Abstract
Heat shock locus V (HslV; also called ClpQ) is the proteolytic core of the ATP-dependent protease HslVU in Escherichia coli. It has sequence similarity with the β-type subunits of the eukaryotic and archaebacterial proteasomes. Unlike these particles, which display 72-point symmetry, it is a dimer of hexamers with 62-point symmetry. The crystal structure of HslV at 3.8-Å resolution, determined by isomorphous replacement and symmetry averaging, shows that in spite of the different symmetry of the particle, the fold and the contacts between subunits are conserved. A tripeptide aldehyde inhibitor, acetyl-Leu-Leu-norleucinal, binds to the N-terminal threonine residue of HslV, probably as a hemiacetal, relating HslV also functionally to the proteasomes of archaea and eukaryotes.
Heat shock leads to increased levels of misfolded proteins. In response, Escherichia coli expresses ATP-dependent chaperones and complexes that appear to combine chaperone-like activity with proteolysis. The discovery of the operon hslVU (heat shock locus VU) (1) in E. coli under transcriptional control of a σ32-dependent heat shock promoter added protease HslVU to this class that already contained the cytosolic proteases Lon (La) and Clp (Ti) (2). Protease HslVU (3) is a hybrid of Clp and the proteasome. The subunits of HslV and the β-subunits of the 20S proteasome of the archaeon Thermoplasma acidophilum are 18% identical in amino acid sequence, and, after processing, they both have a threonine at the N terminus (Fig. 1). The regulatory caps HslU that contain the Walker consensus ATP-binding motif are highly homologous to ClpX from E. coli (4). They seem to play the role of both the α-subunits of the 20S particle and the 19S caps. Unlike the situation in archaea, where the presence of the α-subunits is essential for assembly of the β-subunits (5), HslV assembles in the absence of HslU, and unlike the β-subunits in T. acidophilum, it forms a dimer of hexamers rather than heptamers as first shown by electron microscopy (6, 7). In vitro, it has been shown that HslU stimulates proteolytic activity of HslV against casein (6) and small chromogenic peptides in the presence of ATP (8). The range of small peptide substrates for HslVU appears to be rather limited. Z-Gly-Gly-Leu-AMC (7-amido-4 methylcoumarin) and some, but not all, hydrophobic substrates are degraded (3). Weak peptidase activity has been reported for HslV in the absence of HslU (9). To our knowledge, tryptic and peptidyl-glutamyl-like activities, both present in eukaryotic proteasomes, have not been found in HslV.
Figure 1.
Amino acid sequence alignment of the β-subunits of Thermoplasma acidophilum and HslV. Secondary structures are marked as strands (arrows) and helices (cylinders).
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification.
Oligonucleotides 5′-CGAGGGCCCATATGACAACTATAGTAAGCGTACGCCG-3′ and 5′-GCGTCGACTTATTAGTGGTGGTGATGGTGGTGGAATTCCGCTTTGTAGCCTAATTCTTCGATGGTGTG-3′, derived from the published sequence (1), were used to amplify hslV by PCR from E. coli XL-1 Blue. The amplified fragment coding for HslV-EFHHHHHH was cloned into pET12b (Novagen) using restriction sites NdeI and SalI and was checked for correctness by double-stranded sequencing. For expression, the plasmid was transformed into E. coli BL21(DE3) cells.
