Abstract
Objective: To investigate molecular alterations associating with prostate carcinoma progression and potentially provide information toward more accurate prognosis/diagnosis. Methods: A set of laser captured microdissected (LCM) specimens from 300 prostate cancer (PCa) patients undergoing radical prostatectomy (RP) were defined. Ten patients representing “aggressive” PCa, and 10 representing “non-aggressive” PCa were selected based on prostate-specific antigen (PSA) recurrence, Gleason score, pathological stage and tumor cell differentiation, with matched patient age and race between the two groups. Normal and neoplastic prostate epithelial cells were collected with LCM from frozen tissue slides obtained from the RP specimens. The expressions of a panel of genes, including NPY, PTEN, AR, AMACR, DD3, and GSTP1, were measured by quantitative real-time RT-PCR (TaqMan), and correlation was analyzed with clinicopathological features. Results: The expressions of AMACR and DD3 were consistently up-regulated in cancer cells compared to benign prostate epithelial cells in all PCa patients, whereas GSTP1 expression was down regulated in each patient. NPY, PTEN and AR exhibited a striking difference in their expression patterns between aggressive and non-aggressive PCas (P=0.0203, 0.0284, and 0.0378, respectively, Wilcoxon rank sum test). The lower expression of NPY showed association with “aggressive” PCas based on a larger PCa patient cohort analysis (P=0.0037, univariate generalized linear model (GLM) analysis). Conclusion: Despite widely noted heterogeneous nature of PCa, gene expression alterations of AMACR, DD3, and GSTP1 in LCM-derived PCa epithelial cells suggest for common underlying mechanisms in the initiation of PCa. Lower NPY expression level is significantly associated with more aggressive clinical behavior of PCa; PTEN and AR may have potential in defining PCa with aggressive clinical behavior. Studies along these lines have potential to define PCa-associated gene expression alterations and likely co-regulation of genes/pathways critical in the biology of PCa onset/progression.
Keywords: Prostate cancer, NPY expression, Quantitative real-time reverse-transcript polymerase chain reaction (RT-PCR)
INTRODUCTION
Prostate cancer (PCa) is the most common malignancy and the second leading cause of cancer related deaths in men of Western countries. The adoption of screening based upon the measurement of the serum prostate-specific antigen (PSA) has led to the earlier detection and management of PCa. However, despite these advances, an estimated 30% of all PCa patients suffer from recurrent disease subsequent to radical prostatectomy (Jemal et al., 2007). Thus there is a critical need to distinguish those patients with aggressive PCa from those with non-aggressive ones. Molecular approaches to this problem have found alterations in a number of candidate genes associated with prostate cancer progression, including losses of p53, p27, GSTP1 and PTEN amplification or overexpressions of MYC, HER2/neu, and cyclin D1 (Singh et al., 2002; Kumar-Sinha and Chinnaiyan, 2003). The aim of this study was, using quantitative real-time RT-PCR, to investigate expressions of a panel of genes in two precisely defined sets of laser captured microdissection (LCM) PCa specimens representing “aggressive” and “non-aggressive” PCa, respectively. The relationship between gene panel expression status and clinicopathological parameters including patient outcome was examined. Our hypothesis was that this gene panel has the potential in defining PCa patients at high risk of disease progression.
MATERIALS AND METHODS
Patients and samples
From 300 PCa patients undergoing radical prostatectomy (RP) in Walter Reed Army Medical Center from 1997 to 2002, 20 with primary prostate cancer were selected. Among them, 10 patients were with “aggressive” (AG) PCa, and the remaining 10 were with “non-aggressive” (NA) PCa based on PSA recurrence, Gleason score, tumor cell differentiation, and seminal vesicle invasion status (Table 1). The patient age and race were matched in two groups. All patients in this study were enrolled in the CPDR (Center for Prostate Disease Research) Triservice Multicenter Longitudinal PCa Database. The median follow-up was 6.8 years (range 5~9 years). And a larger patient cohort (the number up to 59) was investigated for NPY gene.
Table 1.
