Abstract
The semi-dominant Br mutation affects presphenoid growth, producing the facial retrognathism and globular neurocranial vault that characterize heterozygotes. We analysed the impact of this mutation on skull shape, comparing heterozygotes to wildtype mice, to determine if the effects are skull-wide or confined to the sphenoid region targeted by the mutation. In addition, we examined patterns of variability of shape for the skull as a whole and for three regions (basicranium, face and neurocranium). We found that the Br mice differed significantly from wildtype mice in skull shape in all three regions as well as in the shape of the skull as a whole. However, the significant increases in variance and fluctuating asymmetry were found only in the basicranium of mutant mice. These results suggest that the mutation has a significant effect on the underlying developmental architecture of the skull, which produces an increase in phenotypic variability that is localized to the anatomical region in which the mean phenotype is most dramatically affected. These results suggest that the same developmental mechanisms that produce the change in phenotypic mean also produce the change in variance.
Keywords: evolution, fluctuating asymmetry, morphometrics, mutation, variability
Introduction
The effects of mutations are commonly conceived as shifts in the phenotypic mean. Less widely recognized is the fact that mutations can also influence variability, or the propensity to vary (Wagner & Altenberg, 1996, p. 969). This realization stems originally from the observations made by early Drosophila geneticists that mutant phenotypes are often more variable than the wildtype (Waddington, 1942; Mather, 1953; Rendel, 1967). It is not known how commonly such effects occur, as phenotypic variances are rarely reported in the developmental biology literature. Furthermore, the developmental–genetic basis for the modulation of variance in developmental systems is very poorly understood.
Hallgrímsson et al. (2002) distinguished three components of variability: canalization, developmental stability and morphological integration. In this paper we address only the first two components. Canalization, first described by Waddington (1942, 1957), refers to the buffering of developmental processes against environmental and mutational perturbations. Schmaulhausen independently developed this same concept under the term ‘autonomization’ (Schmaulhausen 1949, published in 1938 in Russian). Developmental stability is the ability of developmental processes to buffer random developmental noise, such as thermodynamic fluctuations, that arise within the developmental processes themselves (Waddington, 1957; Klingenberg et al. 2003). Canalization and developmental stability are similar in that both suppress phenotypic variation; they differ in that canalization suppresses variation among individuals who vary in genotypes and environments whereas developmental stability suppresses the variation of a single genotype within a single environment. These components are important because the magnitude and structure of phenotypic variation are modulated through them.
Variability is difficult to measure because it refers to the potential for variation, not the variance observed. However, it, and the canalizing processes that suppress it, can be measured by comparing observed variation so long as both genetic variance and environmental effects are controlled. Developmental stability is usually measured by the minor, random, differences between sides in bilaterally symmetric traits, referred to as fluctuating asymmetry (FA) (Van Valen, 1962). Using FA to measure developmental stability is based on the premise that bilaterally symmetric traits develop under the same genetic and environmental conditions, and therefore deviations from perfect symmetry are caused by developmental disruptions arising within an individual. These measures of components of variability tell us little or nothing unless compared with some standard. Often, one population is subjected to an environmental or mutational stress and is compared with a control population: assuming constant genetic variation, with a decrease in variation indicating a greater degree of canalization (Rutherford, 2000). Similarly, the level of FA is compared between populations, with lower measures of FA signifying a greater degree of developmental stability.
In this study we examined the effects of the Brachyrrhine (Br) mutation on skull shape and components of variability. This mutation, an automosmal, semi-dominant lethal, causes obvious craniofacial abnormalities in both homo- and heterozygotes (Lozanoff, 1993). Heterozygotes are characterized by midfacial retrognathia (Fig. 1); homozygotes also exhibit frontonasal dysplasia with midfacial clefting (Lozanoff et al. 1994; Ma & Lozanoff, 1996; McBratney et al. 2003). Mice homozygous for the mutation die shortly after birth, presumably due to the severity of the cleft and their inability to suckle (McBratney et al. 2003). The mutation appears to target the presphenoid (Lozanoff et al. 1994; Ma & Lozanoff, 1996); in heterozygotes the presphenoid is hypoplastic whereas it is completely absent in homozygous mice (McBratney et al. 2003). The Br mutation decreases the rate of chondrocyte proliferation that is localized to the sphenoethmoidal region of the cranial base (Ma & Lozanoff, 1999, 2002). The Br mutation offers an appealing model to test the effects of a mutation on components of variability not only because it has consistent, obvious effects but also because it targets a specific region.
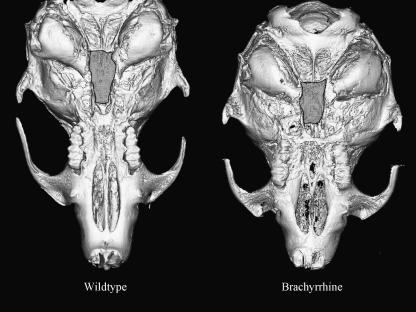
Fig. 1.
Basicranial view of wildtype and Br mouse microtomograph images. The highlighted regions on both skulls illustrate the shortened sphenoid in Br mice as compared with the wildtype.
The objective of this study was to determine whether the Br mutation affects canalization, and developmental stability, and whether these effects are localized to the area targeted by the mutation or distributed throughout the entire skull. We predicted (1) that Br mice would differ significantly from wildtype mice in average shape throughout the cranium; and (2) that Br mice would have lower levels of both canalization and developmental stability than the wildtype mice. The first prediction was based on the central role that the basicranium plays in skull growth (Kasai et al. 1995; Lieberman et al. 2000a, b; Sperber, 2001). We predicted that variation in presphenoid size would therefore be transmitted as shape variation throughout the skull. The second prediction was based on the hypothesis that developmental anomalies disrupt canalization, and would also disrupt developmental stability when these components of variability share a common developmental basis.