In a typical preparation, a 2l culture of these cells in Luria–Bertani broth containing 100 μg/ml ampicillin was grown at 37°C until it reached saturation. Expression was induced by adding isopropyl β-d-thiogalactoside to a final concentration of 1 mM. Cells were harvested 4 hr later, the pellet was resuspended in buffer A (50 mM Tris⋅HCl/300 mM NaCl, pH 7.7) and frozen at −20°C. All subsequent steps were done at 4°C, and all buffers were titrated to pH 7.7 unless stated otherwise. After thawing, 1 mg DNase I and 10 mg hen egg white lysozyme were added, cells were sonified, and cellular debris was removed by centrifugation (100,000 × g, 1 hr). The supernatant was applied to a chelating Sepharose (Pharmacia) column (4.0 cm × 2.5 cm) loaded with loading buffer (5 mg/ml ZnCl2 in 10 mM Tris⋅HCl, pH 6.0) and equilibrated with buffer A. The column was washed extensively, first with buffer A, then with 150 mM imidazole in buffer A. HslV-EFHHHHHH was eluted from the column with 500 mM imidazole in buffer A. As it was prone to aggregation in the absence of EDTA, EDTA was added to a final concentration of 5 mM before concentrating the protein by ultrafiltration with a YM10 membrane (Amicon). The concentrate was applied to a gel filtration column (120 cm × 2.5 cm) packed with Sephacryl S-300 (Pharmacia) and equilibrated with buffer B (20 mM Tris⋅HCl/300 mM NaCl/1 mM EDTA/1 mM NaN3). HslV-EFHHHHHH migrated as a complex.
Crystallization and Data Collection.
Crystals were grown at room temperature within a week to an approximate size of 0.5 mm × 0.3 mm × 0.2 mm by sitting drop vapor diffusion against a reservoir containing 100 mM Hepes/NaOH at pH 7.5, 200 mM sodium acetate, 0.02% NaN3, and between 9% and 14% ethanol. Drops initially contained 2.5 μl protein in buffer B at 10 mg/ml and 2.5 μl reservoir solution. The spacegroup of the crystals was P42212 and cell constants were a = 108.4 Å, b = 108.4 Å, and c = 103.2 Å. Derivatives with useful phasing power were obtained by soaking crystals overnight at room temperature with 2 mg/ml europium chloride and 1 mg/ml sodium ethylmercurithiosalicylate (thiomersal) in reservoir solution. To study inhibitor binding, a crystal was soaked for 18 hr with a saturated solution of acetyl-Leu-Leu-norleucinal (calpain inhibitor I). Data were collected in house with a Mar research imaging plate detector mounted on a Rigaku rotating anode x-ray generator, integrated using mosflm (10), and scaled and merged using programs of the CCP4 suite (11).
Structure Solution and Refinement.
Self-rotation using glrf (12) showed that the 6-fold local axes of the particles pointed in [110] and [11̄0] directions. Crystal packing arguments suggested, and heavy atom positions, as determined and refined using protein (13), confirmed, that the local 6-fold axis and one local 2-fold axis of HslV coincided with 2-fold crystallographic axes, so that three subunits of hslV related by local 3-fold symmetry filled one asymmetric unit. Combined phase information from thiomersal and europium chloride derivatives, after 3-fold averaging and solvent flattening using dm (11) yielded an excellent map that was easily interpretable in terms of the molecular architecture (Fig. 2). frodo (14) was used to build the initial model. Coordinates were refined using x-plor (15), with parameters as described by Engh and Huber (16). Model and derivative phases were merged to compute a new density that was averaged using main (17) applying symmetry operators derived from the refined coordinates. The model was then rebuilt manually and the procedure repeated, including in later stages noncrystallographic symmetry (NCS) restrained refinement of individual isotropic B factors (Table 1).
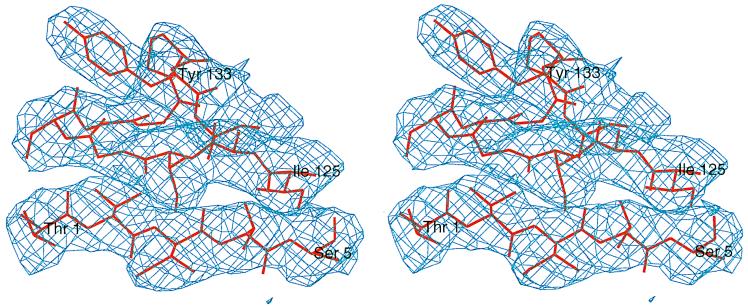
Figure 2.
Electron density calculated with phases from isomorphous replacement after 3-fold averaging, contoured at 1.0 σ around Thr-1 and overlaid with the final model.