Criteria for patient selection
| Group | PRAR | GS | SVI | TCD |
| AG | Yes (2.1~39 months) | 8~9 | Yes | Poor |
| NA | No (>60 months) | 6~7 | No | Moderate to well |
PRAR: PSA recurrence after RP; GS: Gleason score; SVI: Seminal vesicle invasion; TCD: Tumor cell differentiation
Tissue specimens were evaluated immediately by a genitourinary pathologist at the time of specimen acquisition. If a palpable tumor was present, the surface overlying it was painted with black ink and a wedge from the center was immediately embedded in Tissue-Tek OCT (Miles Inc. Diagnostic Division, Elkhart, IN) and snap frozen on dry ice and stored at −70 °C. Sextant 14-gauge true cut biopsies including apex, middle and base of the right and left lobes of the prostate were obtained on each case.
The volume of biopsy specimens was about 1 cm×0.5 cm×0.5 cm. Serial 10 μm frozen sections were cut and achieved at −70 °C. One set of slides was stained with H & E, and read by the urological pathologists to define tumor cells. The pure prostate cancerous cells and normal appearing epithelial cells were collected, respectively, using LCM according to the protocol provided by the manufacturer (Arcturus Engineering, Mountain view, CA). The selected cells adhered to the transfer cap were lifted off the tissue section and placed directly into an Eppendorf tube for RNA extraction.
RNA extraction and cDNA synthesis
Total RNA were extracted from LCM cells by using the micro-isolation kit (Stratagene, TX, USA) according to the manufacturer. Total RNA were reverse-transcribed in a final volume of 20 μl using Omnisensecript RT-kit (Qiagene, USA) according to the manufacture’s protocol.
Gene panel selection
Six PCa related genes, including AMACR, DD3, GSTP1, AR, PTEN, and NPY, were selected based on the literatures (Chen et al., 2004; Bialkowska-Hobrzanska et al., 2006; Murphy et al., 2007; Meiers et al., 2007; Pourmand et al., 2007) and our lab (CPDR) PCa gene discovery efforts (Shaheduzzaman et al., 2006).
Real-time quantitative RT-PCR
1. Primer and probe
The primer and probe for AR, AMACR, GSTP1, and NPY were assays-on-demand gene expression products obtained from PE Applied Biosystems (Foster, CA); DD3 and PTEN were chosen with the assistance of PE Primer Express® software, and were ordered from PE Applied Biosystems (Foster, CA). One of the paired primers or probe was designed to be intron spanning to preclude amplification of genomic DNA. Except DD3 and PTEN probes labeled with TET, all the others were labeled with FAM (6-carboxy-fluorescein). DD3 and PTEN primer and probe sequences are listed in Table 2.
Table 2.
DD3 and PTEN primer/probe sequences used in real-time PCR
| Gene | Oligo-neucleotide | Sequence (from 5′ to 3′) | PCR product size (bp) |
| DD3 | Forward primer | CAC ATT TCC AGC CCC TTT AAA TA | 112 |
| Reverse primer | GGG CGA GGC TCA TCG AT | ||
| Probe |
TET-GGA AGC ACA GAG ATC CCT GGG AGA AAT G-TAMARA |
||
| PTEN | Forward primer | AAG ACA TTA TGA CAC CGC CAA AT | 134 |
| Reverse primer | ATG ATT GTC ATC TTC ACT TAG CCA TT | ||
| Probe | TET-TGC AGA GTT GCA CAA TAT CCT TTT GAA GAC C-TAMARA |
2. PCR amplification
All PCR reactions were performed using an ABI prism 7700 Sequence Detection System PE Applied Biosystems (Foster, CA). For each PCR run, a master-mix was prepared on ice with 1×TaqMan Master Mix, 1×GAPDH primer/probe, and 1×target gene primer/probe (for AMACR, AR, GSTP1, and NPY) or 150~300 nmol/L primer/probe (for DD3 and PTEN). Two μl of each diluted cDNA sample (about 0.1 ng) was added in duplicate to 28 μl of PCR master-mix. The thermal cycling conditions comprised an initial denaturation step at 95 °C for 10 min, 50 cycles at 95 °C for 15 s and 65 °C for 1 min. The expression of house keeping gene, GAPDH, was simultaneously analyzed as the internal control of same batch of cDNA, and each sample was normalized to the internal control. The negative control was RNA samples without reverse transcription.
