Abstract
The solution structure of the complex between α-bungarotoxin (α-BTX) and a 13-residue library-derived peptide (MRYYESSLKSYPD) has been solved using two-dimensional proton–NMR spectroscopy. The bound peptide adopts an almost-globular conformation resulting from three turns that surround a hydrophobic core formed by Tyr-11 of the peptide. The peptide fills an α-BTX pocket made of residues located at fingers I and II, as well as at the C-terminal region. Of the peptide residues, the largest contact area is formed by Tyr-3 and Tyr-4. These findings are in accord with the previous data in which it had been shown that substitution of these aromatic residues by aliphatic amino acids leads to loss of binding of the modified peptide with α-BTX. Glu-5 and Leu-8, which also remarkably contribute to the contact area with the toxin, are present in all the library-derived peptides that bind strongly to α-BTX. The structure of the complex may explain the fact that the library-derived peptide binds α-BTX with a 15-fold higher affinity than that shown by the acetylcholine receptor peptide (α185–196). Although both peptides bind to similar sites on α-BTX, the latter adopts an extended conformation when bound to the toxin [Basus, V., Song, G. & Hawrot, E. (1993) Biochemistry 32, 12290–12298], whereas the library peptide is nearly globular and occupies a larger surface area of α-BTX binding site.
Keywords: protein–peptide complex, library peptide, NMR
The curaremimetic α-neurotoxins, such as α-bungarotoxin (α-BTX; 74 residues), are among the most lethal components found in snake venom. These toxins bind with very high affinities and selectivities to the nicotinic acetylcholine receptor (AcChoR). Several residues that are evolutionary conserved within the neurotoxins, K26, W28, D30, F/H/W32, R36, G37, V/I39, E41, K52, and V/L/I57 (numbering is according to α-BTX), were postulated to be involved either directly or indirectly, in binding to AcChoR (for a review, see ref. 1). The postsynaptic neurotoxins can be classified into two families: short neurotoxins (60–62 residues) and long neurotoxins (66–74 residues). The toxins from both families have been studied by x-ray crystallography (refs. 2–4 and references therein) and NMR spectroscopy (refs. 5–9 and references therein). Both classes exhibit a significant sequence homology and similar overall topologies characterized by a globular head, which is composed of a core of triple-stranded antiparallel β-sheet with three finger-like protruding loops. Differences in the structure are encountered at the end of the middle loop and at the C terminus. The intensive studies of neurotoxins are not only due to the interest in improving existing therapy of snake bite, but also because their affinity and specificity has proven invaluable for the studies of AcChoR.
The AcChoR is a ligand-gated ion channel. It is a pentameric complex composed of four types of subunits. The sensitivity of AcChoR to cholinergic neurotoxins is largely, although not entirely, dependent on its α-subunit (10–13). Though the sequence α181–200 of the AcChoR does not comprise the complete binding site, it specifically binds agonists (for a review, see refs. 14 and 15). This segment contains highly conserved residues: K185, W187, Y189, Y190, C192, C193, P194, D195, P197, and Y198 (for a review, see refs. 14 and 16). A shorter segment that binds α-BTX and which overlaps the acetylcholine-binding site (15) was mapped to residues α185–196 of the receptor (KHWVYYTCCPDT) (17). It was found that a peptide corresponding to this sequence binds α-BTX with an apparent Kd of 35 μM (17), in comparison to 0.4 nM for the AcChoR/α-BTX complex (18). NMR studies of α-BTX complexed with the α185–196 peptide (19) have revealed that the bound peptide adopts an extended conformation. In that complex, 25 intermolecular nuclear Overhauser effect (NOE) cross-peaks were observed.
A phage-displayed epitope library that consists of tens of millions of short peptides can be used to probe the specificity of antibodies, receptors, and enzymes (20, 21). Such selected peptides can in principle mimic not only a sequential determinant (epitope) but also discontinuous or conformational determinants (mimotopes). Despite the widespread use of peptide libraries to identify both sequential as well as discontinuous epitopes, no structure of a peptide mimicing a protein surface recognized by another protein has been reported to date. The only structures of complexes involving library-derived peptides were solved by Schreiber and coworkers (22, 23) and are of Src homology 3 (SH3) domains complexed with proline-rich peptides selected from biased phage-display libraries. As the α-BTX binding seems to involve AcChoR residues distant in sequence, a sequential epitope of the receptor may not fill the entire binding site. Using a 15-residue phage-displayed peptide library, we identified a 13-residue peptide (MRYYESSLKSYPD) that contains the consensus motif YYxSSL, and that binds α-BTX with a 15-fold higher affinity compared with the affinity of the AcChoR peptide (α185–196) (24). In this paper, we report the solution structure of α-BTX complexed with the 13-residue library-derived peptide as studied by two-dimensional 1H-NMR spectroscopy.
