It has been suggested that a phenolate ion, and not merely an aromatic residue, is required at this position (Tyr-190) for interaction with the AcChoR (26). When Tyr-190 was replaced by serine in the Torpedo receptor peptide (residues 187–199) or in the library peptide, interaction with α-BTX was abolished (data not shown). It thus appears that Tyr-190 is required for both agonist and α-BTX binding.
Tyr-189 (or Phe-189) in the AcChoR α-subunit is required for α-BTX binding, but not necessarily for agonist binding. Neuronal AcChoRs that do not bind α-BTX (e.g., α2, α3, α4), as well as muscle AcChoRs from animals that are resistant to α-BTX, do not have an aromatic amino acid at this position. Earlier studies with synthetic peptides from the Torpedo AcChoR binding-site domain also demonstrated that Tyr-189 is essential for α-BTX binding (27, 28). When this tyrosine was replaced by lysine in the muscle AcChoR peptide, or in the corresponding α7 peptide (Fig. 5A), or in the library-derived peptide (Fig. 5B), the binding capacity for α-BTX was abolished. Notably, the only difference between the neuronal α7 peptide, which binds α-BTX, and the neuronal α2 peptide, which does not bind α-BTX, is at position 189. At this position there is a tyrosine in the α7 and a lysine in the α2-derived peptide. This confirms the requirement of an aromatic residue at position 189 for α-BTX binding.
The selected peptide-presenting phages that bound α-BTX with high affinity (HMB 5 × 10−8 M; Table 2) had a consensus motif of YYXSSL. Alignment of the library-derived peptide (MRYYESSLKSYPD) with a sequence in the AcChoR α-subunit (residues 187–199), so that the consensus motif matches residues 189–194, revealed additional similarities to both muscle (α1) and neuronal (α7, α8) α-BTX-binding AcChoRs (see Table 1). The two adjacent serines in the library-derived peptide are at the same relative positions as the two cysteines (192 and 193) in the AcChoR. It has been reported (29) that replacing one of these cysteines with a serine does not affect binding. We demonstrate here that α-BTX binding of the library-derived peptide, in which two serines replace the two cysteines, is similar in extent to that of the corresponding AcChoR-derived peptides (Figs. 3 and 4). Thus, it is likely that the two cysteines are not essential for binding of the toxin, provided that other, sufficiently similar residues (i.e., serines) are present.
We have previously suggested, mainly on the basis of data from studies on the snake and mongoose AcChoR, that two close prolines downstream of the two cysteines are necessary for toxin binding (10, 16, 18). The library-derived peptide, however, has only one proline, located at the same position as the second proline in neuronal AcChoRs (see Table 1). Moreover, the position of the first conserved proline in the toxin-binding muscle AcChoRs (Pro-194) is occupied by a leucine in the library-derived peptide, as is also the case in the α-BTX-resistant snake and mongoose AcChoRs. Studies by Taylor and collaborators (30) have shown that replacement of this proline for leucine in the mouse AcChoR α-subunit did not result in a substantial change in toxin binding.
It is interesting to note that the library-derived peptide seems to share greater homology with the neuronal α7 and α8 subunits than with the muscle α1 subunit of the AcChoR (Table 1). Nevertheless, peptides derived from either the muscle or the neuronal (α7 and α8) receptor were as active as the library-derived peptide in their ability to inhibit the interaction of α-BTX with the AcChoR. It is not clear whether residues in the library-derived peptide correspond linearly (sequentially) to residues in the AcChoR-binding site, or whether the peptide mimics a spatial conformation allowing an appropriate fit to α-BTX, or a combination of both. The conformation in solution of the library-derived peptide in its complex form with α-BTX as revealed by NMR analysis is described in the following paper (23).
Finally, it should be pointed out that the phage library employed is by no means complete, and that not all possible 15-mer peptides are represented in it. Even with this limitation, the library-selected peptides are as potent as the peptides derived from the receptor. It is possible, however, that other peptides, not represented in the library, might have a higher affinity for α-BTX than the receptor-derived peptides.
Acknowledgments
We thank Prof. David Givol for advice and valuable discussions, and Cetus/Chiron Corp. (Emeryville, CA) for the phage peptide library used in this study. This research was supported by grants from the Rashi Foundation, the Ministry of Science and Arts, the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities, the Muscular Dystrophy Association of America, the Association Francaise Contre les Myopathies, and the Leo and Julia Forchheimer Center for Molecular Genetics at the Weizmann Institute of Science.