The relative target gene expression level was presented as “fold change” of matched tumor vs normal cells:
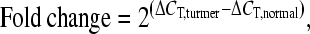 |
where C T is the cycle of threshold, and ΔC T means the target genes’ C T value normalized to GAPDH.
Immunohistochemical staining of NPY
Whole mount fixed tissue sections from the first selected 20 patients with primary PCa were immunohistochemically stained according to the modified ABC method. Briefly, tissue sections were deparaffinized, rehydrolized, and rinsed in PBS with pretreatment by microwave retrieval. The sections were then blocked with 0.3% hydrogen peroxide for 10 min, and then 10% goat serum for 15 min to reduce nonspecific background at room temperature. Then sections were incubated sequentially with a rabbit polyclonal anti-NPY antibody (1:100, Abcam, Cambridge, UK) overnight at 4 °C, biotinylated secondary antibody for 30 min at 40 °C, and ABC complex for 30 min. Antibody binding sites were visualized using 3,3′-diaminobenzidine as the chromogenic substrate, and tissue sections were lightly counterstained with hematoxylin. Nerve fibers in prostate tissue were used as internal positive control. Negative controls were treated with PBS instead of primary antibodies.
Statistical analysis
Wilcoxon rank sum test was performed to analyze the relationship between each gene relative expression level and categorical clinical factors (such as pathological T stage, tumor differential grade, capsule status, surgical margin status and seminal vesicle invasion, PSA recurrence and risk groups, etc.). Pearson correlation analysis was used to calculate the correlation coefficients between each gene and a continuous clinical variable, which included PSA doubling time, diagnostic age, biopsy Gleason sum, pathological Gleason sum, tumor number, etc. Finally, univariate generalized linear model (GLM) was used to analyze the relationship between each log transformed gene expression ratio and a categorical clinical factors, and multivariate GLM to analyze the relationship between a log transformed gene expression ratio and multi-clinical factors.
RESULTS
Dynamic range of each gene real-time RT-PCR assays (sensitivity and efficiency test)
To determine the dynamic range and efficiency of assay of each gene, several standard curves were constructed with serially diluted cDNA of normal prostate tissue (Clontech). Wide dynamic ranges were obtained with samples containing as much as 10 ng or as little as 10~100 pg of normal prostate cDNA. A strong linear relationship was always obtained (r 2≥0.99). The reaction efficiency (E) calculated as E=101/m−1, where m is absolute value of the slope of the calibration curve, was always higher than 90%.
Consistent alteration of AMACR, DD3 and GSTP1 in PCa
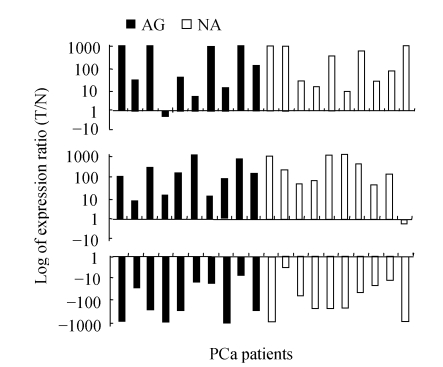
AMACR, DD3 and GSTP1 were most consistently altered in PCa. Nineteen out of 20 (95%) and 15 out of 20 (75%) of PCa patients had over-expressed AMACR and DD3 in tumor cells, respectively, while all the 20 PCa patients had down-regulated GSTP1 in tumor cells (Fig.1). Additionally, none of these three genes was found to have significant difference between AG and NA groups (all three P values were greater than 0.05, Wilcoxon rank sum test).
Fig. 1.