MATERIALS AND METHODS
Peptide Synthesis.
The selection and the synthesis of the 13-residue library peptide was described in the previous paper of this issue of the Proceedings (24). Crude peptide was purified in two steps: (i) gel filtration chromatography on a Sephadex G-25 column equilibrated with 30% acetic acid in water, and (ii) reverse-phase HPLC with a gradient of increasing concentration of acetonitrile in water containing 0.12% trifluoroacetic acid. Better than 98% purity was obtained. The amino acid composition of the purified peptide was verified by NMR sequential assignment.
NMR Sample Preparation.
α-BTX was purchased from Sigma. A reducing SDS/PAGE revealed a single monomeric protein, and the toxin was used without further purification. To form a 1:1 complex, aliquots of the lyophilized peptide were added to the toxin. The stoichiometry of binding was monitored by following the disappearance of a well-resolved resonance of the free toxin molecule at 6.39 ppm. Samples of the complex were prepared in either 90% H2O/10% 2H2O or 99.9% 2H2O, at a concentration of 5 mM, with 10 mM sodium phosphate buffer and at pH 5.8 or 4.8 (uncorrected for isotope effects) containing 0.05% azide.
NMR Spectroscopy.
1H-NMR spectra were recorded on either a Bruker (Karlsruhe, Germany) AM500 or DMX600 spectrometers. NMR spectra of the complex in 2H2O were recorded at 30 and 37°C, and included double-quantum filtered correlation spectroscopy (DQF-COSY; ref. 25), two-dimensional NOE spectroscopy (NOESY; ref. 26) with a 150-ms mixing period and homonuclear Hartmann–Hahn spectroscopy (HOHAHA; ref. 27) with a 70-ms spin-lock time. Spectra recorded in 90% H2O/10% 2H2O included DQF-COSY and rotating-frame Overhauser effect spectroscopy (ROESY) (45-ms mixing time) at 30°C, NOESY (45-, 60-, 120-, and 150-ms mixing times) and HOHAHA (35-, 70-, and 140-ms spin-lock times) which were measured at 22, 30, 37, and 42°C. Water suppression was achieved either by presaturation during the relaxation delay on the Bruker AM500, or using WATERGATE (WATER suppression by GrAdient-Tailored Excitation) suppression scheme on the DMX600 (28). Spectra were acquired with 2K or 4K complex data points in t2 and 512 points in t1, with a spectral width of 14 ppm. To identify slowly exchanging amide protons, the complex was lyophilized from H2O and redissolved in 99.8% 2H2O. Amide protons that were still observed after 24 h in 2H2O at room temperature, were considered to be in slow exchange with the solvent.
Experimental Restraints.
The cross-peak intensities in NOESY spectra of the complex recorded with several mixing times were compared to eliminate the possibility of spin-diffusion effects. The NOE intensities obtained from the spectrum recorded with a mixing time of 150 ms were used for structure calculation. To relate these NOE data with interproton distances, the standard distance of 2.3 Å for the interstrand Hα-Hα distances in a β-sheet was used. NOEs were divided into three groups, corresponding to upper-bound distances of 2.7, 3.5 and 5.0 Å. The lower-bound distance was 1.8 Å in all cases. A correction factor of 0.5 Å was added to the upper bound of the constraints involving methyl groups. Backbone hydrogen bonds within the antiparallel β-sheet were identified from amide proton exchange data and interstrand NOEs. For each hydrogen bond, two constraints were used: 1.7–2.3 Å for rNH-O and 2.4–3.3 Å for rN-O. Additional restraints were included to define the five known disulfide bridges of the toxin (3/23, 16/44, 29/33, 48/59, and 60/65) with bounds 1.9–2.1 Å (Sγ-Sγ) and 2.9–3.1 Å (Cβ-Sγ cross-bridge). The φ angle restraints were derived from 3JHN-Hα coupling constants measured from DQF-COSY spectra. For coupling constants between 8 and 10 Hz, the φ angle was restricted to −120 ± 50°, for couplings >10 Hz the angle was restricted to −120 ± 20° and for couplings <5 Hz the φ angle was restricted to the range between −90° and −40°. Stereospecific assignments were based on the analysis of both 3JHα-Hβ (determined from DQF-COSY measured in 2H2O), and HN-Hβ and Hα-Hβ NOEs (measured from NOESY spectra recorded with a 45-ms mixing time) (29). For stereospecific assignments of valine methyl groups as well as for determination of χ1 angles for isoleucine and threonine, the intensities of Hα-CγH3 and HN-CγH3 were used. χ1 angles were restrained to 60 ± 60°, 180 ± 60°, or −60 ± 60°.