We discuss these results in the light of other similar studies and suggest specific developmental mechanisms that may be responsible for the observed phenotypic effects of the mutation.
Materials and methods
Composition of the sample
The Br mutation initially arose through experiments designed to determine the effects of irradiation on chromosome structure (Searle, 1966). The mutation is carried on a C3H/He × 101/H (3H1) background and is autosomal semi-dominant (Lozanoff, 1993). Homozygous Br mice (Br/Br) die postnatally presumably due to the severity of the midline cleft that characterizes these mice (Ma & Lozanoff, 1993; McBratney et al. 2003). Heterozygous (Br/+) mice were used for comparison with wildtype 3H1 (+/+) mice in this study.
The sample consisted of the macerated skulls of 52 Br/+ mice and 40 3H1 +/+ mice. Breeding and husbandry protocols followed McBratney et al. (2003). The sexes of the individuals used are largely unknown, so we could not control for sexual dimorphism; however, sexual dimorphism is unlikely given that Hallgrímsson et al. (2004a, b) found no sexual dimorphism for the first two principal components of shape for three different strains of mice. Therefore, we assumed that sex was not a major confounding factor in this study.
The mice used in this study vary in age, which could make samples heterogeneous in shape due to allometry and thus inflate the within-sample variance. Previous work (Zelditch et al. 2003) has shown that after 30 days, allometric trajectories stabilize in the growth of the mouse skull, and all individuals included in the present study are known to be of more than 30 days of age. Given the linear relationship between age, size and shape after 30 days, variation in age within samples can be statistically controlled using size as a proxy for age (see below).
Data acquisition
Skulls were scanned using a Skyscan 1072 100-kV microtomograph using a protocol optimized for macerated adult mouse skulls (no filter, 0.9 rotation step, three-frame averaging, 5.9-ms exposure time, 19.43-mm resolution). Before each scanning session, flat field corrections were performed and post-alignment corrections and global threshold values were manually verified before two-dimensional (2D) reconstruction. Noise was then minimized for the 2D reconstructed slices by running a three-kernel median filter using a custom plug-in written for ImageJ.
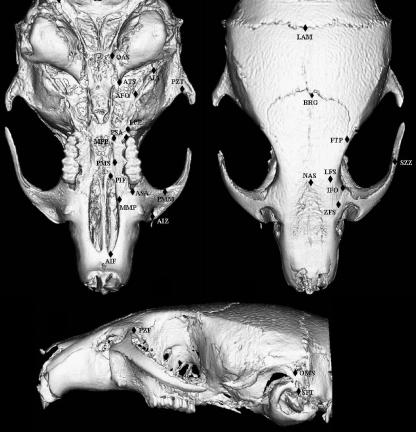
Three-dimensional reconstructions were generated from the filtered image stacks using Analyze 3D 5.0. Using a dual monitor setup, 24 bilateral and three midline 3D landmarks were digitized directly from the 3D reconstructions. Figure 2 shows the landmarks used in this study and Table 1 provides anatomical descriptions for each landmark.
Fig. 2.
Landmarks collected from mouse skulls from lateral, superior and basicranial views. Landmarks are only shown for one side of the skull, but were digitized bilaterally.
Table 1.
List of landmarks and their definitions used in this study (see Fig. 1)
| Landmark | Definition |
|---|---|
| AIF | Anterior margin of incisive foramen |
| AIZ | Anterior inferior zygomatic |
| PMM | Point of greatest curvature on the posterior margin of the malar process |
| ASA | Anterior superior alveoli |
| PIF | Posterior incisive foramen |
| PMS | Point along palatine–maxillary suture |
| PSA | Posterior superior alveoli |
| LPP | Lateral palatal–pterygoid junction |
| AFO | Anterior foramen ovale |
| AIA | Anterior inferior auditory bulla |
| PZT | Point of greatest curvature along posterior edge of zygomatic process of temporal bone |
| OAS | Occipital–auditory–sphenoid junction |
| ATS | Auditory–temporal–sphenoid junction |
| MPP | Medial palatal–pterygoid junction |
| MMP | Medial maxilla–premaxilla junction |
| ZFS | Anteriormost point along lateral zygomatic–frontal suture |
| NAS | Nasalis |
| LFS | Lateral point along frontal suture |
| IFO | Intersection of frontal suture with orbital rim |
| SZZ | Superior margin of suture of temporal and zygomatic processes of zygomatic arch |
| FTP | Frontal–temporal–parietal junction |
| BRG | Bregma |
| LAM | Lambda |
| SPT | Superior posterior extremity of tympanic ring |
| PZF | Posterior zygomatic–frontal junction |
| OMS | Point along occipitomastoid suture |
Each individual was digitized three times on separate days by the same observer in order to assess measurement error. Scatterplots of superimposed Procrustes data were used to detect gross outliers visually resulting from mislabelling of landmarks, side reversal or misplacement due to pseudoforamina. Individuals with such gross errors were redigitized.
Data analysis
Data used for all analyses were reflected and superimposed using the procrustes generalized least squares method; however, the programs used to obtain reflected and superimposed data to calculate object FA are different from those used for all other analyses. The resulting data from these two methods were the same. We first describe how we processed our raw data for the majority of the analyses and leave the description of how we calculated object FA for the section on FA analysis.