Expression of AMACR (top), DD3 (middle) and GSTP1 (bottom) in matched tumor (T) and normal (N) prostate epithelial cells
X-axis is 20 cases of prostate carcinoma patients. Y-axis is relative gene expression level on a log scale. The maximum and minimum were set at +1000/−1000 since ratio more than 1000 was regarded as artificial (the same in the following figures)
NPY differentially regulated in AG and NA PCa
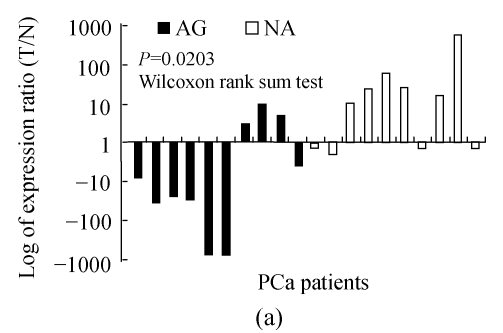
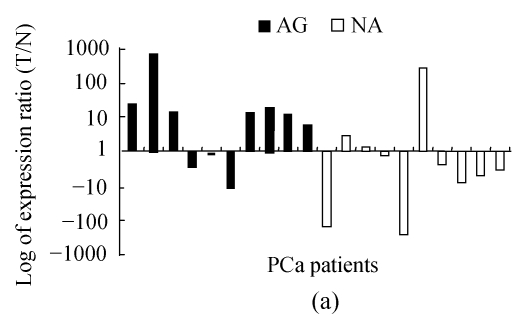
Six out of ten (60%) non-aggressive tumors showed an up-regulated NPY expression, but only 30% (3/10) of aggressive tumors had an NPY overexpression (Fig.2a). Wilcoxon rank sum test analysis showed the difference was significant (P=0.0203).
Fig. 2.
NPY was differentially regulated in aggressive and non-aggressive PCa cells
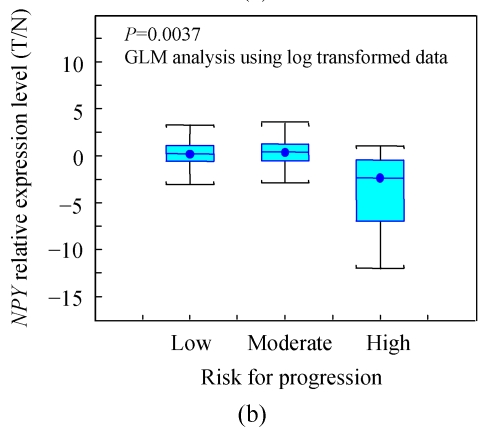
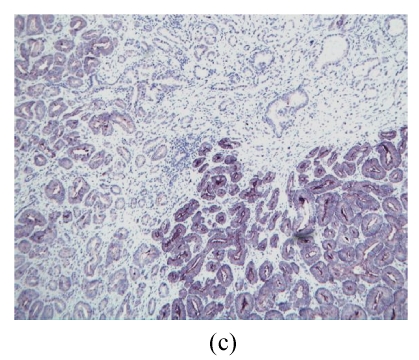
(a) Relative expression level of NPY in 20 PCa patients, 7 out of 10 AG cases had down-regulated NPY in tumor cells, while 6 of 10 NA cases had up-regulated NPY in tumor cells (P=0.0203); (b) The relative expression level of NPY in 59 cases of PCa was stratified as low, moderate and high risk according D′Amico (2001)’s definition. NPY was significantly lower in high risk group than that in low and moderate risk group (P=0.0037). X-axis is risk group, Y-axis is relative expression level presented as log transformed ratio of tumor (T) vs normal (N); (c) Immunohistochemical staining of NPY in one representative PCa tissue section. Well-differentiated cancerous glands showed stronger intensity than that of poor-differentiated areas (ABC method, original magnification 100×)
In order to further validate this observation, a larger PCa patient cohort (59 cases) stratified by risk (D′Amico (2001)’s definition) was analyzed using the same approach (TaqMan qRT-PCR).
The relative expression level of NPY in PCa was lower in the high-risk group than that in the low and moderate risk group (Fig.2b). Due to the big deviation of fold change, NPY fold change was log transformed to be near normal distributed, and univariate generalized linear model analysis showed the difference was highly significant (P=0.0037).
The NPY immunostaining shows heterogeneous intensity and percentage in both benign and cancerous epithelial cells. Due to the staining heterogeneity in each case, they cannot be simply quantified into 3 or 4 degrees. However, a trend had been observed that there are stronger signals in well-differentiated tumor cells than those in poor-differentiated ones, even in the same case (Fig.2c).