Table 1.
Summary of crystallographic data
| Derivative
|
||||
|---|---|---|---|---|
| Native | Ac-LLnL | Thiomersal | EuCl3 | |
| Data collection and isomorphous replacement statistics | ||||
| Resolution limit, Å | 3.8 | 3.9 | 4.5 | 4.0 |
| Measured reflections | 36,796 | 18,945 | 24,833 | 23,751 |
| Unique reflections | 6,404 | 5,813 | 3,956 | 5,180 |
| Overall completeness, % | 100 | 96 | 98 | 95 |
| Rmerge* | 7.3 | 10.1 | 7.8 | 7.2 |
| Weighted Rcullis† | — | — | 0.44 | 0.64 |
| Phasing power‡ | — | — | 2.9 | 1.5 |
| Refinement statistics | ||||
| Rcryst,§ % | 25.6 | 28.3 | ||
| Rfree,¶ % | 31.5 | 28.5 | ||
| Rback,‖ % | 18.6 | — | ||
| rms bond length, Å | 0.013 | — | ||
| rms bond angles, ° | 1.8 | — | ||
| rms NCS protein,** Å | 0.06 | — | ||
| Average B, Å2 | 51 | |||
| rms bonded B, Å2 | 5.9 | |||
| rms NCS B, Å2 | 3.5 | |||
Space group P42212; cell constants 108.4 Å × 108.4 Å × 103.2 Å × 90.0° × 90.0° × 90.0°. Ac-LLnL, acetyl-Leu-Leu-norleucinal.
Rmerge: ∑h∑i|I(hi) − 〈I(h)〉|/∑h∑iI(hi), where I(hi) is the intensity of reflection h as determined by the ith measurement and 〈I(h)〉 is the average value.
Weighted Rcullis: [∑hkl∥FD,calc| − |FD,obs∥/(σN2 + σD2)]/[∑hkl∥FD,obs| − |FN,obs∥/(σN2 + σD2)], where FD,calc = |FN,obs|exp(iϕcalc) + FH,calc. Indices N, D, and H denote native, derivative, and heavy atom data and σ denotes a standard deviation.
Phasing power: (∑hkl|FH,calc|2)1/2/[∑hkl (|FD,calc| − |FD,obs|)2]1/2, where FD,calc = |FN,obs|exp(i ϕcalc) + FH,calc. Indices N, D, and H denote native, derivative, and heavy atom data and σ denotes a standard deviation.
Rcryst: ∑hkl∥Fobs| − |Fcalc∥/∑hkl|Fobs|, where |Fobs| is the observed and |Fcalc| the calculated structure factor amplitude of reflection hkl.
Rfree: calculated as Rcryst for a test set comprising 2% of the reflections of the whole data set that were excluded in the refinement.
Rback: ∑hkl∥Fobs| − |Fcalc, back∥/∑hkl|Fobs|, where |Fobs| is the observed and |Fcalc, back| is the calculated structure factor amplitude after backtransformation of the averaged electron density.
The rms deviation between positions related by noncrystallographic symmetry.
RESULTS
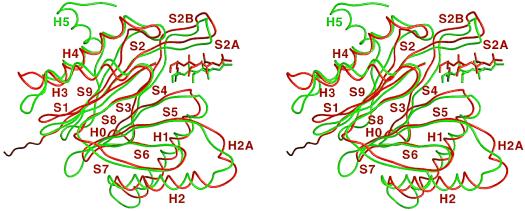
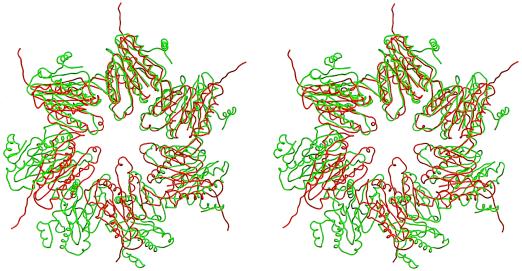
The crystal structure of HslV shows that the dodecamer forms a proteolytic chamber (Figs. 3 and 4). The subunit fold is the same as in T. acidophilum (18) (Figs. 5 and 6). A look along the 6-fold axis shows that secondary structure elements in each subunit cluster in four planes roughly perpendicular to the 6-fold axis. Helices H1 and H2 protrude furthest toward the observer. β-strands S3, S4, S5, S6, and S7 and β-strands S9, S2, S1, and S8 form the second and third layer, respectively. β-strands are antiparallel within one layer and tilted with respect to the β-strands in the other layer. The last layer, at the interface of the two rings, is made of helices H3 and H4. The only major differences in structure between the T. acidophilum β-subunits and HslV are due to deletions and insertions in the amino acid sequence of HslV.