Structure Calculations.
Three-dimensional structures of the complex were calculated on the basis of the NMR data with a distance geometry/dynamical simulated annealing protocol (30) using the xplor version 3.1 program (31). Distance restraints were used with a square-well potential and NOE force constant of 50 kcal mol−1⋅Å−2. The inverse sixth-power distance averaging (〈r−6〉) (32) was used for stereospecifically unresolved resonances.
RESULTS
Sequential Assignment.
The sequence-specific resonance assignment of the α-BTX/peptide complex was done according to the well-established procedure developed by Wüthrich (33). The rather strong Hαi-Hδi+1 NOE cross-peaks of all prolines observed in the 2H2O spectra indicate that all proline residues are in the trans configuration. The appearance of Oγ1H resonances of both T5 and T6 of the toxin in the spectra of the complex at 6.32 and 6.71 ppm, respectively, indicates that these protons are buried at the interior of the protein. Indeed, in the subsequent structure calculation it was found that T5 and T6 are 90% and 98% buried in the interior of the complex, respectively. Due to the very weak and relatively broad resonances of toxin residues S34, S35, and R36, their assignment was completed only toward the end, and was based on the very weak Hαi-HNi+1 connectivities. With the exception of the amide proton resonance of C33, all the resonances of both the bound toxin and the bound peptide were identified.
The resonances of residues in all the three loops (H4, T6, A7, S9, I11, S12, M27, W28, C29, D30, A31, F32, R36, G37, K38, V39, V40, E41, K52, Y54, and E56), as well as at the C-terminal region of the toxin (H68, P69, and K70) experience considerable changes in chemical shift upon peptide binding. The largest perturbations (>0.4 ppm) are found for the amide protons of W28, D30, F32, K38, V40, K52, and K70, for the α proton of V40 and for side-chain protons of T6, I11, R36, H68, and P69. In the calculated structure of the complex these residues are near or at the binding site.
Structure Calculations.
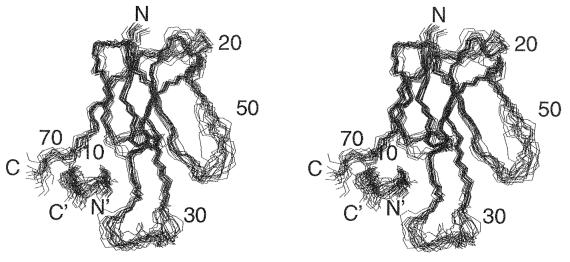
For the final refinements, a total of 1,101 distance restraints were used: 941 intramolecular interactions in the bound toxin (including 291 long-range interactions between protons that are more than 5 residues apart), 98 intrapeptide interactions (including 7 long-range interactions), and 62 intermolecular interactions between the toxin and the library peptide. The φ angle restraints included 48 angles for the toxin and 5 for the bound peptide. Also, 30 χ1 angle restraints and 23 stereospecific assignments of β-methylene protons were introduced. Twenty final structures (Fig. 1) that satisfy the experimental distance restraints with no restraint violation larger than 0.46 Å and that show good covalent geometry (Table 1) were obtained. The atomic rms deviation (rmsd) between the individual structures and the energy-minimized average structure is 0.81 Å for the backbone atoms of α-BTX residues 1–29 and 39–72, and 0.84 Å for toxin residues 1–29 and 39–68 and peptide residues 2–11. The backbone rmsd for all 13 residues of the bound peptide is 0.88 Å, and for all 87 residues of the complex is 1.35 Å. Minimization of individual structures with the charmm force-field (34) gave negative Lennard–Jones energies (Table 1), indicating good nonbonded contacts.
Figure 1.
Stereoview showing the superposition of backbone atoms for the 20 solution structures of the complex of α-BTX with library-derived peptide. N, N′, C, and C′ denote the N and C termini of the toxin and the peptide, respectively.
Table 1.
Structural statistics and rms deviation
| Structural statistics | 〈SA〉* |
|---|---|
| rmsd from experimental distance restraints, Å | 0.046 ± 0.001 |
| rmsd from experimental dihedral restraints, degree | 0.87 ± 0.10 |
| rmsd from idealized covalent geometry: | |
| Bonds, Å | 0.004 ± 0.000 |
| Angles, degree | 0.73 ± 0.01 |
| Impropers, degree | 0.59 ± 0.01 |
| x-plor potential energies, kcal⋅mol−1 | |
| EL-J† | −492.1 ± 48.1 |
| Etot | 430.7 ± 12.5 |
| Erepel‡ | 44.2 ± 5.1 |
| Eimp | 36.3 ± 1.8 |
| Enoe | 120.3 ± 6.7 |
| Ecdih | 3.8 ± 0.8 |
〈SA〉 is the ensemble of 20 NMR structures of the complex.