Both AR and PTEN differentially regulated in AG and NA PCa cells
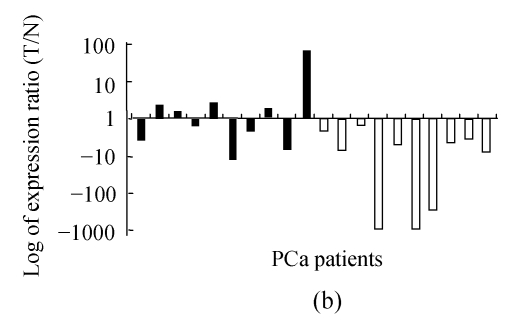
AR was overexpressed in 70% (7/10) of AG tumors; but only 30% (3/10) of NA tumors had overexpressed AR (P=0.0378, Wilcoxon rank sum test) (Fig.3a).
Fig. 3.
Relative expression levels of AR (a) and PTEN (b)
Both AR and PTEN were differentially regulated in AG and NA PCa cells, though they were of more deviation compared to the other genes (P=0.0378 and 0.0284, respectively, Wilcoxon rank sum test)
All NA tumors and 50% of AG cases showed down-regulated PTEN, while the rest 5 cases of AG group up-regulated, and the difference between AG and NA was significant (P=0.0284, Wilcoxon rank sum test) (Fig.3b).
DISCUSSION
Recent years, cDNA microarray analysis, differential display analysis and serial analysis of gene expression (SAGE) have identified a number of molecular markers specially altered in PCa. We comprehensively examined the expression patterns of 6 genes in the carefully selected PCa tissues including aggressive and non-aggressive PCas. Among them, AMACR and DD3 were reported prostate specific and over-expressed in PCa (Bialkowska-Hobrzanska et al., 2006; Murphy et al., 2007), and the loss of expression (silencing) of the GSTP1 gene was the most common (>90%) genetic alteration reported to date in PCa (Meiers et al., 2007). In agreement with the literature, our study showed the expressions of AMACR and DD3 were consistently up-regulated in cancer cells compared to benign prostate epithelial cells in almost all PCa patients, whereas GSTP1 expression was down-regulated in each patient, which implied the high quality of the specimens used in this study. And most importantly, despite widely noted heterogeneous nature of PCa, AMACR, DD3, and GSTP1, their expression alterations in PCa cells suggest common underlying mechanisms in the initiation of PCa.
Strikingly, NPY, PTEN and AR were found to be differentially expressed in aggressive and non-aggressive PCa cells. NPY was selected in this study based on our genechip data analysis (Shaheduzzaman et al., 2006). NPY is a member of a family of 36 amino acid long peptides, including NPY, PYY (peptide YY), and PP (pancreatic polypeptide). Its main function is not that of an endocrine or gut hormone, but that of a neurotransmitter, a vasoconstrictor, and increasing the actions of noradrenalin (Silva et al., 2002). The role of NPY in human tumor is not clear, and the study results are controversial. Reubi et al.(2001) found that NPY can inhibit the growth of SK-N-MC cells, a neuroblastoma cell line expressing NPY receptor Y1. Another study found lower degrees of proNPY processing to NPY in tumor tissue were correlated to advanced clinical stages and poor outcome in neuroblastomas (Bjellerup et al., 2000). However, Knerr et al.(2001) investigated a series of intracranial tumors and demonstrated that well-differentiated tumors (such as grade I astrocytomas) exhibit higher levels of NPY than anaplastic tumors. The importance of NPY in the prostate carcinoma is rarely investigated. NPY immunoreactivity has been reported in prostate neuroendocrine and secretory cells, nerve fibers, and carcinoma cells, but the role of NPY contained in prostate glandular cells is unknown (Mack et al., 1997). Most recently, Rasiah et al.(2006) and Ruscica et al.(2006; 2007) found higher NPY immunostaining in HGPIN (high grade prostate intraepithelial neoplasia) and PCa than that in benign epithelium. NPY immunostaining of PCa was independently associated with relapse, after adjusting for traditional prognostic factors. Their studies support the concept that NPY may directly regulate PCa cell growth via Y1-R, NPY and the related receptors are overexpressed in PCa and may play a relevant role in PCa progression at both androgen dependent and independent stages. In this study, we demonstrated that NPY mRNA expression level is significantly higher in non-aggressive PCa cells than that in aggressive PCa. In order to further validate our exciting data on NPY, we analyzed NPY expression in a larger cohort of PCa patients. According to Dr. D′Amico (2001)’s definition of PCa risk group, this cohort included 24 of high risk (HR), 23 of middle risk (MR) and 12 of low risk (LR) patients. As shown in the boxplot (Fig.2b), the tumor (T)/normal (N) expression ratio of NPY is significantly lower in HR group than that in LR and MR group. In addition, the immunostaining of NPY in the first 20 cases showed that the well differentiated tumor area had stronger staining intensity and/or higher percentage, which was partly consistent with RT-PCR results. But due to the heterogenous staining and limited cases, we cannot draw any definite conclusion from the immunohistochemical result. However, multivariate GLM analysis on NPY mRNA expression level in the larger cohort showed NPY can be regarded as an independent prognostic marker for PCa progression.