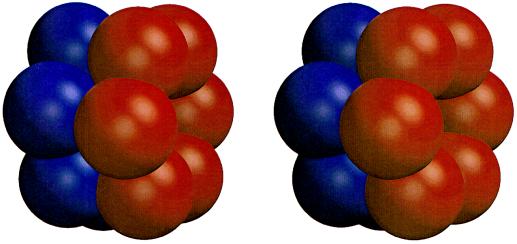
Figure 3.
Sphere model of HslV showing the dodecamer of two stacked hexameric rings in a staggered arrangement.
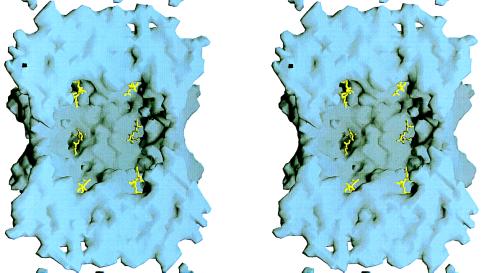
Figure 4.
Space-filling representation of HslV drawn with grasp (20). HslV is cut open along the cylinder axis to show the hydrolytic chamber and the bound calpain inhibitor molecules.
Figure 5.
Overlay of HslV (red) with the T. acidophilum β-subunit (green) with bound calpain inhibitors. The secondary structural elements are labeled.
Figure 6.
Overlay of one hexameric ring of HslV (red) with one heptameric ring of T. acidophilum β-subunits (green).
A deletion of six residues in the turn between helices H1 and H2 around residue 70 tightens this turn. Similarly, the transition between strands S5 and S6 is short-cut in HslV by a deletion of three residues. Most prominently, HslV completely lacks strand S10 and helix H5. Helix H5 is also absent in two β-type subunits of the yeast proteasome (19) and much shorter in a third one. The extension of strand S9 in HslV seen in Figs. 5, 6 and 7 is a cloning artifact. In the crystal lattice, the extra eight residues of the tag form a parallel β-sheet with neighboring strands.
Figure 7.
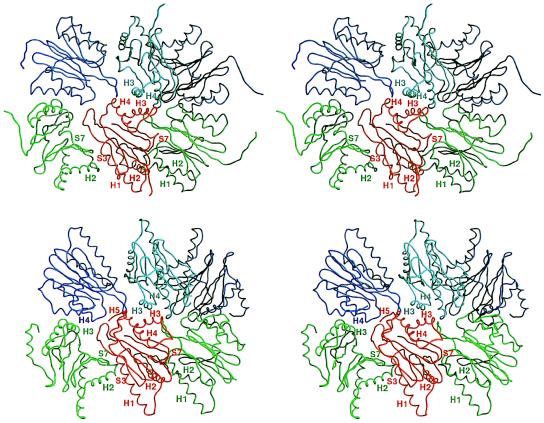
Side view of a substructure of the stacked hexameric rings of HslV (Upper) and the heptameric rings of T. acidophilum (Lower), respectively. The red subunits are in identical orientations. The topology of the intersubunit contacts is conserved.