The Lennard–Jones potential was not used during the refinement stage.
The x-plor Frepel function was used to simulate van der Waals interactions with atomic radii set to 0.75 times their charmm (34) values.
As emerges from Fig. 1, toxin residues located at the C terminus and at the flexible loops at the tips of the three fingers are poorly defined and exhibit the highest rmsds relative to the average structure. Residues 17–20 that form the solvent-exposed turn connecting fingers I and II also show a higher than average rmsd. In agreement with the crystal structure of the α-BTX (2), no long-range interactions are observed for the residues in these highly exposed segments of the toxin. Residues at the N terminus and particularly at the C terminus of the peptide exhibit the highest rmsds in comparison with the other residues of the bound peptide.
It should be noted that for other long neurotoxins such as α-cobratoxin (7) or LsiII (9) the coordinates of the last 5–6 residues at the C terminus were not defined at all since only sequential constraints were found in this region. In our structure only the last two toxin residues are not defined.
Structure of Bound α-BTX.
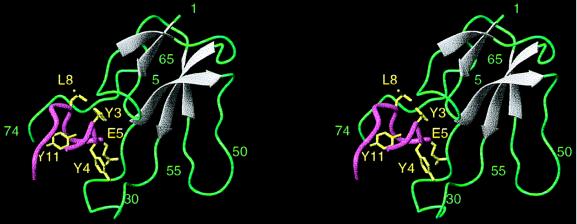
Fig. 2 shows a schematic ribbon diagram of the NMR-derived structure of the α-BTX/library peptide complex. The conformation of α-BTX is stabilized by four disulfide bridges (3/23, 16/44, 48/59, 60/65) close to each other in the tangle-like region of the molecule. Three long fingers and a tail protrude from this region. Two β-hairpins are formed by fingers I and II of the toxin. The antiparallel β-strands in finger I, consisting of residues 2–5 and 12–16, were not observed in other members of the family, except for neurotoxin 1 (4). The two strands are connected by a 6-residue loop (residues 6–11). The antiparallel β-strands of the second β-hairpin consist of residues 22–27 and 40–45. The loop of the second β-hairpin comprising residues 29–38 (a 14-residue loop) forms two β-turns (residues 30–33 and 33–37). This loop is stabilized by a fifth disulfide bridge (residues 29/33). Intramolecular interactions between W28, C29, and V39 at the base of the flexible loop in finger II, observed in the complex but not in the free toxin, indicate that in the bound toxin the β-strands are longer. A third strand comprising residues 57–60 of finger III and the two antiparallel β strands of finger II form a triple-stranded antiparallel β-sheet that is found in all neurotoxins of the family and create the core of the molecule. Type II β-turns appear at the tip of finger III (residues 52–55) and at its C-terminal end (residues 62–65). Two additional turns appear in the toxin and comprise residues 66–72 and 17–20. While the first turn exhibits several hydrogen bonds (N66/H68, H68/K70, and K70/R72) no restraints were observed for the second turn. Finger I residues that include the four N-terminal residues and I11 form multiple contacts with five C-terminal residues: T62, K64, N66, P67, and H68. A hydrogen bond between the side chain of T6 from finger I and the amide proton of L42 from finger II keeps finger I close to finger II.
Figure 2.
Ribbon plot of the complex of α-BTX with the library-derived peptide. α-BTX residues are shown in grey (for β-sheet regions) and in green. Peptide backbone is shown in pink; side chains of peptide residues interacting with the toxin (Y3P, Y4P, E5P, L8P) as well as of Y11P which forms the hydrophobic core of the peptide, are shown in yellow. The figure was generated using the program molmol (35).
Structure of the Bound Library Peptide.
The peptide folds into a compact and nearly globular conformation that is formed by three turns comprising residues 2–4, 4–6, and 7–10 (Fig. 2). These turns exhibit multiple HNi-HNi+1 NOE connectivities between residues 2/3, 3/4, 4/5, 5/6, 6/7, and 8/9 of the bound peptide. The first two residues of the first turn interact with the last residue of the third turn. The α and β protons of S10P interact with the amide proton of R2P, the aromatic ring of Y3P interact with the amide proton of S10P, and two hydrogen bonds are formed between R2P and K9P. The peptide wraps itself over the aromatic ring of Y11P which forms the core for the folding of the bound peptide. Y11P interacts with the amide protons of both R2P and Y4P. The turns are stabilized by the intramolecular interactions R2P(Hβ)/E5P(HN), Y4P(Hβ)/S6P(HN), and Y4P(HN)/S7P(HN), as well as by the intramolecular hydrogen bonds R2P(O)/Y4P(HN), Y4P(O)/S6P(HN), and S7P(Oγ1)/K9P(HN).