Raw data were reflected by multiplying the x coordinate by -1, and then the paired landmarks were relabelled to match their left or right counterpart, creating a mirror image of the dataset. These raw reflected data were then divided into landmark configurations that outlined the entire skull as well as specific regions of the skull, including the basicranium, neurocranium and facial skeleton (Table 2, Fig. 3). Procrustes superimposed data were obtained from raw data, including both the original and the reflected datasets using the program Morpheus (Slice, 1994–1999). Procrustes data were obtained for both wildtype and mutant groups, and were collected separately for each landmark configuration as subsets of Procrustes data are not independent of one another. Grubb's test for outliers was performed within groups to test for robustness of the data. Outliers for both measurement error and asymmetry that were significant using Bonferroni adjustment (P = 0.001) were removed from further analysis. It is important to remove these data, as outliers for measurement error due to entry or gross measurement errors could mask FA, and outliers for asymmetry due to specimen damage could artificially inflate measures of FA.
Table 2.
Landmarks that correspond with specific regions of the skull used in this study
| Basicranium | Face | Neurocranium |
|---|---|---|
| LPP | AIF | IFO |
| AIA | ASA | FTP |
| PZT | PIF | OMS |
| OAS | PSA | NAS |
| ATS | MMP | BRG |
| LFS | LAM | |
| SZZ |
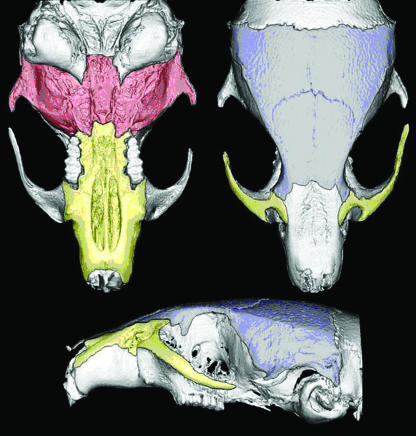
Fig. 3.
Visual description of the three subregions of the skull used in this analysis. The area outlined in blue depicts the neurocranium, the red area represents the basicranium and the yellow area outlines the facial region.
To remove the heterogeneity due to varying ages within the samples, we first determined that size could be used as a proxy for age by regressing centroid size on age. Centroid size and age were all significantly correlated (P < 0.01) with R2 ranging from 0.6 to 0.8 for individuals over 30 days old. We then regressed shape on size to remove the variation due to allometry; the residuals from that regression were added to the values for the expected shape at the mean size and calculations were done using the program 3DStand (Sheets, 2004b).
Mean shape
Differences in mean shape between Br and wildtype mice for all four regions of the skull were tested using Goodall's F-test, implemented by program Simple3D (Sheets, 2004a). Principal component analyses were performed for each cranial region to aid visualization of the differences in mean shape between groups. Deformations of wireframes were also used to visualize the differences in mean shape along the first principal component using Morphologika (O'Higgins & Jones, 1998).
Among-individual variance
Among-individual variances were calculated for both wildtype and mutant mouse groups for the entire skull as well as the three subregions of the cranium. We calculated variance of the hemi-skulls only, using the coordinates of right side and midline coordinates; hemi-skulls were then averaged across trials and reflection for each individual and coordinate. Overall variances for each group were calculated using the standard metric for variance in Procrustes-based studies:
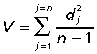 |
where dj is the Procrustes distance of individual j from the mean shape for its group and n is the sample size for the mouse group. The Procrustes distance is approximately the square root of the summed squared distances between homologous landmarks of two optimally superimposed forms, in this case between those of individual j and the mean of the sample. That same measure of variance is incorporated in Goodall's F-test (which is used to test for differences in mean shape), and also in the FA analysis. Levene's test was used to determine if differences between group variances were significant for each region of the skull.
Fluctuating asymmetry
Fluctuating asymmetry was calculated using the object FA method outlined by Klingenberg et al. (2002). Object FA measures the difference in whole landmark configurations with their mirror image (Klingenberg et al. 2002). This method is based on the two-way mixed model anova design recommended by Palmer & Strobeck (1986, 2003), but it includes median landmarks allowing the detection of asymmetry in the midline. In this model, reflection (side) is the fixed factor and individual and trial are random factors. Specifically, Palmer and Strobeck's FA10 index was used:
where MSsj is the mean square for sides–individuals interaction, MSm is the mean square for measurement error and M is the number of trials. This measure of FA was chosen as it accounts for measurement error. Directional asymmetry (DA) was calculated by an F-test, with F = MSs/MSsj, where MSs is the mean square for sides and MSsj is again the mean square for sides–individuals interaction. The signed asymmetry distributions were inspected for evidence of bimodality to check for potential anti-symmetry because it is not possible to differentiate fluctuating asymmetry from anti-symmetry given the between-sides variance measure (MSsj) (Palmer & Strobeck, 2003). These analyses were carried out separately within wildtype and Br mouse groups for each of the regions of the skull using the program Sage3D (Marquez, 2004). The program is designed to reflect raw coordinate data, and therefore in addition to calculating object FA, Sage3D was also used to reflect the data for this analysis only.
Object FA was calculated to ensure that there was significant FA for both mouse groups for all regions of the skull. In order to determine if the differences in FA between wildtype and Br mice were significant we used a multivariate calculation for FA. For this test, FA was calculated as the average of the squared deviation between the original and reflected data for each individual for each Procrustes coordinate. The sum of this deviation was then averaged across coordinates and divided by the number of landmarks. Levene's tests were performed on this averaged sum of the deviation between original and reflected data to determine if FA is significantly different between mutant and wildtype groups for each region of the skull.
