Abstract
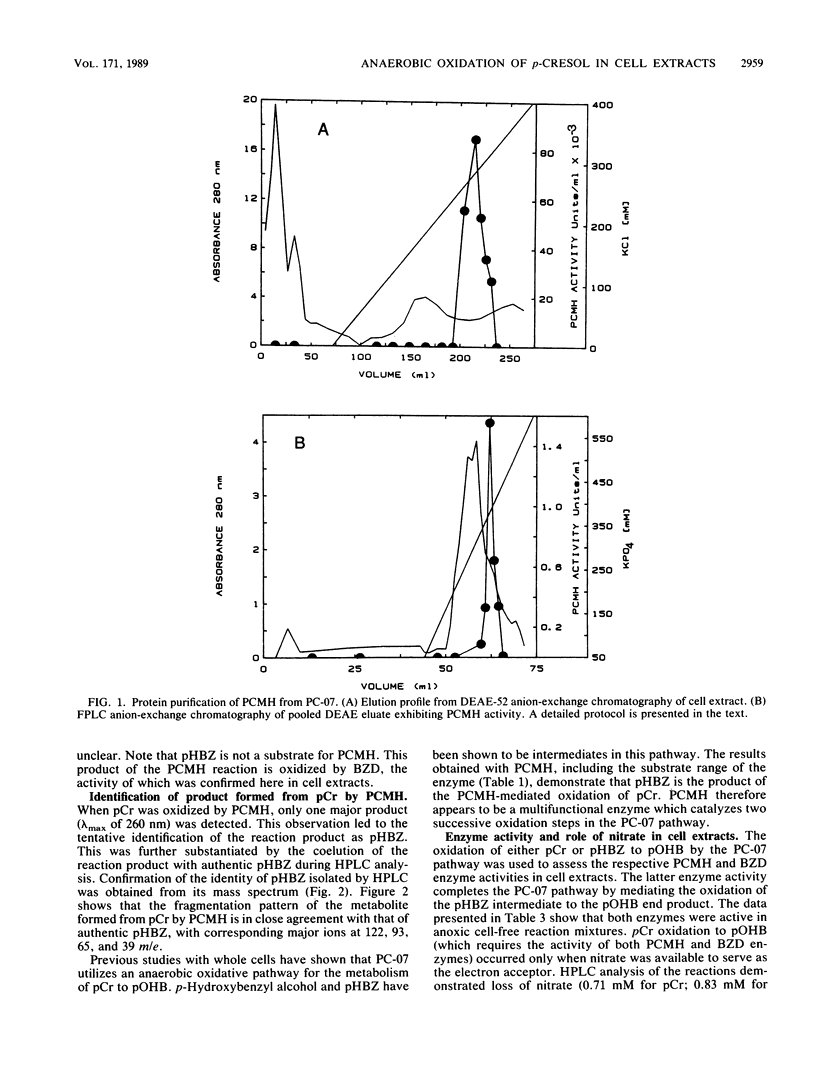
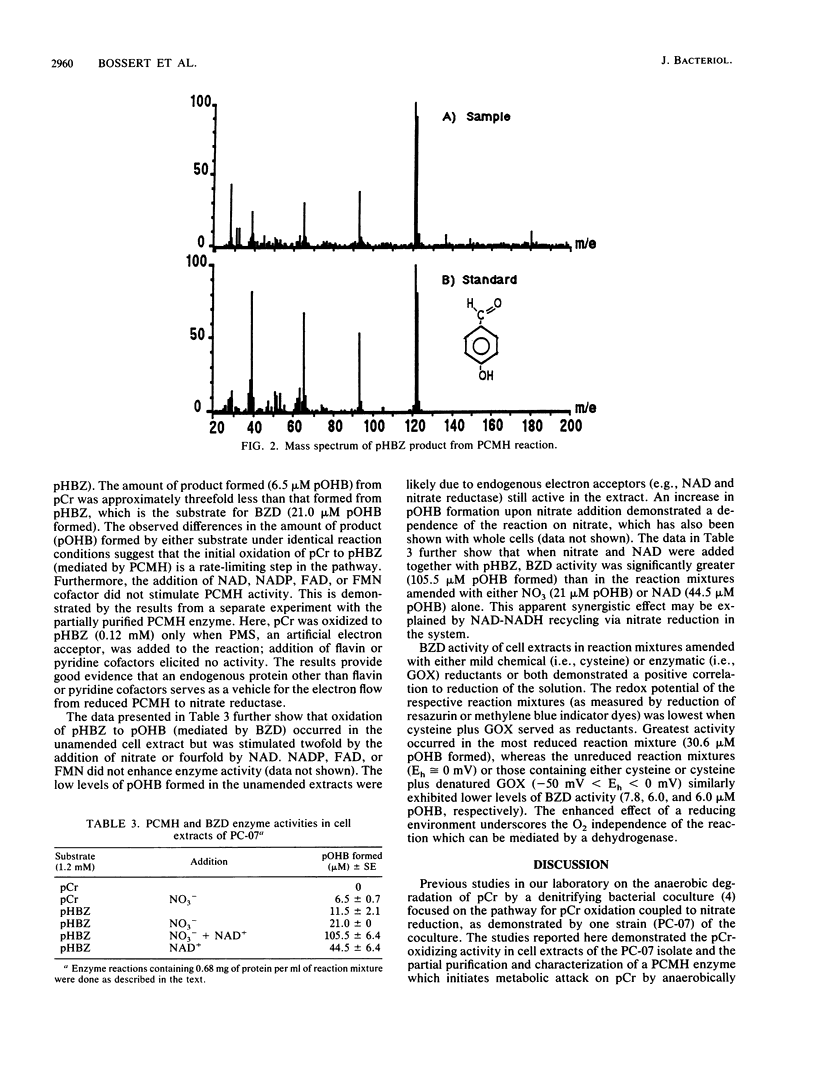
Anoxic cell extracts of a denitrifying bacterial isolate (PC-07) were shown to oxidize p-cresol to p-hydroxybenzoate. Oxidation of the substrate was independent of molecular oxygen and required nitrate as the natural terminal electron acceptor. Two enzyme activities were implicated in the pathway utilized by PC-07. A p-cresol methylhydroxylase mediated the oxidation of p-cresol to p-hydroxybenzaldehyde, which was further oxidized to p-hydroxybenzoate by an NAD+-dependent dehydrogenase. The PC-07 methylhydroxylase was partially purified by anion-exchange chromatography. The protein appeared to be a multifunctional flavocytochrome, which first oxidized p-cresol to p-hydroxybenzyl alcohol, which was then oxidized to p-hydroxybenzaldehyde. The identity of the aldehyde was confirmed by mass spectroscopy. The PC-07 methylhydroxylase had a limited substrate range and required an alkyl-substituted phenolic ring with a hydroxyl group in the para position. From the available evidence, p-cresol, a naturally occurring phenol, exhibited the greatest affinity to the enzyme and therefore may be its natural substrate.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Brown L. H. The amino acid sequence of Pseudomonas fluorescens azurin. Biochem J. 1967 Sep;104(3):784–825. doi: 10.1042/bj1040784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert I. D., Young L. Y. Anaerobic oxidation of p-cresol by a denitrifying bacterium. Appl Environ Microbiol. 1986 Nov;52(5):1117–1122. doi: 10.1128/aem.52.5.1117-1122.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque D., Bisaillon J. G., Beaudet R., Sylvestre M., Ishaque M., Morin A. Microbiological Degradation of Malodorous Substances of Swine Waste under Aerobic Conditions. Appl Environ Microbiol. 