Binding Interactions and the Contact Surface.
The NMR-derived intermolecular interactions in the complex are listed in Table 2. Twelve residues of the toxin interact with six peptide residues: T6, A7, and I11 of finger I; D30, R36, K38, V39, and V40 of finger II; and H68, K70, Q71, and R72 at the C terminus of the toxin interact with R2P, Y3P, Y4P, E5P, S7P, and L8P of the peptide. Y3P and Y4P are the most prominent residues in the binding and form the largest number of intermolecular interactions with the toxin. While Y3P forms contacts with toxin residues located at fingers I and II and at the C terminus, Y4P interacts only with finger II of α-BTX.
Table 2.
Intermolecular interactions and contact areas of library peptide residues
| Peptide residues | Interacting α-BTX residues | No. of interactions | Contact area, Å2 |
|---|---|---|---|
| R2 | V39 | 1 | 46 |
| Y3 | T6, A7, I11, V39, H68 | 17 (13*) | 135 |
| Y4 | D30, R36, V39, V40 | 21 (16*) | 125 |
| E5 | K38, V39, V40, H68 | 8 | 104 |
| S7 | H68, R72 | 3 | 32 |
| L8 | I11, H68, K70, Q71 | 14 | 97 |
Interactions that involve side-chain atoms of the peptide.
Hydrophobic interactions in the complex are observed between the aromatic protons of Y3P and toxin residues T6 and I11, and between the aromatic protons of Y4P and toxin residues D30(Hβ), R36(Hβ and Hγ), and V39 (Table 2). In the calculated structures, intermolecular hydrogen bonds are observed between the amide proton of D30 and the OH group of Y4P. Although not restrained by NMR data, intermolecular electrostatic interactions between the side chains of R36 and D13P appear in the calculated structures of the complex.
Accessible Surface Area.
The total decrease in the accessible surface area of peptide residues upon complex formation is 917 Å2, which is a decrease of 55%. The surface of the peptide that forms contact with α-BTX has an area of 620 Å2. Therefore about 300 Å2 of the peptide surface area became buried due to intramolecular interactions within the bound peptide, thus providing an additional indication for the compact folding of the peptide. Y3P and Y4P contribute more than 20% each to the total peptide contact area, and E5P and L8P contribute to the contact area to a lower extent. The loss of the accessible surface area of α-BTX upon complex formation is 530 Å2. Nearly 50% of the toxin’s total contact area is contributed by residues located at the second finger, 35% by residues located at the C terminus, and the rest by residues from loop I.
DISCUSSION
Peptides bound to proteins are frequently found in an extended conformation, but may also form β-turns or α-helices. A remarkable feature of the bound conformation of the library-derived peptide is its compactness and almost globular form. This is also manifested by the relatively large decrease in its accessible surface area (917 Å2) upon binding. This compactness is achieved by the three turns and the folding around a central core created by the aromatic ring of Y11P. This residue does not have any interactions with the toxin and has only a structural role.
Comparison with the Structures of Other α-Neurotoxins.
As shown in Fig. 2, α-BTX forms three loops emerging like fingers of a hand, knotted together by four disulfide bridges. This structure is similar to that of other short and long members of the α-neurotoxin family (2–4, 7–9). However, a comparison between the solution structure of α-BTX when bound to the library peptide and when bound to the AcChoR α185–196 peptide (19) reveals two striking differences. In the complex with the library peptide, fingers I and II of the toxin are closer to each other, and only their loops contribute to the creation of a deep concave binding pocket (Fig. 2). This is in contrast to the complex with the α185–196 peptide, in which a deep linear groove penetrates between fingers I and II. Furthermore, in our structure the C-terminal region is an integral part of the binding pocket, while in the complex with the AcChoR peptide the C-terminal segment is in the “back” of the molecule and behind loop II.
A comparison between the average structure of α-BTX in complex with the library-derived peptide and the crystal structure of α-BTX at 2.5-Å resolution (2) reveals several noticeable differences between the two structures. In both our structure and the x-ray structure (2), the C terminus is located “behind” loop I. However, while in solution the C-terminal segment forms one of the walls of a binding-pocket, in the crystal structure it partially fills the pocket. In comparison with the x-ray structure, in our structure, loop II of the toxin is bent and twisted, thus enabling the interactions of D30, K38, V39, and V40 with the peptide. Two other differences distinguish the crystal structure from the two NMR structures of α-BTX (19), and from the crystal as well as NMR structures of other members of the α-neurotoxin family (3, 4, 8, 9): (i) due to the dimerization of the α-BTX in the crystal lattice, the extent of intramolecular β-sheet structure in finger II decreases at the expense of intermolecular β-sheet formation, and (ii) the invariant residue W28 that in our structure is located on the concave side of α-BTX and interacts with the side-chain of V39, points to the opposite direction and to the back of the molecule in the x-ray structure.
