Abstract
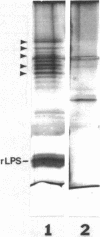
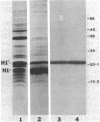
Overexpression of the divalent cation-regulated outer membrane protein H1 of Pseudomonas aeruginosa is associated with resistance to polymyxin B, aminoglycosides, and EDTA. Protein H1 is believed to act by replacing divalent cations at binding sites on lipopolysaccharide, thereby preventing disruption of the sites and subsequent self-promoted uptake of the antibiotics. Protein H1 purified by two cycles of anion-exchange chromatography was apparently associated with lipopolysaccharide. Lipopolysaccharide-free protein H1 was purified in high yield by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis and was subjected to N-terminal amino sequencing. Complementary oligodeoxyribonucleotides were used to clone the structural gene for protein H1, oprH, into Escherichia coli. Successful cloning was confirmed by nucleotide sequence analysis. Southern hybridization suggested that oprH was present as a single-copy gene in P. aeruginosa. The deduced amino acid sequence revealed that H1 was a slightly basic polypeptide of 178 residues, with a leader sequence typical of an exported procaryotic protein. It had little similarity, however, to other bacterial surface proteins for which sequence data were available. No expression of protein H1, from its own or the lac promoter, was detected in E. coli. We concluded that, as for some other regulated Pseudomonas genes, expression of oprH, at least under some conditions, is blocked in E. coli.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balland A., Courtney M., Jallat S., Tessier L. H., Sondermeyer P., de la Salle H., Harvey R., Degryse E., Tolstoshev P. Use of synthetic oligonucleotides in gene isolation and manipulation. Biochimie. 1985 Jul-Aug;67(7-8):725–736. doi: 10.1016/s0300-9084(85)80160-7. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. S., Georgopapadakou N. H. Routes of quinolone permeation in Escherichia coli. Antimicrob Agents Chemother. 1988 Apr;32(4):438–442. doi: 10.1128/aac.32.4.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagon R. G., Phibbs P. V., Jr Kinetics of transport of glucose, fructose, and mannitol by Pseudomonas aeruginosa. Can J Biochem. 1971 Sep;49(9):1031–1041. doi: 10.1139/o71-151. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984 Jun;158(3):1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray G. L., Smith D. H., Baldridge J. S., Harkins R. N., Vasil M. L., Chen E. Y., Heyneker H. L. Cloning, nucleotide sequence, and expression in Escherichia coli of the exotoxin A structural gene of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1984 May;81(9):2645–2649. doi: 10.1073/pnas.81.9.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Bell A. Antibiotic uptake into gram-negative bacteria. Eur J Clin Microbiol Infect Dis. 1988 Dec;7(6):713–720. doi: 10.1007/BF01975036. [DOI] [PubMed] [Google Scholar]
- Hancock R. E., Carey A. M. Outer membrane of Pseudomonas aeruginosa: heat- 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979 Dec;140(3):902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Irvin R. T., Costerton J. W., Carey A. M. Pseudomonas aeruginosa outer membrane: peptidoglycan-associated proteins. J Bacteriol. 1981 Jan;145(1):628–631. doi: 10.1128/jb.145.1.628-631.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E., Raffle V. J., Nicas T. I. Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1981 May;19(5):777–785. doi: 10.1128/aac.19.5.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Kelly N. M., Bell A., Hancock R. E. Surface characteristics of Pseudomonas aeruginosa grown in a chamber implant model in mice and rats. Infect Immun. 1989 Feb;57(2):344–350. doi: 10.1128/iai.57.2.344-350.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski A. M., Lewis V., Berry D. Effect of growth temperature on the lipids, outer membrane proteins, and lipopolysaccharides of Pseudomonas aeruginosa PAO. J Bacteriol. 1987 May;169(5):1960–1966. doi: 10.1128/jb.169.5.1960-1966.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lathe R. Synthetic oligonucleotide probes deduced from amino acid sequence data. Theoretical and practical considerations. J Mol Biol. 1985 May 5;183(1):1–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B., Van Alphen L. Molecular architecture and functioning of the outer membrane of Escherichia coli and other gram-negative bacteria. Biochim Biophys Acta. 1983 Mar 21;737(1):51–115. doi: 10.1016/0304-4157(83)90014-x. [DOI] [PubMed] [Google Scholar]
- Moore R. A., Bates N. C., Hancock R. E. Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob Agents Chemother. 1986 Mar;29(3):496–500. doi: 10.1128/aac.29.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutharia L. M., Hancock R. E. Surface localization of Pseudomonas aeruginosa outer membrane porin protein F by using monoclonal antibodies. Infect Immun. 1983 Dec;42(3):1027–1033. doi: 10.1128/iai.42.3.1027-1033.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicas T. I., Hancock R. E. Alteration of susceptibility to EDTA, polymyxin B and gentamicin in Pseudomonas aeruginosa by divalent cation regulation of outer membrane protein H1. J Gen Microbiol. 1983 Feb;129(2):509–517. doi: 10.1099/00221287-129-2-509. [DOI] [PubMed] [Google Scholar]
- Nicas T. I., Hancock R. E. Outer membrane protein H1 of Pseudomonas aeruginosa: involvement in adaptive and mutational resistance to ethylenediaminetetraacetate, polymyxin B, and gentamicin. J Bacteriol. 1980 Aug;143(2):872–878. doi: 10.1128/jb.143.2.872-878.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H., Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985 Mar;49(1):1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Parr T. R., Jr, Poole K., Crockford G. W., Hancock R. E. Lipopolysaccharide-free Escherichia coli OmpF and Pseudomonas aeruginosa protein P porins are functionally active in lipid bilayer membranes. J Bacteriol. 1986 Feb;165(2):523–526. doi: 10.1128/jb.165.2.523-526.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard A. E., Vasil M. L. Nucleotide sequence and expression of a phosphate-regulated gene encoding a secreted hemolysin of Pseudomonas aeruginosa. J Bacteriol. 1986 Jul;167(1):291–298. doi: 10.1128/jb.167.1.291-298.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J., Thom J. R. Export of protein: a biochemical view. Annu Rev Microbiol. 1987;41:507–541. doi: 10.1146/annurev.mi.41.100187.002451. [DOI] [PubMed] [Google Scholar]
- Reithmeier R. A., Bragg P. D. Cross-linking of the proteins in the outer membrane of Escherichia coli. Biochim Biophys Acta. 1977 Apr 18;466(2):245–256. doi: 10.1016/0005-2736(77)90222-x. [DOI] [PubMed] [Google Scholar]
- Travers A. A. Structure and function of E. coli promoter DNA. CRC Crit Rev Biochem. 1987;22(3):181–219. doi: 10.3109/10409238709101483. [DOI] [PubMed] [Google Scholar]
- Tsai C. M., Frasch C. E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982 Jan 1;119(1):115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- West S. E., Iglewski B. H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988 Oct 11;16(19):9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L. S. Treatment of infections due to gram-negative bacilli: a perspective of past, present, and future. Rev Infect Dis. 1985 Nov-Dec;7 (Suppl 4):S572–S578. doi: 10.1093/clinids/7.supplement_4.s572. [DOI] [PubMed] [Google Scholar]