The HslV sequence contains one short two-residue insertion, helix H2A, that lines the entrance to the particle. Its 19-Å diameter, as measured between Cα carbons, is slightly larger than the 17-Å diameter of the α-ring and significantly smaller than the 27-Å diameter of the β-ring opening of the archaebacterial proteasome. The loops that line the entrance are stabilized by cis-intersubunit contacts (Thr-89, Asp-90, and Met-93). A second site of cis-contacts is between the N terminus of S7 (Gly-117 and Asp-118) and the N terminus of helix H1 (Thr-49 and Ala-50), a third one between the C-termini of strands S2B (Lys-28, Gly-29, and Asn-30) and S7 (Val-120, Gln-121, and Glu-123). Trans-contacts are H3-H3 and H3-H4 contacts and contacts around the dyads (Asp-24, Thr-25, and Val-26 with Asp-164, Ile-165, and Ile-167). These contacts are very reminiscent of the ones in T. acidophilum (Fig. 7), and again there are narrow side windows in the wall of the particle at the site of binding of calpain-inhibitor I, acetyl-Leu-Leu-norleucinal.
The bound tripeptide aldehyde (Figs. 2 and 5) extends strand S1 and runs antiparallel to strand S2A to which it is hydrogen-bonded in an arrangement similar to the one seen in the proteasome complexes of T. acidophilum and yeast. The electron density is compatible with the formation of a hemiacetal with Thr-1 as had been shown in the yeast proteasome at higher resolution. In HslV, the S1 pocket, which accommodates the norleucine side chain at P1 has an apolar character by Phe 45, Val-31, and Thr-49. It is spacious to accept large side chains. P2 makes no contacts with the central chamber, P3 binds in a characteristic pocket well suited for basic residues that is formed by Met-27 and by Thr-114, Asn-116, and Asp-118 of a neighboring subunit in the same hexamer.
DISCUSSION
HslV differs prominently from archaebacterial and eukaryotic 20S proteasomes in subunit assembly. Unlike these proteasomes, that are four-ring structures displaying 7-fold or pseudo-7-fold symmetries, HslV is a dimer of two hexameric rings stacked head to head.
We attribute assembly into a dimer of hexamers and not into a dimer of heptamers to several extrinsic and intrinsic factors. In the T. acidophilum proteasome, the β-subunits assemble only in the presence of the α-subunits and the 7-fold symmetry could be enforced by the latter. It is intriguing that the very residues 68 to 73 that are missing in HslV make the most prominent contacts between the α- and the β-subunits in T. acidophilum. In proteasomes of T. acidophilum, the peripheral depressions of the heptameric β-rings are filled by the C-terminal helices in an antiparallel arrangement. Their absence in HslV might allow a tighter packing and a smaller hexameric ring size (Fig. 6). Other more sequence specific contacts will contribute to stabilize the different quaternary architectures.
The entrance to the proteolytic chamber of HslV is slightly wider than the one to the α-chambers and more constricted than the one to the β-chambers of the T. acidophilum proteasome. Helix H2A, that lines the channel into HslV, is absent in the β-subunits of the T. acidophilum proteasome and corresponds to an insertion also seen in the α-subunits of the T. acidophilum proteasome. The extra helix is probably necessary to protect normal proteins from entry into the proteolytic core, especially because HslV lacks the antichamber that is present in the archaebacterial proteasome.
Binding of calpain inhibitor I in the proteolytic chamber of HslV occurs at Thr-1, in line with results obtained for the T. acidophilum proteasome, that identify Thr-1 Oγ as the nucleophile. Thr-1 Oγ is in close proximity to Lys-33 Nζ, suggesting a role for this conserved residue in the activation of the nucleophile. The effect is probably indirect and mediated electrostatically, if, as suggested for the T. acidophilum (18) and yeast (19) proteasomes, the proton acceptor for the Thr-1 Oγ is Thr-1 N. Thr-1 N appears to be hydrogen-bonded to Ser-129 Oγ. The serine, that is conserved from eubacteria to eukaryotes, may be necessary to control protonation of Thr-1 N, or it may be there to stabilize the orientation of the Thr-1 toward the substrate binding pocket. Thr-2, in contrast, points away from the inhibitor. Mutational studies (9) show that mutation Thr-1 → Ala leads to a reduction of proteolytic activity, whereas mutation Thr-2 → Ala abolishes activity, suggesting that the latter substitution results in drastic structural alterations.