Our results are in a good agreement with previous studies of highly homologous neurotoxin that showed disorder in regions 7–10, 17–20, 30–38, and 49–55, and the C terminus, as revealed by higher rmsds for the NMR structures of neuronal-bungarotoxin (36) and LsiII (9), and larger B-factor for the crystal structures of neurotoxin 1 (4) and α-cobratoxin (3).
Contribution of Invariant Residues to the Neurotoxins Binding Site.
Site-directed mutagenesis (37) and chemical modification experiments (for a review, see ref. 1) have demonstrated the importance of finger II residues for the toxicity of neurotoxins. We have found that residues D30, R36, and V39 interact with the library peptide (Table 2), and the highly conserved residues W28, F32, G37, and E41 changed their chemical shift upon complex formation.
Comparison with the Structure of the Complex of α-BTX with an α-AcChoR Peptide.
A common motif between the sequences of the 13-residue library-derived peptide and the 12-residue AcChoR peptide (α185–196) studied by Basus et al. (19) is two adjacent tyrosine residues (positions 3 and 4 in the library peptide, and 189 and 190 in the AcChoR). In the α-BTX complex with the AcChoR peptide, 25 intermolecular NOE interactions were observed (19). Five residues of the AcChoR peptide (H186, W187, V188, Y189, and Y190; the numbering is according to AcChoR) were found to interact with 10 α-BTX residues (T5, T6, A7, I11, Y24, D30, V39, V40, G43, and H68). In contrast to the nearly globular conformation adopted by the bound library peptide, which fits very well into the toxin’s binding pocket, the bound AcChoR peptide adopts an extended conformation that fits an elongated cleft created by loop I and the second finger, with H68 at the C terminus forming the floor of the binding cleft. Although the overall topology of the α-BTX binding site differs between the complex with the library peptide and the complex with the α185–196 peptide, four intermolecular interactions are common to both complexes, and they involve the pair of tyrosine residues: the first tyrosine interacts with T6, I11, and H68, and the second with D30 of the toxin. It should be noted that overall, α-BTX residues T6, A7, I11, D30, V39, V40, and H68 interact with the ligand in both complexes, although with different residues of the two peptides. A noticeable difference between the two complexes concerns R36 that was previously postulated to be involved in AcChoR binding (37). This residue interacts with the library-derived peptide (Table 2), but not with the AcChoR α185–196 peptide (19). Our NMR data that reveal many more peptide–toxin interactions (62 vs. 25) and a 20% increase in the number of residues involved in these interactions (Table 2), might explain the 15-fold higher association constant for the library-derived peptide in comparison to the AcChoR peptide (24).
Recently, Hawrot and coworkers (38) used NMR spectroscopy to study the complex between the toxin and an 18-residue peptide of the AcChoR (α181–198) which binds the toxin with an apparent Kd of 65 nM. However, the lack of a complete proton resonance assignment for most of the 18-mer peptide, as well as for the bound toxin (38), did not allow calculation of the three-dimensional solution structure of the complex. Nonetheless, studies of the chemical-shift changes of the assigned resonances indicate that the toxin binding site or its vicinity contains residues I1, H4, T5, T6, A7, M27, W28, D30, F32, G37, V39, V40, E41, K51, V57, and N66 (38).
Only one structure of a complex of a three-finger toxin with its target molecule was reported to date, and this is the structure of fasciculin complex with acetylcholine esterase (39). In this complex finger I, finger II, and the C-terminal residue of fasciculin interact with the enzyme. This is in accordance with our findings that the first two fingers and the C-terminal region of α-BTX interact with the library-derived peptide.
Implications for Binding to the AcChoR.
The library peptide has several structural elements that are similar to those found in the AcChoR α-subunits. The consensus motif YYxSS (peptide residues 3–7) is similar to the sequence YYxCC (AcChoR α189–193). Analysis of the decrease in solvent exposure, the number of interactions and the contact area contributed by the various amino acid residues of the peptide upon complex formation, revealed that Y3P and Y4P are the most prominent residues in the binding. These findings are in accord with the previous findings (24) in which it had been shown that substitution of these aromatic residues by aliphatic amino acids leads to loss of binding of the modified peptide with α-BTX. Further indication for the importance of Y3P, Y4P, and D13P in α-BTX binding comes from a single-residue substitutions study which demonstrated the significance of the side chains of AcChoR Y189, Y190, and D195 in toxin binding (40). Peptide residues E5P and L8P that remarkably contribute to the contact area with the toxin and are in the neighborhood of the tandem aromatic residues (Y3P, Y4P), are present in all the library-derived peptides that bind strongly to α-BTX (41).