Results
Mean shape
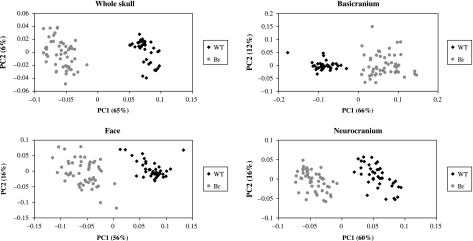
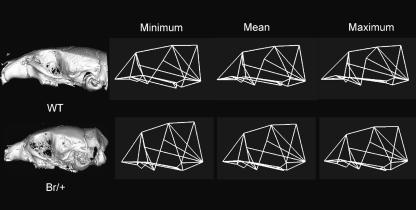
Significant differences in mean shape were found between mutant and wildtype groups for all regions of the skull; however, this distance is greatest for the landmark configuration outlining the basicranium (Table 3). Principal component analyses likewise illustrate that the mean shape for mutant and wildtype mice separate quite clearly along the first principal component (Fig. 4). Wireframe deformations along the first principal component show that the cranial base and facial skeleton of mutant mice are markedly shortened compared with wildtype mice and display a more globular neurocranium than is characteristic of wildtype mice (Fig. 5).
Table 3.
Results from Goodall's F-test between wildtype and Br mice using Simple 3D (Sheets, 2004a) for the entire skull, basicranium, face and neurocranial regions
| Region of skull | d.f.1 | d.f.2 | Distance | F score | P value |
|---|---|---|---|---|---|
| Whole skull | 71 | 6461 | 0.01791 | 749.23 | < 0.001 |
| Basicranium | 8 | 720 | 0.03026 | 132.03 | < 0.001 |
| Face | 14 | 1260 | 0.01730 | 143.74 | < 0.001 |
| Neurocranium | 11 | 990 | 0.01717 | 59.55 | < 0.001 |
Fig. 4.
Scatterplots of the first two principal components for the entire skull, basicranium, face and neurocranium showing differences in mean shape between wildtype and Br mouse groups.
Fig. 5.
Variation along the first principal component in lateral view for landmarks outlining the entire skull. The wireframe linking the points used in the analysis was arbitrarily constructed using Morphologika (O'Higgins & Jones, 1998) to aid visualization of both the magnitude and the direction of mean shape change in the skull.
Among-individual variance
The variance of mutant skull shape is apparently higher throughout the skull (Table 4); however, the difference between variances is statistically significant only for the configuration of the entire skull and the basicranial region (Table 4).
Table 4.
Overall among-individual variances for wildtype and mutant mouse groups using Procrustes distances. Levene's test for significant differences in among-individual variance was likewise calculated from Procrustes distances for all regions of the skull
| Region and group | Variance | F score | P value |
|---|---|---|---|
| Whole skull | 20.16 | < 0.001 | |
| WT | 0.00275 | ||
| Br | 0.00436 | ||
| Basicranium | 16.43 | < 0.001 | |
| WT | 0.00950 | ||
| Br | 0.01711 | ||
| Face | 2.58 | > 0.05 | |
| WT | 0.00486 | ||
| Br | 0.00667 | ||
| Neurocranium | 0.46 | > 0.05 | |
| WT | 0.01470 | ||
| Br | 0.00869 |
Fluctuating asymmetry
Object FA was significant for both groups for all regions of the skull (Table 5). Directional asymmetry was significant for both wildtype and Br groups for the whole skull, wildtype only for the basicranium and both groups for the neurocranium (Table 5). Anti-symmetry was not detected for either group in any of the cranial regions. The level of fluctuating asymmetry, as measured by FA10, is higher for mutant mice both for the skull as a whole and for the basicranium and face regions (Table 5). Wildtype mice apparently have a higher level of neurocranial FA but the difference is not statistically significant (Tables 5 and 6). Statistically significant differences between the two groups were found for the whole skull, basicranium and facial skeleton, with the basicranium showing the largest and most highly significant difference (Table 6).
Table 5.
Procrustes anova table for object asymmetry analysis for the entire skull as well as the three cranial subregions. Object FA was calculated as recommended by Klingenberg et al. (2002) and was calculated using the program Sage3D
| Group and region | d.f. | SS | MS | F | P value | FA10 |
|---|---|---|---|---|---|---|
| Whole skull | ||||||
| Wildtype 3H1 (+/+) | ||||||
| Individual | 2840 | 0.496730 | 0.00017490 | |||
| Reflection | 69 | 0.013946 | 0.00020212 | 4.50 | < 0.001 | |
| Individual × reflection | 2760 | 0.124110 | 0.00004497 | 10.95 | < 0.001 | 0.002943 |
| Measurement error | 11480 | 0.046756 | 0.00000407 | |||
| Br 3H1 (Br/+) | ||||||
| Individual | 3621 | 0.874430 | 0.00024149 | |||
| Reflection | 69 | 0.074450 | 0.00107900 | 20.83 | < 0.001 | |
| Individual × reflection | 3519 | 0.182430 | 0.00005184 | 8.82 | < 0.001 | 0.003122 |
| Measurement error | 14560 | 0.085558 | 0.00000588 | |||
| Basicranium | ||||||
| Wildtype 3H1 (+/+) | ||||||
| Individual | 320 | 1.470900 | 0.00459670 | |||
| Reflection | 8 | 0.000858 | 0.00010721 | 0.08 | < 0.05 | |
| Individual × reflection | 320 | 0.456900 | 0.00142780 | 4.75 | < 0.001 | 0.015468 |
| Measurement error | 1312 | 0.394530 | 0.00030071 | |||
| Br 3H1 (Br/+) | ||||||
| Individual | 408 | 3.560200 | 0.00872600 | |||
| Reflection | 8 | 0.018014 | 0.00251700 | 0.79 | > 0.05 | |
| Individual × reflection | 408 | 1.298000 | 0.00318140 | 12.54 | < 0.001 | 0.024929 |
| Measurement error | 1664 | 0.422250 | 0.00025375 | |||
| Face | ||||||
| Wildtype 3H1 (+/+) | ||||||
| Individual | 560 | 0.940460 | 0.00167940 | |||
| Reflection | 14 | 0.004518 | 0.00032271 | 0.98 | > 0.05 | |
| Individual × reflection | 560 | 0.184290 | 0.00032910 | 12.66 | < 0.001 | 0.008021 |
| Measurement error | 2296 | 0.059689 | 0.00002600 | |||
| Br 3H1 (Br/+) | ||||||
| Individual | 714 | 1.553800 | 0.00217620 | |||
| Reflection | 14 | 0.006831 | 0.00048792 | 1.01 | > 0.05 | |
| Individual × reflection | 714 | 0.344420 | 0.00048238 | 8.53 | < 0.001 | 0.009507 |
| Measurement error | 2912 | 0.164760 | 0.00005658 | |||
| Neurocranium | ||||||
| Wildtype 3H1 (+/+) | ||||||
| Individual | 440 | 0.616020 | 0.00140010 | |||
| Reflection | 9 | 0.155900 | 0.01732300 | 43.08 | < 0.001 | |
| Individual × reflection | 360 | 0.144730 | 0.00040204 | 15.05 | < 0.001 | 0.008926 |
| Measurement error | 1640 | 0.043825 | 0.00002672 | |||
| Br 3H1 (Br/+) | ||||||
| Individual | 560 | 0.741630 | 0.00132200 | |||
| Reflection | 9 | 0.203610 | 0.02262400 | 63.66 | < 0.001 | |
| Individual × reflection | 459 | 0.163120 | 0.00035538 | 10.30 | < 0.001 | 0.008253 |
| Measurement error | 2080 | 0.071735 | 0.00003449 |
Table 6.