AR signaling pathway is very important in the progression of PCa. It has been reported that approximately one-third of prostate carcinomas recurring during endocrine therapy contain an AR gene amplification (Chen et al., 2004; Montironi et al., 2004). In this study, AR was overexpressed in 70% (7/10) of AG tumors, but only in 30% (3/10) of NA tumors. The higher expression of AR may promote the growth of PCa cells and give rise of aggressive clinical manifestation. Therefore, AR might be regarded as another prognostic marker for PCa progression. The further assay of AR expression in a larger patient cohort is now underway in our group.
PTEN, a tumor suppressor gene encoding a phosphatase active against both proteins and lipid substrates, is a common target for somatic alteration during PCa progression. Many studies stated that PTEN mutation and the loss of expression is an indicator of more advanced disease at surgery, and is predictive of a shorter time to biochemical recurrence of disease (Pourmand et al., 2007). Herein we showed that all NA cases and 5 out of 10 AG cases had down-regulated expression of PTEN, which was consistent with the published studies. However, we also unexpectedly found that PTEN was overexpressed at mRNA level in 50% (5/10) of AG tumors, though the degree of overexpression was very low except one case. The difference between AG and NA groups was statistically significant. According to the accumulated data, which showed high frequency of PTEN mutation in advanced PCa (Yoshimoto et al., 2007), it is reasonable to postulate that the higher mRNA level of PTEN may be due to different PTEN mutations or other changes in aggressive PCa from that in non-aggressive PCa.
In summary, we investigated the expressions of a panel of genes in a set of LCM derived cancerous cells, representing aggressive and non-aggressive PCa behavior, and normal matched prostate epithelial cells. This is a proof of principle study and the consistent gene expression alterations of AMACR, GSTP1, and DD3 in PCa cells suggest that certain biologic pathways are frequently altered in PCa. Most significantly, NPY, but also PTEN and AR, were found to be differentially expressed in AG and NA PCa cells. Lower NPY expression level is significantly associated with more aggressive clinical behavior of PCa; PTEN and AR may have potential in defining PCa with aggressive clinical behavior; however, this needs a further investigation.
Footnotes
Project supported by the Center for Prostate Disease Research, and the Henry M. Jackson Foundation for the Advancement of Military Medicine, Rockville, MD, USA
References
- 1.Bialkowska-Hobrzanska H, Driman DK, Fletcher R, Harry V, Razvi H. Expression of human telomerase reverse transcriptase, survivin, DD3 and PCGEM1 messenger RNA in archival prostate carcinoma tissue. Can J Urol. 2006;13(1):1967–1974. [PubMed] [Google Scholar]
- 2.Bjellerup P, Theodorsson E, Jornvall H, Kogner P. Limited neuropeptide Y precursor processing in unfavourable metastatic neuroblastoma tumours. Br J Cancer. 2000;83(2):171–176. doi: 10.1054/bjoc.2000.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Molecular determinants of resistance to antiandrogen therapy. Nature Medicine. 2004;10(1):33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 4.D′Amico AV. Combined-modality staging for localized adenocarcinoma of the prostate. Oncology (Huntington) 2001;15(8):1049–1059. [PubMed] [Google Scholar]