Ten percent of the total intermolecular NOEs observed in the spectra of the α-BTX/library peptide complex are between backbone atoms, while more than 50% are between side-chain atoms of the two molecules. Both backbone and side-chain atoms of Y3P, Y4P, E5P, and L8P participate in α-BTX binding, whereas R2P and S7P are involved in intermolecular interactions through solely side-chain or backbone atoms, respectively. In this study, the bound library peptide folds into a nearly globular conformation, and the importance of specific side-chain recognition is further demonstrated by the fact that backbone atoms of the peptide are involved in only 20% of the intermolecular interactions.
In conclusion, we have shown that the use of a peptide selected from a phage–peptide library to study and characterize the receptor’s binding site, contributes to a more precise definition of the structural requirements for the toxin binding site of the AcChoR. Although the peptide used in this study was selected from a nonconstrained library, it does bind the toxin while adopting a unique nearly globular conformation. It is possible that a peptide selected from a constrained peptide library that is designed according to our results, would bind α-BTX with a higher affinity, and could be used as an antidote for α-BTX.
Acknowledgments
We are highly indebted to Prof. M. Levitt for fruitful suggestions, and to Mr. V. Tugarinov and Mrs. A. Zvi for most fruitful discussions, and to Mrs. Aliza Weinberg for her help in editing this manuscript. This work was supported by grants from the U.S.–Israel Binational Science Foundation (95–246) (to J.A.), the Rashi Foundation (to E.K.-K.), the Abisch–Frenkel Foundation (to T.S.), and the Israeli Science Foundation founded by the Israel Academy of Sciences and Humanities (to E.K.-K. and to S.F.).
ABBREVIATIONS
- α-BTX
α-bungarotoxin
- DQF-COSY
double-quantum filtered correlation spectroscopy
- HOHAHA
homonuclear Hartmann–Hahn spectroscopy
- AcChoR
nicotinic acetylcholine receptor
- NOE
nuclear Overhauser effect: NOESY, two-dimensional NOE spectroscopy
- rmsd
rms deviation. A superscript P (XP) is used to mark library–peptide residues
References
- 1.Endo T, Tamiya N. Pharmacol Ther. 1987;34:403–451. doi: 10.1016/0163-7258(87)90002-7. [DOI] [PubMed] [Google Scholar]
- 2.Love R A, Stroud R M. Protein Eng. 1986;1:37–46. doi: 10.1093/protein/1.1.37. [DOI] [PubMed] [Google Scholar]
- 3.Betzel C, Lange G, Pal G P, Wislon K S, Maelicke A, Saenger W. J Biol Chem. 1991;266:21530–21536. doi: 10.2210/pdb2ctx/pdb. [DOI] [PubMed] [Google Scholar]
- 4.Nickitenko A V, Michailov A M, Betzel Ch, Wilson K S. FEBS Lett. 1993;320:111–117. doi: 10.1016/0014-5793(93)80073-4. [DOI] [PubMed] [Google Scholar]
- 5.Inagaki F, Hiders R C, Hodges S J, Drake A F. J Mol Biol. 1985;183:575–590. doi: 10.1016/0022-2836(85)90173-1. [DOI] [PubMed] [Google Scholar]
- 6.Basus V J, Billeter M, Love R A, Stroud R M, Kuntz I D. Biochemistry. 1988;27:2763–2771. doi: 10.1021/bi00408a016. [DOI] [PubMed] [Google Scholar]
- 7.Le Goas R, LaPlante S R, Mikou A, Delsuc M A, Guitter E, Robin M, Charpentier I, Lallemand J Y. Biochemistry. 1992;31:4867–4875. doi: 10.1021/bi00135a018. [DOI] [PubMed] [Google Scholar]
- 8.Zinn-Justin S, Roumestand C, Gilquin B, Bontems F, Ménez A, Toma F. Biochemistry. 1992;31:11335–11347. doi: 10.1021/bi00161a011. [DOI] [PubMed] [Google Scholar]
- 9.Connolly P J, Stern A S, Hoch J C. Biochemistry. 1996;35:418–426. doi: 10.1021/bi9520287. [DOI] [PubMed] [Google Scholar]