Levene's test for significant differences in FA between wildtype and mutant mice for the whole skull, basicranium, face and neurocranium
| Region of skull | F score | P value |
|---|---|---|
| Whole skull | 6.97 | < 0.01 |
| Basicranium | 16.29 | < 0.001 |
| Face | 5.38 | < 0.05 |
| Neurocranium | 0.11 | > 0.05 |
Discussion
This study looked at the effects of a mutation known to disrupt the craniofacial phenotype in mice on mean shape, canalization and developmental stability. We found that mutant mouse skulls exhibited dramatically altered shape, as well as significantly higher levels of among-individual variance and fluctuating asymmetry compared with wildtype mice. The results indicate that the Br mutation exerts its effects on mean shape throughout the skull, whereas the effects on variability are localized to the area targeted by the mutation, namely the basicranium.
Most studies that have looked at mutational effects have focused on changes in mean shape. Of the studies that have looked at mutational effects on variability, far more studies have analysed their impact on canalization than on developmental stability. As with our study, it has frequently been shown using a variety of model organisms and characters that among-individual variance is increased in mutant populations as compared with wildtype mice (Dunn & Fraser, 1958, 1959; Rendel, 1959; Scharloo, 1991; Tanaka et al. 1997). Indeed, increased variance in the presence of mutation is so common that canalization is often defined by this response. That is, canalization is inferred if a trait displays greater variance in the mutant than in the wildtype background (Rutherford, 2000).
Comparatively few studies have examined the impact of mutations on developmental stability. Nonetheless, the general trend of these studies is that levels of FA are higher in mutants and therefore developmental stability is lowered by mutation (Sakai & Shimamoto, 1965; Clarke & McKenzie, 1987; McKenzie & Clarke, 1988; Clarke, 1997; Indrasamy et al. 2000). However, despite the apparent generality of the rule that mutations increase variance or FA, there are exceptions. These should be expected because canalization is causally heterogeneous (Scharloo, 1991), as is probably also true of developmental stability. The causal heterogeneity of canalization is indicated by the differential effects of mutations or environmental perturbations on different genetic backgrounds and different traits. For example, Gibson & van Helden (1997) found that the Ultrabithorax (Ubx) mutation of Drosophila did not increase haltere variation or FA, which is surprising because Ubx is a homeotic regulatory gene and was therefore expected to perturb development. Rutherford & Lindquist (1998) found that a mutation inhibiting the function of the Hsp90 protein increased the variance of a number of traits in Drosophila, but had no effect on FA (Rutherford, 2000). The differential phenotypic effects found between studies that have looked at mutations and variability suggest that developmental stability and canalization are emergent byproducts of regulatory complexity and redundancy in developmental systems (Siegal & Bergman, 2002). We therefore attempt to explain our results in terms of potential underlying developmental mechanisms.
Potential developmental causes for patterns of variance
The Br mutation has been mapped to the distal region of chromosome 17 (McBratney et al. 2003) and the phenotypic effects of this mutation have been widely studied using a variety of techniques. The Br mutation causes midfacial retrognathia in heterozygotes (Lozanoff et al. 1994; Ma & Lozanoff, 1996; McBratney et al. 2003) and appears to target the presphenoid (McBratney et al. 2003) by decreasing the number of proliferating chondrocytes in the sphenoethmoidal area (Ma & Lozanoff, 1999). The response of sphenoethmoidal chondrocytes to epidermal growth factor (EGF) is reduced by the Br mutation (Ma & Lozanoff, 2002). The Br mutation also affects the kidney; compared with the wildtype, Br mice have small kidneys, deficient cortical tissue and display multifocal cyst formation (Ma & Lozanoff, 1993; Lozanoff et al. 2001). The reduced chondrocyte proliferation found in the sphenoethmoidal area in Br mice (Ma & Lozanoff, 1999) probably accounts for their hypoplastic presphenoid (McBratney et al. 2003) and may also be related to the increased variance in the basicranial region found in this study. Chondrocyte proliferation in the nasal septal region of the basicranium was not affected by the mutation (Ma & Lozanoff, 1999). Perhaps this suggests that only cells in the sphenoid express the Br gene, or perhaps that the mutation only exerts its effects during a certain temporal window, one coinciding with chondrocyte proliferation within the sphenoid alone, leaving the rest of the basicranium unaffected.