To conclude, our results support a Thr-1-dependent catalytic mechanism for HslV. They show that HslV and the T. acidophilum proteasome share a similar subunit structure and similar intersubunit contacts. They confirm that HslV can be regarded as the eubacterial ancestor of archaebacterial and eukaryotic 20S proteasomes and that protease HslVU is an attractive model system for studies of ATP-dependent proteolysis in proteasome-related particles.
Acknowledgments
We thank J. Lowe and W. Reuter for discussions, M. Schneider for help with the figures, and the Deutsche Forschungsgemeinschaft for finanical support.
ABBREVIATIONS
- HslV
heat shock locus V
- HslU
heat shock locus U.
Footnotes
Data deposition: The atomic coordinates and structure factors in this paper have been deposited in the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (reference number 1NED).
References
- 1.Chuang S-E, Burland V, Plunkett G, III, Daniels D L, Blattner F R. Gene. 1993;134:1–6. doi: 10.1016/0378-1119(93)90167-2. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg A L. Eur J Biochem. 1992;203:9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- 3.Rohrwild M, Coux O, Huang H-C, Moerschell R P, Yoo S J, Seol J H, Chung C H, Goldberg A L. Proc Natl Acad Sci USA. 1996;93:5808–5813. doi: 10.1073/pnas.93.12.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schirmer E C, Glover J R, Singer M A, Lindquist S. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 5.Zwickl P, Kleinz J, Baumeister W. Nat Struct Biol. 1994;1:765–770. doi: 10.1038/nsb1194-765. [DOI] [PubMed] [Google Scholar]
- 6.Kessel M, Wu W-F, Gottesman S, Kocsis E, Steven A C, Maurizi M R. FEBS Lett. 1996;398:274–278. doi: 10.1016/s0014-5793(96)01261-6. [DOI] [PubMed] [Google Scholar]
- 7.Rohrwild M, Pfeifer G, Santarius U, Müller S A, Huang H-C, Engel A, Baumeister W, Goldberg A L. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- 8.Yoo S J, Seol J H, Shin D H, Rohrwild M, Kang M-S, Tanaka K, Goldberg A L, Chung C H. J Biol Chem. 1996;271:14035–14040. doi: 10.1074/jbc.271.24.14035. [DOI] [PubMed] [Google Scholar]
- 9.Missiakis D, Schwager F, Betton J-M, Georgopoulos C, Raina S. EMBO J. 1996;15:6899–6906. [PMC free article] [PubMed] [Google Scholar]
- 10.Leslie A G W. In: Crystallographic Computing 5. Moras D, Podjarny A D, Thiery J C, editors. Oxford: Oxford Univ. Press; 1991. pp. 50–61. [Google Scholar]
- 11.Collaborative Computational Project, No. 4. Acta Cryst. 1994;D50:760–763. [Google Scholar]
- 12.Tong L, Rossmann M G. Acta Cryst. 1990;A46:783–792. doi: 10.1107/s0108767390005530. [DOI] [PubMed] [Google Scholar]
- 13.Steigemann W. In: Crystallographic Computing 5. Moras D, Podjarny A D, Thiery J C, editors. Oxford: Oxford Univ. Press; 1991. pp. 115–125. [Google Scholar]
- 14.Jones T A. J Appl Cryst. 1978;11:268–272. [Google Scholar]
- 15.Bruenger A T. x-plor Version 3.1: A System for Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 16.Engh R A, Huber R. Acta Cryst. 1991;A47:392–400. [Google Scholar]
- 17.Turk D. Ph.D. thesis. Munich: Technische Universität München; 1992. [Google Scholar]
- 18.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 19.Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik H D, Huber R. Nature (London) 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 20.Nicholls A. grasp: Graphical Representation and Analysis of Surface Properties. New York: Columbia Univ.; 1992. [Google Scholar]