- 10.Gershoni J M, Hawrot E, Lentz T L. Proc Natl Acad Sci USA. 1983;80:4973–4977. doi: 10.1073/pnas.80.16.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neumann D, Gershoni J M, Fridkin M, Fuchs S. Proc Natl Acad Sci USA. 1985;82:3490–3493. doi: 10.1073/pnas.82.10.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neumann D, Barchan D, Safran A, Gershoni J M, Fuchs S. Proc Natl Acad Sci USA. 1986;83:3008–3011. doi: 10.1073/pnas.83.9.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Changeux J-P, Galzi J L, Devillers-Thiery A, Bernard D. Q Rev Biophys. 1992;25:395–432. doi: 10.1017/s0033583500004352. [DOI] [PubMed] [Google Scholar]
- 14.Conti-Tronconi B M, MacLane K E, Raftery M A, Grando S A, Pia Protti M. Crit Rev Biochem Mol Biol. 1994;29:69–123. doi: 10.3109/10409239409086798. [DOI] [PubMed] [Google Scholar]
- 15.Fraenkel Y, Shalev D E, Gershoni J, Navon G. Crit Rev Biochem Mol Biol. 1996;31:273–301. doi: 10.3109/10409239609106586. [DOI] [PubMed] [Google Scholar]
- 16.Karlin A. Curr Opin Neurobiol. 1993;3:299–309. doi: 10.1016/0959-4388(93)90121-e. [DOI] [PubMed] [Google Scholar]
- 17.Neumann D, Barchan D, Fridkin M, Fuchs S. Proc Natl Acad Sci USA. 1986;83:9250–9253. doi: 10.1073/pnas.83.23.9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson P T, Lentz T L. Biochemistry. 1988;27:6667–6674. doi: 10.1021/bi00418a004. [DOI] [PubMed] [Google Scholar]
- 19.Basus V, Song G, Hawrot E. Biochemistry. 1993;32:12290–12298. doi: 10.1021/bi00097a004. [DOI] [PubMed] [Google Scholar]
- 20.Scott J K, Smith G P. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 21.Devlin J J, Panganiban L C, Devlin P E. Science. 1990;249:404–406. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- 22.Yu H, Chen J K, Feng S, Dagarno D C, Brauer A W, Schreiber S L. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 23.Feng S, Kasahara C, Rickles R J, Schreiber S L. Proc Natl Acad Sci USA. 1995;92:12408–12415. doi: 10.1073/pnas.92.26.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balass M, Katchalski-Katzir E, Fuchs S. Proc Natl Acad Sci USA. 1997;94:6054–6058. doi: 10.1073/pnas.94.12.6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piantini U, Sørenson O, Ernst R R. J Am Chem Soc. 1982;104:6800–6801. [Google Scholar]
- 26.Macura S, Ernst R R. Mol Phys. 1981;41:95–117. [Google Scholar]
- 27.Davis D G, Bax A. J Am Chem Soc. 1985;107:2820–2821. [Google Scholar]
- 28.Piotto M, Saudek V, Sklenar V. J Biomol NMR. 1992;2:661–669. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 29.Wagner G, Braun W, Havel T F, Schaumann T, Go N, Wüthrich K. J Mol Biol. 1987;196:611–639. doi: 10.1016/0022-2836(87)90037-4. [DOI] [PubMed] [Google Scholar]
- 30.Nilges M, Clore G M, Gronenborn A M. FEBS Lett. 1988;229:317–324. doi: 10.1016/0014-5793(88)81148-7. [DOI] [PubMed] [Google Scholar]
- 31.Brünger A T. The xplor 3.1 Manual. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 32.Brünger A T, Clore G M, Gronenborn A M, Karplus M. Proc Natl Acad Sci USA. 1986;83:3801–3805. doi: 10.1073/pnas.83.11.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 34.Brooks B R, Bruccoleri R E, Olafson B O, States D J, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 35.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 36.Sutcliffe M J, Dobson C M, Oswald R E. Biochemistry. 1992;31:2962–2970. doi: 10.1021/bi00126a017. [DOI] [PubMed] [Google Scholar]
- 37.Pillet L, Tremeau O, Ducancel F, Drevet P, Zinn-Justin S, Pinkasfeld S, Boulain J C, Ménez A. J Biol Chem. 1993;268:909–916. [PubMed] [Google Scholar]
- 38.Gentile L, Basus V J, Shi Q L, Hawrot E. Ann NY Acad Sci. 1995;757:222–237. doi: 10.1111/j.1749-6632.1995.tb17479.x. [DOI] [PubMed] [Google Scholar]
- 39.Tzartos S J, Remoundos M S. J Biol Chem. 1990;265:21462–21467. [PubMed] [Google Scholar]
- 40.Harel M, Kleywegt G J, Ravelli R, Silman I, Sussman J. Structure (London) 1995;3:1355–1366. doi: 10.1016/s0969-2126(01)00273-8. [DOI] [PubMed] [Google Scholar]
- 41.Balass M, Morag E, Bayer E A, Fuchs S, Wilcheck M, Katchalski-Katzir E. Anal Biochem. 1996;243:264–269. doi: 10.1006/abio.1996.0515. [DOI] [PubMed] [Google Scholar]