We suggest that increased FA in mutant mice is expected given the effects of the Br mutation on chondrocyte proliferation. The effects of reduced chondrocyte proliferation would cause localized variance in cartilaginous structures or structures that are formed by cartilaginous precursors such as the basicranium. Differences in the number of chondrocytes in discrete regions of a character could lead to random left–right differences in variance and therefore increase FA. Trotta et al. (2005) also found that genes regulating cell proliferation increase FA precisely where they affect Drosophila wing phenotype. However, unlike with our results, FA was also increased in the regions neighbouring the target. The Drosophila wing is a compact structure and there are several common factors that influence the development of the entire wing, not just regions, increasing the integration among different areas within the wing. The mammalian skull, by contrast, is composed of several modules that develop relatively independently of each other. Therefore, as with the Br mutation, if chondrocyte proliferation is reduced in the sphenoid only, we might expect a more localized effect on FA.
The increased among-individual variance in the Br group suggests that the mutation might have incomplete penetrance, whereby the mutation will exert a stronger effect on some individuals than on others. Br individuals might also vary more than wildtype mice in their ability to buffer against perturbations. These mice are highly inbred and therefore the genetic variance in both groups is very low. However, the Br mutation may lead to expression of existing variation that is cryptic in the wildtype. Variation in the ability of individuals to compensate for the reduction in chondrocyte proliferation may be due to genetic variation in related pathways that is normally not translated into phenotypic variation.
Another mutation that targets basicranial cartilage also causes changes in mean cranial shape and increases variance much like the Br mutation. However, unlike that found for Br mice, the mutation does not cause an increase in FA, suggesting that the pattern of variance observed in Br mice is not simply an artefact of basicranial disruption. Rather, the contrast between patterns of variance of these two mutants is presumably due to the different processes affected by these two genes. The Brachymorph (Bm) mutant mouse has a remarkably similar phenotype to that of the Br mouse with a shortened midface, and neurocranial vaulting. The Bm mutation is autosomal recessive and affects the phosphoadenosine phosphosulfate synthetase 2 gene (Papps2) (Hallgrímsson et al. 2006). The Papps2 mutation causes under-sulfation of glycosaminoglycans (GAGs) in the cartilage matrix, resulting in fewer and smaller proteoglycan aggregates and therefore a reduction in cartilage growth (Orkin et al. 1976). Hallgrímsson et al. (2006) looked at the effects of the Papps2 mutation on mouse skull variability. As in this study, mean shape was found to be significantly altered in the Bm group compared with the wildtype and that among-individual variance was increased in the mutant group compared with the wildtype. However, a significant increase in FA was not detected in the Bm mice. The difference in effects on FA by these two mutations (Br and Papps2) probably arises from the developmental effects of each mutation. Unlike the Br mutation, the Papps2 mutation affects tissue properties, creating global changes in cartilaginous structures, not localized differences that may create unilateral changes in structure. Hallgrímsson et al. (2006) suggest that differences in among-individual variance between the mutant and wildtype groups results from a threshold effect or a non-linear relationship between the degree of sulfation and growth of cartilage. They hypothesize that variation in the degree of sulfation of GAGs in wildtypes is normally sufficient and so does not influence cartilage growth. However, the dramatic reduction of mean sulfation in Bm mutants results in a situation in which sulfation is insufficient and therefore in which variation in the degree of sulfation is translated into variation in the growth of cartilage. Degree of sulfation is thus an element of variation that is cryptic in the wildtype with respect to cranial shape, but is exposed as a source of cranial shape variation by the Bm mutation.
Above the cellular level, there are several other potential functional and developmental mechanisms that might be responsible for the patterns of variance observed in the Br mice. One interpretation is that the effects of the mutation on mean cranial shape away from the presphenoid are epigenetic. That is, the changes in regions away from the basicranium are due to secondary effects. Such effects could include the consequences of altered growth of the presphenoid for physically adjacent elements with cascading effects throughout the skull. Spatial packing of the brain could also play a role for neurocranial and facial shape due to the reduction in basicranial length. To the extent that the Br mutation shortens the basicranium, the need to accommodate the unaltered brain will produce changes in the shape of the cranial vault. Finally, altered mechanics of masticatory muscles may well result from these secondary shape changes, and mechanically mediated modelling and remodelling of bone may produce further epigenetic alterations in shape. By contrast, the increase in variability that is local to the basicranium could result from the disruption of particular localized developmental pathways. This would be consistent with the idea that variation in canalization and developmental stability emerge from the same developmental mechanisms that produce the primary changes in the mean phenotype. By contrast, the epigenetic alterations to shape elsewhere in the skull may not produce the kinds of developmental changes necessary to alter phenotypic variability. Alternatively, the impact of the mutation on variability may be a function of the magnitude of its impact on the phenotype. The changes in shape away from the basicranium may not be of sufficient magnitude to produce an alteration in variability.
Implications
The finding that variability is localized to regions directly affected by a mutation while average shape is affected by epigenetic factors associated with that mutation yields interesting implications. One is that patterns of variability could be used to determine the specific regions directly targeted by a mutation and could aid in piecing together the developmental pathways specifically affected by that mutation. That strategy could provide insights into the developmental cause of many congenital malformations or syndromes, which often affect several traits; patterns of variability might allow us to pinpoint the traits directly affected by the mutation, distinguishing them from those displaying epigenetic effects. Obviously, we would need to understand better the developmental basis for variation in phenotypic variability and determine whether the pattern observed here holds for other developmental contexts before confidently interpreting such patterns.