ABBREVIATIONS
- AcCho
acetylcholine
- AcChoR
AcCho receptor
- α-BTX
α-bungarotoxin
- bio-α-BTX
biotinylated α-BTX
- HMB
half maximal binding
- T/P
Tween 20 in PBS
References
- 1.Karlin A. Curr Opin Neurobiol. 1993;3:299–309. doi: 10.1016/0959-4388(93)90121-e. [DOI] [PubMed] [Google Scholar]
- 2.Changeux J-P, Galzi J-L, Devillers-Thiery A, Bertrand D. Q Rev Biophys. 1992;25:395–431. doi: 10.1017/s0033583500004352. [DOI] [PubMed] [Google Scholar]
- 3.Karlin A, Akabas M H. Neuron. 1995;15:1231–1244. doi: 10.1016/0896-6273(95)90004-7. [DOI] [PubMed] [Google Scholar]
- 4.Kao P N, Dwork A J, Kaldany R R, Silver M L, Wideman J, Stein S, Karlin A. J Biol Chem. 1984;259:11662–11665. [PubMed] [Google Scholar]
- 5.Dennis M, Giraudat J, Kotzyba-Hibert F, Goeldner M, Hirth C, Chang J Y, Lazure C, Chretien M, Changeux J-P. Biochemistry. 1988;27:2346–2357. doi: 10.1021/bi00407a016. [DOI] [PubMed] [Google Scholar]
- 6.Abramson S M, Li Y, Culver P. J Biol Chem. 1989;264:12666–12672. [PubMed] [Google Scholar]
- 7.Galzi J-L, Revah F, Black D, Goeldner M, Hirth C, Changeux J-P. J Biol Chem. 1990;265:10430–10437. [PubMed] [Google Scholar]
- 8.Cohen J B, Sharp S D, Liu W S. J Biol Chem. 1991;266:23354–23364. [PubMed] [Google Scholar]
- 9.Middleton R E, Cohen J B. Biochemistry. 1991;30:6987–6997. doi: 10.1021/bi00242a026. [DOI] [PubMed] [Google Scholar]
- 10.Barchan D, Kachalsky S, Neumann D, Vogel Z, Ovadia M, Kochva E, Fuchs S. Proc Natl Acad Sci USA. 1992;89:7717–7721. doi: 10.1073/pnas.89.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatuverdi V, Donnelly-Roberts D L, Lentz T. Biochemistry. 1992;31:1371–1375. doi: 10.1021/bi00120a012. [DOI] [PubMed] [Google Scholar]
- 12.Wilson P T, Lentz T L, Hawrot E. Proc Natl Acad Sci USA. 1985;82:8790–8794. doi: 10.1073/pnas.82.24.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neumann D, Barchan D, Safran A, Gershoni J M, Fuchs S. Proc Natl Acad Sci USA. 1986;83:3008–3011. doi: 10.1073/pnas.83.9.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann D, Barchan D, Fridkin M, Fuchs S. Proc Natl Acad Sci USA. 1986;83:9250–9253. doi: 10.1073/pnas.83.23.9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basus V J, Song G, Hawrot E. Biochemistry. 1993;32:12290–12298. doi: 10.1021/bi00097a004. [DOI] [PubMed] [Google Scholar]
- 16.Neumann D, Barchan D, Horowitz M, Kochva E, Fuchs S. Proc Natl Acad Sci USA. 1989;86:7255–7259. doi: 10.1073/pnas.86.18.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barchan D, Ovadia M, Kochva E, Fuchs S. Biochemistry. 1995;34:9172–9176. doi: 10.1021/bi00028a029. [DOI] [PubMed] [Google Scholar]
- 18.Kachalsky S G, Jensen B S, Barchan D, Fuchs S. Proc Natl Acad Sci USA. 1995;92:10801–10805. doi: 10.1073/pnas.92.23.10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott J K, Smith G P. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 20.Devlin J J, Panganiban L C, Devlin P E. Science. 1990;249:404–406. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- 21.Balass M, Heldman Y, Cabilly S, Givol D, Katchalski-Katzir E, Fuchs S. Proc Natl Acad SciUSA. 1993;90:10638–10642. doi: 10.1073/pnas.90.22.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barchan D, Balass M, Souroujon M, Katchalski-Katzir E, Fuchs S. J Immunol. 1995;155:4264–4269. [PubMed] [Google Scholar]
- 23.Scherf T, Balass M, Fuchs S, Katchalski-Katzir E, Anglister J. Proc Natl Acad Sci USA. 1997;94:6059–6064. doi: 10.1073/pnas.94.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aharonov R, Tarrab-Hazdai R, Silman I, Fuchs S. Immunochemistry. 1977;14:129–137. doi: 10.1016/0019-2791(77)90291-9. [DOI] [PubMed] [Google Scholar]
- 25.Merrifield R B. Science. 1965;150:178–185. doi: 10.1126/science.150.3693.178. [DOI] [PubMed] [Google Scholar]
- 26.Tomaselli G F, McLaughlin J T, Jurman M E, Hawrot E, Yellen G. Biophys J. 1991;60:721–727. doi: 10.1016/S0006-3495(91)82102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzartos S J, Remoundos M S. J Biol Chem. 1990;265:21462–21467. [PubMed] [Google Scholar]
- 28.Ohana B, Fraenkel Y, Navon G, Gershoni J M. Biochem Biophys Res Commun. 1991;179:648–654. doi: 10.1016/0006-291x(91)91421-8. [DOI] [PubMed] [Google Scholar]
- 29.Mishina M, Tobimatsu T, Imoto K, Tona-Ka K, Fujita Y, Fukuda K, Kurasaki M, Takahashi H, Morimoto Y, Hirose T, Inayama S, Takahashi T, Kuno M, Numa S. Nature (London) 1985;313:364–369. doi: 10.1038/313364a0. [DOI] [PubMed] [Google Scholar]
- 30.Kreinkamp H J, Sine S M, Maeda R K, Taylor P. J Biol Chem. 1994;269:1–7. [PubMed] [Google Scholar]