1987 Jan;53(1):137–141. doi: 10.1128/aem.53.1.137-141.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bühler M., Giesel H., Tischer W., Simon H. Occurrence and the possible physiological role of 2-enoate reductases. FEBS Lett. 1980 Jan 14;109(2):244–246. doi: 10.1016/0014-5793(80)81096-9. [DOI] [PubMed] [Google Scholar]
- D'Ari L., Barker H. A. p-Cresol formation by cell-free extracts of Clostridium difficile. Arch Microbiol. 1985 Dec;143(3):311–312. doi: 10.1007/BF00411256. [DOI] [PubMed] [Google Scholar]
- Hopper D. J. Incorporation of [18O]water in the formation of p-hydroxybenzyl alcohol by the p-cresol methylhydroxylase from Pseudomonas putida. Biochem J. 1978 Oct 1;175(1):345–347. doi: 10.1042/bj1750345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J., Jones M. R., Causer M. J. Periplasmic location of p-cresol methylhydroxylase in Pseudomonas putida. FEBS Lett. 1985 Mar 25;182(2):485–488. doi: 10.1016/0014-5793(85)80359-8. [DOI] [PubMed] [Google Scholar]
- Hopper D. J., Taylor D. G. The purification and properties of p-cresol-(acceptor) oxidoreductase (hydroxylating), a flavocytochrome from Pseudomonas putida. Biochem J. 1977 Oct 1;167(1):155–162. doi: 10.1042/bj1670155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keat M. J., Hooper D. J. Aromatic aldehyde dehydrogenase from Pseudomonas putida N.C.I.B. 9869. Biochem Soc Trans. 1975;3(3):385–386. doi: 10.1042/bst0030385. [DOI] [PubMed] [Google Scholar]
- Smolenski W. J., Suflita J. M. Biodegradation of cresol isomers in anoxic aquifers. Appl Environ Microbiol. 1987 Apr;53(4):710–716. doi: 10.1128/aem.53.4.710-716.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama M. T., Carlson J. R. Production of Skatole and para-Cresol by a Rumen Lactobacillus sp. Appl Environ Microbiol. 1981 Jan;41(1):71–76. doi: 10.1128/aem.41.1.71-76.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]