Building on previous work (Lozanoff, 1993; Ma & Lozanoff, 1993, 1996, 1999, 2002; Lozanoff et al. 1994; McBratney et al. 2003), we have shown that the Br mutation produces an integrated pattern of mean phenotypic change in the mouse skull but a localized change in phenotypic variability. This finding increases our understanding of how developmental mechanisms interplay to produce changes in the mean phenotype as well as the variance about that mean both for evolutionary change and for dysmorphology. Currently, work is being done to identify the Br gene. Once the Br gene is known it will be possible to relate Br expression with specific developmental events and link these events to phenotypic changes. As our understanding of the developmental mechanisms that underlie the phenotypic changes improves, continued merging of morphological studies with developmental and molecular biology will be the key to unravelling the complex relationship between development and evolution.
Acknowledgments
We are grateful to David Sheets and Eladio Marquez for their assistance with, and their impromptu programming of, statistical software used for this study. We would also like to thank two anonymous reviewers whose comments greatly improved the manuscript, and Dan Lieberman for several insightful and stimulating discussions on this subject. This work was supported by National Science and Engineering Research Council grant 238993-02, Canadian Foundation for Innovation grant #3923, and Alberta Innovation and Science grant #URSI-01-103-R1 (to B.H.), National Institute of Health #R01 DK064752-01 (to S.L.) as well as the William H. Davies Student Fellowship, and Medical Science Graduate Research Fellowship, University of Calgary Graduate Studies (to K.E.W.). This work was supported by Genome Alberta, in part through Genome Canada, a not-for-profit corporation which is leading a national strategy on genomics with $375 million in funding from the Government of Canada
References
- Clarke GM, McKenzie JA. Developmental stability of insecticide resistant phenotypes in blowfly; a result of canalizing selection. Nature. 1987;325:345–346. [Google Scholar]
- Clarke GM. The genetic and molecular basis of developmental stability: the Lucilla story. Trends Ecol Evol. 1997;12:89–91. doi: 10.1016/s0169-5347(97)01029-x. [DOI] [PubMed] [Google Scholar]
- Dunn RB, Fraser AS. Selection for an invariant character- ‘vibrissae number’-in the house mouse. Nature. 1958;181:1018–1019. doi: 10.1038/1811018a0. [DOI] [PubMed] [Google Scholar]
- Dunn RB, Fraser AS. Selection for an invariant character, vibrissae number in the house mouse. Aust J Biol Sci. 1959;12:506–523. doi: 10.1038/1811018a0. [DOI] [PubMed] [Google Scholar]
- Gibson G, van Helden S. Is function of the Drosophila homeotic gene Ultrabithorax canalized? Genetics. 1997;147:1155–1168. doi: 10.1093/genetics/147.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrímsson B, Willmore K, Dorval C, Cooper DM. Craniofacial variability and modularity in macaques and mice. J Exp Zool Part B Mol Dev Evol. 2004a;302:207–225. doi: 10.1002/jez.b.21002. [DOI] [PubMed] [Google Scholar]
- Hallgrímsson B, Willmore K, Hall BK. Canalization, developmental stability, and morphological integration in primate limbs. Am J Phys Anthropol Supplement. 2002;35:131–158. doi: 10.1002/ajpa.10182. Yearbook. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrimsson B, Dorval CJ, Zelditch ML, German RZ. Craniofacial variability and morphological integration in mice susceptible to cleft lip and palate. J Anat. 2004b;205:501–517. doi: 10.1111/j.0021-8782.2004.00356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrímsson B, Brown JJY, Ford-Hutchinson A, Sheets HD, Zelditch ML, Jirik FR. The brachymorph mouse and the developmental–genetic basis for canalization and morphological integration. Evol Dev. 2006;8:61–73. doi: 10.1111/j.1525-142X.2006.05075.x. [DOI] [PubMed] [Google Scholar]
- Indrasamy H, Woods RE, McKenzie JA, Batterham P. Fluctuating asymmetry for specific bristle characters in Notch mutants of Drosophila melanogaster. Genetica. 2000;109:151–159. doi: 10.1023/a:1017535930657. [DOI] [PubMed] [Google Scholar]
- Kasai K, Moro T, Kanazawa E, Iwasawa T. Relationship between cranial base and maxillofacial morphology. Eur J Orthod. 1995;17:403–410. doi: 10.1093/ejo/17.5.403. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Barluenga M, Meyer A. Shape analysis of symmetric structures: quantifying variation among individuals and asymmetry. Evolution. 2002;56:1909–1920. doi: 10.1111/j.0014-3820.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Mebus K, Auffray JC. Developmental integration in a complex morphological structure: how distinct are the modules in the mouse mandible? Evol Dev. 2003;5:522–531. doi: 10.1046/j.1525-142x.2003.03057.x. [DOI] [PubMed] [Google Scholar]
- Lieberman DE, Pearson OM, Mowbray KM. Basicranial influence on overall cranial shape. J Hum Evol. 2000a;38:291–315. doi: 10.1006/jhev.1999.0335. [DOI] [PubMed] [Google Scholar]
- Lieberman DE, Ross CF, Ravosa MJ. The primate cranial base: ontogeny, function, and integration. Am J Phys Anthropol Supplement. 2000b;31:117–169. doi: 10.1002/1096-8644(2000)43:31+<117::aid-ajpa5>3.3.co;2-9. (Yearbook) [DOI] [PubMed] [Google Scholar]
- Lozanoff S. Midfacial retrusion in adult brachyrrhine mice. Acta Anat (Basel) 1993;147:125–132. doi: 10.1159/000147492. [DOI] [PubMed] [Google Scholar]