- 5.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. C A Cancer J Clin. 2007;57(1):43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 6.Knerr I, Schuster S, Nomikos P, Buchfelder M, Sotsch J, Schoof E, Fahlbusch R, Rascher W. Gene expression of adrenomedullin, leptin, their receptors and neuropeptide Y in hormone-secreting and non-functioning pituitary adenomas, meningiomas and malignant intracranial tumors in humans. Neuropathol Appl Neurobiol. 2001;27(3):215–222. doi: 10.1046/j.0305-1846.2001.00324.x. [DOI] [PubMed] [Google Scholar]
- 7.Kumar-Sinha C, Chinnaiyan AM. Molecular markers to identify patients at risk for recurrence after primary treatment for prostate cancer. Urology. 2003;62(Suppl. 6B):19–35. doi: 10.1016/j.urology.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 8.Mack D, Hacker GW, Hauser-Kronberger C, Frick J, Dietze O. Vasoactive intestinal polypeptide (VIP) and neuropeptide tyrosine (NPY) in prostate carcinoma. Eur J Cancer. 1997;33(2):317–318. doi: 10.1016/S0959-8049(96)00402-9. [DOI] [PubMed] [Google Scholar]
- 9.Meiers I, Shanks JH, Bostwick DG. Glutathione S-transferase pi (GSTP1) hypermethylation in prostate cancer: review 2007. Pathology. 2007;39(3):299–304. doi: 10.1080/00313020701329906. [DOI] [PubMed] [Google Scholar]
- 10.Montironi R, Scarpelli M, López Beltran A. Carcinoma of the prostate: inherited susceptibility, somatic gene defects and androgen receptors. Virchows Arch. 2004;444(6):503–508. doi: 10.1007/s00428-004-0996-2. [DOI] [PubMed] [Google Scholar]
- 11.Murphy AJ, Hughes CA, Lannigan G, Sheils O, O'Leary J, Loftus B. Heterogeneous expression of alpha-methylacyl-CoA racemase in prostatic cancer correlates with Gleason score. Histopathology. 2007;50(2):243–251. doi: 10.1111/j.1365-2559.2007.02572.x. [DOI] [PubMed] [Google Scholar]
- 12.Pourmand G, Ziaee AA, Abedi AR, Mehrsai A, Alavi HA, Ahmadi A, Saadati HR. Role of PTEN gene in progression of prostate cancer. Urol J. 2007;4(2):95–100. [PubMed] [Google Scholar]
- 13.Rasiah KK, Kench JG, Gardiner-Garden M, Biankin AV, Golovsky D, Brenner PC. Aberrant neuropeptide Y and macrophage inhibitory cytokine-1 expression are early events in prostate cancer development and are associated with poor prognosis. Cancer Epidemiol Biomarkers Prev. 2006;15(4):711–716. doi: 10.1158/1055-9965.EPI-05-0752. [DOI] [PubMed] [Google Scholar]
- 14.Reubi JC, Gugger M, Waser B, Schaer JC. Y1-mediated effect of NPY in cancer: breast carcinomas as targets. Cancer Res. 2001;61(11):4636–4641. [PubMed] [Google Scholar]
- 15.Ruscica M, Dozio E, Boghossian S, Bovo G, Martos Riaño V, Motta M, Magni P. Activation of the Y1 receptor by neuropeptide Y regulates the growth of prostate cancer cells. Endocrinology. 2006;147(3):1466–1473. doi: 10.1210/en.2005-0925. [DOI] [PubMed] [Google Scholar]
- 16.Ruscica M, Dozio E, Motta M, Magni P. Modulatory actions of neuropeptide Y on prostate cancer growth: role of MAP kinase/ERK 1/2 activation. Adv Exp Med Biol. 2007;604(1):96–100. doi: 10.1007/978-0-387-69116-9_7. [DOI] [PubMed] [Google Scholar]
- 17.Shaheduzzaman S, Gao CL, Wang Z, et al. Prostate Epithelial Cell Specific Gene Expression Signatures Define Prostate Cancer Patients at High or Moderate Risk of Disease Progression; Washington DC, USA. 2006. Presented at 97th AACR Annual Meeting. [Google Scholar]
- 18.Silva AP, Cavadas C, Grouzmann E. Neuropeptide Y and its receptors as potential therapeutic drug targets. Clinica Chimica Acta. 2002;326(1-2):3–25. doi: 10.1016/S0009-8981(02)00301-7. [DOI] [PubMed] [Google Scholar]
- 19.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D'Amico AV, Ritchie JP, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1(2):203–209. doi: 10.1016/S1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 20.Yoshimoto M, Cunha IW, Coudry RA, Fonseca FP, Torres CH, Soares FA, Squire JA. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97(5):678–685. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]