- Lozanoff S, Jureczek S, Feng T, Padwal R. Anterior cranial base morphology in mice with midfacial retrusion. Cleft Palate Craniofac J. 1994;31:417–428. doi: 10.1597/1545-1569_1994_031_0417_acbmim_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- Lozanoff S, Johnston J, Ma W, Lie Saux CJ. Immunohistochemical localization of Pax2 and associated proteins in the developing kidney of mice with renal hypoplasia. J Histochem Cytochem. 2001;49:1081–1097. doi: 10.1177/002215540104900903. [DOI] [PubMed] [Google Scholar]
- Ma W, Lozanoff S. External craniofacial features, body size, and renal morphology in prenatal brachyrrhine mice. Teratology. 1993;47:321–332. doi: 10.1002/tera.1420470409. [DOI] [PubMed] [Google Scholar]
- Ma W, Lozanoff S. Morphological deficiency in the prenatal anterior cranial base of midfacially retrognathic mice. J Anat. 1996;188:547–555. [PMC free article] [PubMed] [Google Scholar]
- Ma W, Lozanoff S. Spatial and temporal distribution of cellular proliferation in the cranial base of normal and midfacially retrusive mice. Clin Anat. 1999;12:315–325. doi: 10.1002/(SICI)1098-2353(1999)12:5<315::AID-CA2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Ma W, Lozanoff S. Differential in vitro response to epidermal growth factor by prenatal murine cranial-base chondrocytes. Arch Oral Biol. 2002;47:155–163. doi: 10.1016/s0003-9969(01)00105-4. [DOI] [PubMed] [Google Scholar]
- Marquez E. Sage3D. Ann Arbor, MI: University of Michigan Museum of Zoology; 2004. [Google Scholar]
- Mather K. Genetical control of stability in development. Nature. 1953;7:297–336. [Google Scholar]
- McBratney BM, Margaryan E, Ma W, Urban Z, Lozanoff S. Frontonasal dysplasia in 3H1Br/Br mice. Anat Rec. 2003;271A:291–302. doi: 10.1002/ar.a.10034. [DOI] [PubMed] [Google Scholar]
- McKenzie JA, Clarke GM. Diazinon resistance in the Australian sheep blowfly, Lucilia cuprina. Genetics. 1988;120:213–220. doi: 10.1093/genetics/120.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Higgins P, Jones N. Morphologika. London: University College; 1998. [Google Scholar]
- Orkin RW, Pratt RM, Martin GR. Undersulfated chondroitin sulfate in the cartilage matrix of brachymorph mice. Dev Biol. 1976;50:82–94. doi: 10.1016/0012-1606(76)90069-5. [DOI] [PubMed] [Google Scholar]
- Palmer AR, Strobeck C. Fluctuating asymmetry: measurement, analysis, patterns. Ann Rev Ecol Syst. 1986;17:391–421. [Google Scholar]
- Palmer R, Strobeck C. Fluctuating Asymmetry Analysis Unplugged. In: Polak M, editor. Developmental Instability (DI): Causes and Consequences. Oxford: Oxford University Press; 2003. pp. 279–319. [Google Scholar]
- Rendel JM. Canalization of the scute phenotype of Drosophila. Evolution. 1959;13:425–439. [Google Scholar]
- Rendel JM. Canalization and Gene Control. London: Logos Press; 1967. [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Rutherford SL. From genotype to phenotype: buffering mechanisms and the storage of genetic information. Bioessays. 2000;22:1095–1105. doi: 10.1002/1521-1878(200012)22:12<1095::AID-BIES7>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Sakai K-I, Shimamoto Y. Developmental instability in leaves and flowers of Nicotiana tabacum. Genet Res. 1965;6:93–103. doi: 10.1093/genetics/51.5.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharloo W. Canalization: genetic and developmental aspects. Ann Rev Ecol Syst. 1991;22:65–93. [Google Scholar]
- Schmalhausen II. Factors of Evolution. Chicago: University of Chicago Press; 1949. [Google Scholar]
- Searle A. New mutants. Mouse Letter. 1966;35:27. [Google Scholar]
- Sheets HD. Simple3D. Buffalo: Canisius College; 2004a. [Google Scholar]
- Sheets HD. 3DStand. Buffalo: Canisius College; 2004b. [Google Scholar]
- Siegal ML, Bergman A. Waddington's canalization revisited: developmental stability and evolution. Proc Natl Acad Sci USA. 2002;99:10528–10532. doi: 10.1073/pnas.102303999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slice DE. Morpheus. Stony Brook, NY: State University of New York; 1994–1999. [Google Scholar]
- Sperber GH. Craniofacial Development. Hamilton: BC Decker Inc; 2001. [Google Scholar]
- Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein–Taybi syndrome. Proc Natl Acad Sci USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta V, Garoia F, Guerra D, Pessoli MC, Grifoni D, Cavicchi S. Developmental instability of the Drosophila wing as an index of genomic perturbation and altered cell proliferation. Evol Dev. 2005;7:234–243. doi: 10.1111/j.1525-142X.2005.05026.x. [DOI] [PubMed] [Google Scholar]
- Van Valen LM. A study of fluctuating asymmetry. Evolution. 1962;16:125–142. [Google Scholar]
- Waddington CH. The canalisation of development and the inheritance of acquired characters. Nature. 1942;150:563. [Google Scholar]
- Waddington CH. Strategy of the Genes. New York: MacMillan; 1957. [Google Scholar]
- Wagner GP, Altenberg L. Complex adaptations and the evolution of evolvability. Evolution. 1996;50:967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- Zelditch ML, Lundrigan BL, Sheets HD, Garland T. Do precocial mammals development at a higher rate? A comparison of rates of skull development in Sigmodon fulviventer and Mus musculus domesticus. J Evol Biol. 2003;16:708–720. doi: 10.1046/j.1420-9101.2003.00568.x. [DOI] [PubMed] [Google Scholar]